Introduction
Drug discovery is a tedious and costly process that
requires extensive pre-clinical and clinical evaluations. Although
numerous drugs exhibit promising pre-clinical efficacies, their
clinical efficacies may be unfavorable, resulting in the omission
of pre-clinically favorable drug candidates in clinical drug
development settings (1,2). Drug repurposing approaches identify
novel pharmacological targets of drug candidates that have already
obtained regulatory approval for clinical use (3). As the safety, pharmacodynamic and
pharmacokinetic profiles of clinically approved drug candidates are
already established, the exploration of novel pharmacological
properties or targets provides a number of benefits in terms of the
cost associated with drug discovery approaches and opens up new
arenas in modern drug discovery landscapes (3).
Bempedoic acid was approved by the Food and Drug
Administration (FDA) in 2020 for the treatment of refractory
hypercholesterolemia (4). Bempedoic
is a pro-drug that is activated in the liver by the enzyme very
long-chain acyl-CoA synthetase (ACSVL1). The active form of
bempedoic acid (bempedoic acid attached to coenzyme A) is an ATP
lyase inhibitor (5). ATP lyase
plays a key role in cholesterol biosynthesis (6). By inhibiting the activity of ATP
lyase, bempedoic acid upregulates the expression of low-density
lipoprotein (LDL) cholesterol receptors, decreasing LDL-cholesterol
by increasing cholesterol uptake and clearance in the liver
(6).
According to epigenetics, chromatin-bound
information, other than the information available in DNA sequences,
is responsible for the regulation of gene expression (7). Notably, dysregulated epigenetic events
are commonly observed in human diseases, rendering them attractive
pharmacological targets (8). Of the
various epigenetic events, histone acetylation has been
well-characterized. Histone acetyltransferases (HATs) and histone
deacetylases (HDACs) are involved in the acetylation of histones.
HATs mediate acetylation reactions, while HDACs mediate
deacetylation reactions in a well-balanced and reversible manner
(7). Thus far, 18 different HDACs
have been identified in humans (9).
There is ample evidence to indicate that HDAC6 plays a key role in
a wide range of human diseases, including cancer, neurological
diseases, inflammatory diseases and metabolic diseases, thus
rendering it an attractive drug target (10,11).
To date, various HDAC6 inhibitors have been identified from natural
and synthetic sources. The HDAC inhibitory effects of fatty acids
have been well-established and numerous fatty acids have been shown
to exert HDAC inhibitory effects at millimolar concentrations
(i.e., effective concentration for inhibiting HDAC activity)
(9,12,13).
The chemical structure of bempedoic acid resembles
an α,ω-dicarboxylic acid (14). In
a recent study, the authors identified a series of odd-chain fatty
acids as HDAC6 inhibitors, which motivated the exploration of the
HDAC6 inhibitory effects of new fatty acid candidates (15). Considering the recent findings
related to the HDAC6 inhibitory potential of odd-chain fatty acids
and the chemical structure of bempedoic acid, it was hypothesized
that bempedoic acid can exert HDAC6 inhibitory effects. This
hypothesis was tested using a cell-free HDAC6 enzyme assay and
confirmed by in silico analysis.
Materials and methods
Chemicals
Bempedoic acid was purchased from Cayman Chemical
Company (item no. 26409).
HDAC6 enzyme inhibitory assay
A BioVision HDAC6 Enzyme Cell-Free Inhibitory Assay
kit (cat. no. K465; BioVision, Inc.) was used to evaluate the HDAC6
inhibitory potential of bempedoic acid. The assay procedures
provided by the manufacturer were followed when conducting the
enzyme assays. This HDAC6 enzyme inhibitory assay kit includes
HDAC6 synthetic acetylated peptide substrate, human HDAC6 enzyme
and a developer. Prior to the assay, bempedoic acid was dissolved
in ethanol to yield a dilution series starting from 2 to 0.0078 mM.
Fluorescence was measured at excitation/emission wavelengths of
380/490 nm using a microplate reader (Sunrise; Tecan Group,
Ltd).
Molecular docking
The crystal structure of HDAC6 comprising
nexturastat A was obtained from the Protein Data Bank (PDB ID:
5G0J). Prior to docking, nexturastat A was removed from the active
site of HDAC6. The three-dimensional (3D) structure of bempedoic
acid (CID: 10472693) was obtained from the PubChem database and
energetically pre-optimized using the universal force field (PyRx
Python Prescription, version 0.8; The Scripps Research Institute,
2008). The amino acids in the active site were determined using the
Computed Atlas for Surface Topography of Proteins (16) and BIOVIA Discovery Studio 4.5
(Dassault Systèmes BIOVIA, Discovery Studio Modeling Environment,
Version 4.5. San Diego; Dassault Systèmes; 2015). The docking
experiments were performed using the AutoDock Vina module
(Molecular Graphics Lab, The Scripps Research Institute). Based on
the binding energies, the best-docked pose was selected and 3D
images were generated using PyMOL (The PyMOL Molecular Graphics
System, Ver.2.5.0; Schrodinger, LLC). The docked complex of HDAC6
was further optimized, validated and explored using the Discovery
Studio visualizer (version 21.1.0.20298). The hydrogen bonds and
hydrophobic interactions between bempedoic acid and HDAC6 were
analyzed using the LigPlot program (17).
Data analysis
The enzyme inhibitory assay was conducted in
triplicate. The results are presented as the mean ± standard
deviation (SD). GraphPad Prism software was used to generate enzyme
inhibitory graphs and to obtain the half maximal inhibitory
concentration (IC50) of bempedoic acid.
Results
HDAC6 enzyme inhibitory effects of
bempedoic acid
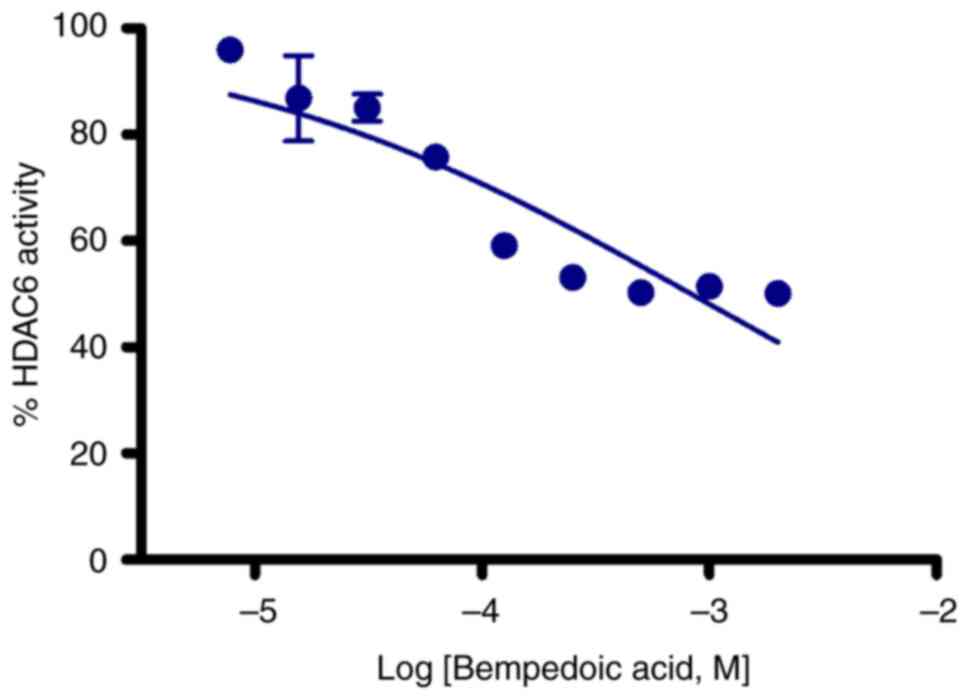
The effect of bempedoic acid on HDAC6 activity was
assessed in vitro. As illustrated in Fig. 1, bempedoic acid exerted a
concentration-dependent HDAC6 inhibitory effect with an
IC50 value of ~0.8 mM.
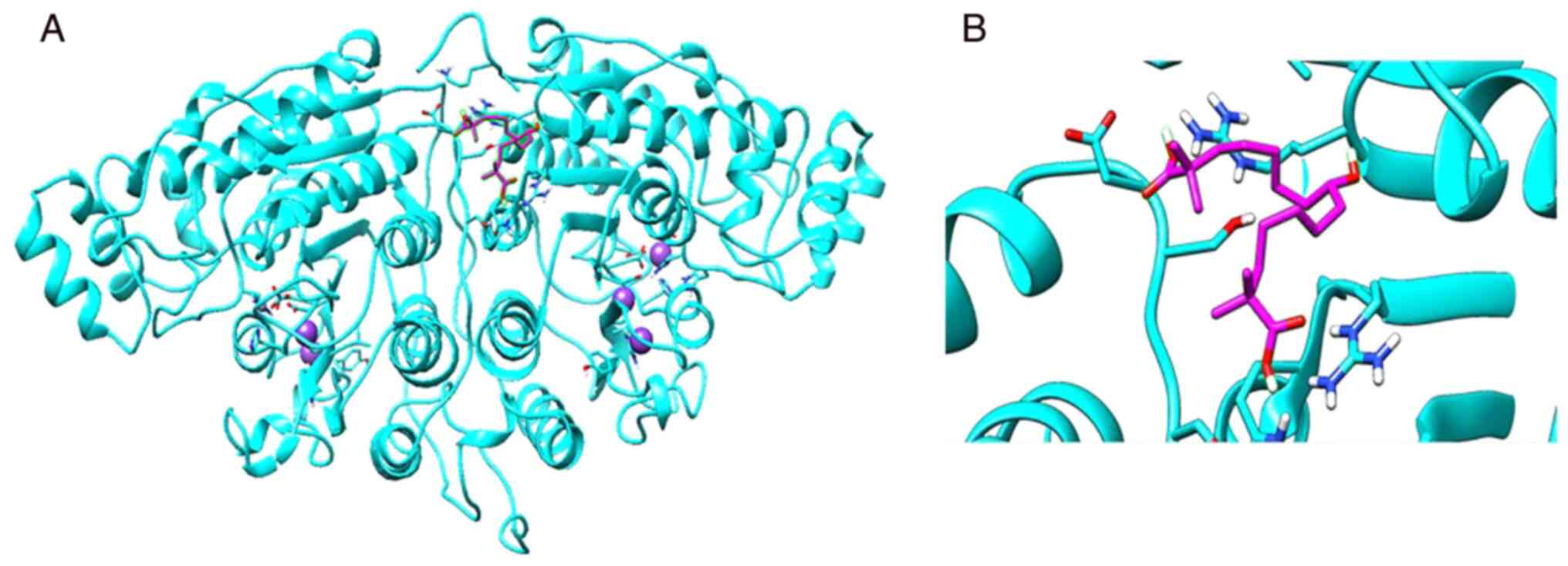
Molecular docking
For in silico analysis, the 3D structure of
Danio rerio HDAC6, 5G0J.pdb, was used as the 3D structure of
Homo sapiens HDAC6 (containing two catalytic domains) has
not been deposited in the PDB (15). The basic molecular structure of
HDAC6 contains two catalytic domains (CD1 and CD2) (18). Among these catalytic domains, CD2
exhibits broad substrate specificity and has been used exclusively
to develop HDAC6-selective inhibitors (19). The docking of bempedoic acid to
HDAC6 exhibited binding energies ranging from -4.7 to -5.1
kcal/mol. The complex with the lowest binding energy was selected
for further analysis. As shown in Fig.
2, bempedoic acid is docked into the active pocket of the
catalytic domain of HDAC6.
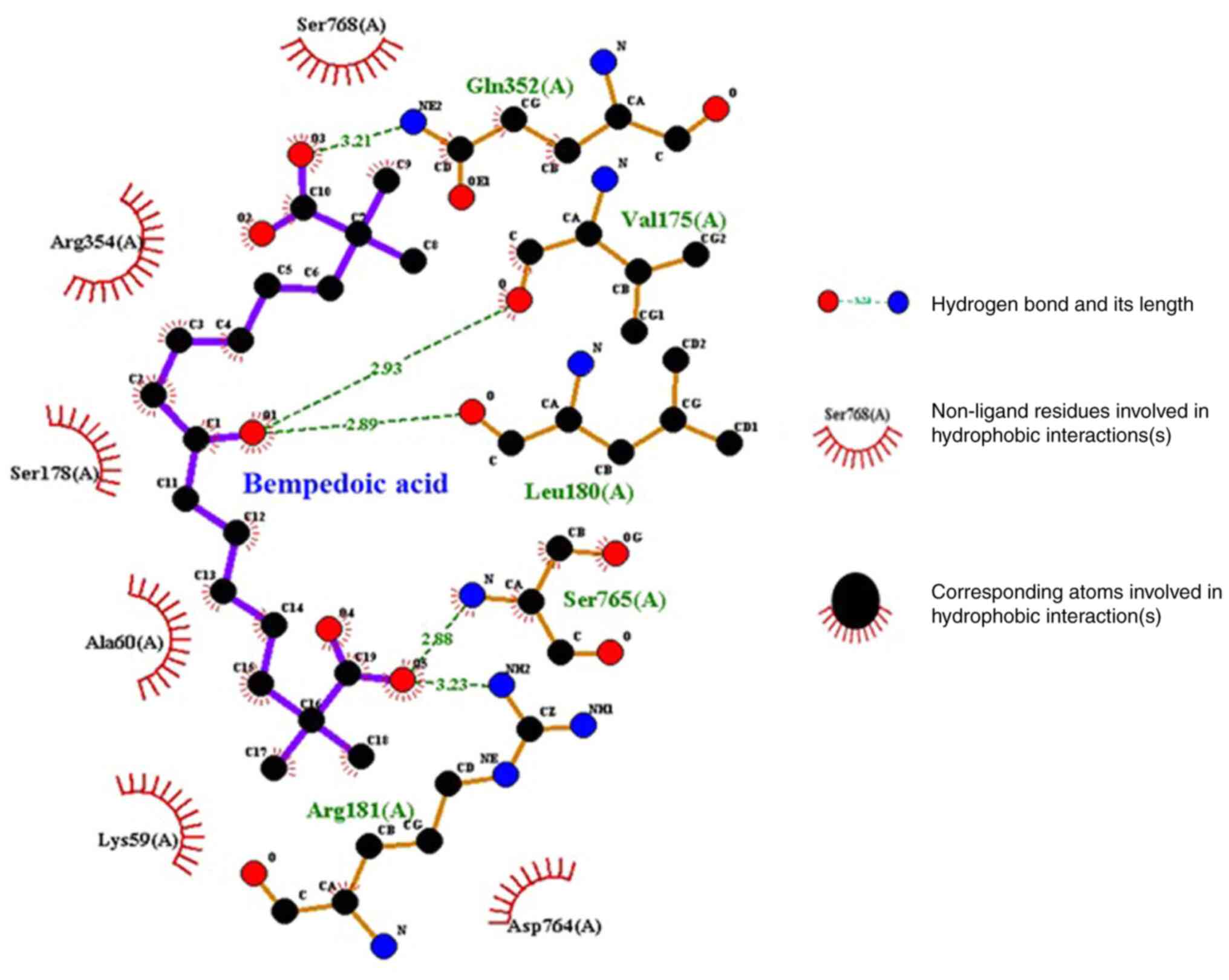
LigPlot analysis revealed molecular interactions
(hydrogen bonds and hydrophobic interactions) between bempedoic
acid and the catalytic domain of HDAC6 (Fig. 3). Furthermore, the LigPlot analysis
revealed that five residues, Gln352, Val175, Leu180, Ser765 and
Arg181, are involved in hydrogen bonds, and six residues, Ser768,
Arg354, Ser178, Ala60, Lys59 and Asp764, are involved in
hydrophobic interactions with bempedoic acid. These hydrogen bonds
and hydrophobic interactions may play an essential role in lead
optimization and may thus enhance the affinity of bempedoic acid
for HDAC6.
Discussion
In the present study, for the first time, to the
best of our knowledge, bempedoic acid was repurposed as a HDAC6
inhibitor. Bempedoic acid is an FDA-approved cholesterol-lowering
agent (4). In the liver, with the
aid of the enzyme ACSVL1, bempedoic acid is converted to its active
form, which is an ATP citrate lyase inhibitor (5). The dysregulated activity or
overexpression of HDAC6 have been reported in a range of human
diseases, rendering HDAC6 an interesting drug target (10,11).
A large number of isoform-specific and pan-HDAC
inhibitor proteins belonging to four major chemical classes have
been identified: Benzamides, fatty acids, hydroxamates and cyclic
tetrapeptides (7). A recent study
by the authors identified a series of odd-chain fatty acids,
including valeric acid (C5:0), heptanoic acid (C7:0), nonanoic acid
(C9;0), undecanoic acid (C11:0) and pentadecanoic acid (C15:0) as
HDAC6 inhibitors (15). Of these,
pentadecanoic acid exerted more potent HDAC6 inhibitory effects
with binding energies ranging from -3.95 to -2.90 kcal/mol. The
HDAC3 and HDAC7 enzyme inhibitory effects of short-chain fatty
acids (namely, valeric, propionic, butyric, caproic and
4-methylvaleric acids) have been previously investigated (20). Butyric acid was identified as the
most potent HDAC3 inhibitor among the fatty acids tested. Fass
et al (21) demonstrated the
class I and class IIa HDAC inhibitory potentials of the short-chain
fatty acids, butyric acid and valporic acid. The branched-chain
fatty acids 2,2-dimethylbutyric acid, 2-ethylbutyric acid, and
valproic acid, and the un-branched fatty acids propionic acid,
valeric acid and butyric acid, were previously identified as weak
and equipotent HDAC inhibitors, respectively (22). In a recent study, valeric acid was
identified as a HDAC3 inhibitor (23).
In the present study, the in vitro HDAC6
enzyme inhibitory assay indicated that bempedoic acid can inhibit
the activity of HDAC6 at millimolar concentrations. According to
the in silico findings, bempedoic acid docked into the
active pocket of the catalytic domain of HDAC6 and was involved in
the formation of hydrogen bonds and hydrophobic interactions with
HDAC6 residues, which may explain the HDAC6 inhibitory effects of
bempedoic acid. Notably, bempedoic acid exhibited a lower binding
energy (-4.7 to -5.1 kcal/mol) compared to pentadecanoic acid,
which was identified as the most potent HDAC6 inhibitor in a recent
by the authors (15).
Bempedoic acid is rapidly absorbed in the small
intestine and reaches a maximum plasma concentration of 20.6±6.1
µg/ml (0.059 mM) following multiple-dose administration at 180
mg/day (24). According to the
enzyme assay results of the present study, bempedoic acid at
concentrations near ~0.059 mM also exerted HDAC6 inhibitory
effects, indicating that the HDAC6 inhibitory action of bempedoic
acid is likely to occur in body tissues. According to the FDA drug
label for bempedoic acid, bempedoic acid and its conjugates are
detected in plasma, with bempedoic acid being the most prominent
compound, accounting for almost 46% of the area under the curve
(AUC0-48 h) (25).
Although the pre-clinical findings are meaningful, it is critical
to assess the HDAC isoform specificities of bempedoic acid, its
active form and conjugates. Moreover, the preliminary findings of
the present study may provide an important foundation with which to
rationalize the HDAC6 inhibitory effects of bempedoic acid and
warrant additional investigations to explore the detailed
epigenetic mechanisms associated with bempedoic acid and its active
form in cell-based systems. A recent study demonstrated that
bempedoic acid can reprogram the epigenetic and transcriptional
machineries of ATP citrate lyase-associated genes in hepatocytes
(26), further supporting the
current preliminary experimental findings. Taken together, the
present study opens up new perspectives for the exploration of the
epigenetic effects of bempedoic acid in human diseases, as HDAC6
activity and expression are frequently altered in a range of human
diseases (10,11).
In conclusion, to the best of our knowledge, the
present study reports for the first time that bempedoic acid, an
FDA-approved cholesterol-lowering drug, functions as an HDAC6
enzyme inhibitor. Similar to several other fatty acids, bempedoic
acid exerts HDAC6 inhibitory effects at millimolar
concentrations.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by grants from the
Basic Science Research Program through the National Research
Foundation of Korea funded by the Ministry of Education (no.
2016R1A6A1A03012862) and the UNESCO-TWAS and the Swedish
International Development Cooperation Agency (Sida; no. 22-140
RG/BIO/AS_I).
Availability of the data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
MKE and SKC designed the study. PR and MKE performed
the experiments. MKE and PR analyzed the data and wrote the
manuscript. SKC supervised the study and revised the manuscript.
All authors have read and approved the final manuscript. MKE and
SKC confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hughes JP, Rees S, Kalindjian SB and
Philpott KL: Principles of early drug discovery. Br J Pharmacol.
162:1239–1249. 2011.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Ediriweera MK, Tennekoon KH and Samarakoon
SR: In vitro assays and techniques utilized in anticancer drug
discovery. J Appl Toxicol. 39:38–71. 2019.PubMed/NCBI View
Article : Google Scholar
|
|
3
|
Pushpakom S, Iorio F, Eyers PA, Escott KJ,
Hopper S, Wells A, Doig A, Guilliams T, Latimer J, McNamee C, et
al: Drug repurposing: Progress, challenges and recommendations. Nat
Rev Drug Discov. 18:41–58. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Marrs JC and Anderson SL: Bempedoic acid
for the treatment of dyslipidemia. Drugs Context.
9(2020-6-5)2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Ray KK, Bays HE, Catapano AL, Lalwani ND,
Bloedon LT, Sterling LR, Robinson PL and Ballantyne CM: CLEAR
Harmony Trial. Safety and efficacy of bempedoic acid to reduce LDL
cholesterol. N Engl J Med. 380:1022–32. 2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Feng X, Zhang L, Xu S and Shen AZ:
ATP-citrate lyase (ACLY) in lipid metabolism and atherosclerosis:
An updated review. Prog Lipid Res. 77(101006)2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Ediriweera MK, Tennekoon KH and Samarakoon
SR: Emerging role of histone deacetylase inhibitors as
anti-breast-cancer agents. Drug Discov Today. 24:685–702.
2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Hatchwell E and Greally JM: The potential
role of epigenomic dysregulation in complex human disease. Trends
Genet. 23:588–95. 2007.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Ediriweera MK and Cho SK: Targeting miRNAs
by histone deacetylase inhibitors (HDACi): Rationalizing
epigenetics-based therapies for breast cancer. Pharmacol Ther.
206(107437)2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Pulya S, Amin SA, Adhikari N, Biswas S,
Jha T and Ghosh B: HDAC6 as privileged target in drug discovery: A
perspective. Pharmacol Res. 163(105274)2021.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Batchu SN, Brijmohan AS and Advani A: The
therapeutic hope for HDAC6 inhibitors in malignancy and chronic
disease. Clinical Sci. 130:987–1003. 2016.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Waldecker M, Kautenburger T, Daumann H,
Busch C and Schrenk D: Inhibition of histone-deacetylase activity
by short-chain fatty acids and some polyphenol metabolites formed
in the colon. J Nutr Biochem. 19:587–593. 2008.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Xu WS, Parmigiani RB and Marks PA: Histone
deacetylase inhibitors: Molecular mechanisms of action. Oncogene.
26:5541–5552. 2007.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Oniciu DC and Myers JL: Bempedoic acid and
the fraudulent fatty acid family: The gold rush to cardiovascular
therapies in the new millennium. Org Process Res Dev. 25:365–372.
2021.
|
|
15
|
Ediriweera MK, To NB, Lim Y and Cho SK:
Odd-chain fatty acids as novel histone deacetylase 6 (HDAC6)
inhibitors. Biochimie. 186:147–156. 2021.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Tian W, Chen C, Lei X, Zhao J and Liang J:
CASTp 3.0: Computed atlas of surface topography of proteins.
Nucleic Acids Res. 46:W363–W367. 2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Laskowski RA and Swindells MB: LigPlot+:
Multiple ligand-protein interaction diagrams for drug discovery. J
Chem Inf Model. 51:2778–2786. 2011.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Osko JD and Christianson DW: Structural
determinants of affinity and selectivity in the binding of
inhibitors to histone deacetylase 6. Bioorg Med Chem Lett.
30(127023)2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Osko JD and Christianson DW: Structural
basis of catalysis and inhibition of HDAC6 CD1, the enigmatic
catalytic domain of histone deacetylase 6. Biochemistry.
58:4912–4924. 2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Ho RH, Chan JC, Fan H, Kioh DY, Lee BW and
Chan EC: In silico and in vitro interactions between short chain
fatty acids and human histone deacetylases. Biochemistry.
56:4871–4878. 2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Fass DM, Shah R, Ghosh B, Hennig K, Norton
S, Zhao WN, Reis SA, Klein PS, Mazitschek R, Maglathlin RL, et al:
Effect of inhibiting histone deacetylase with short-chain
carboxylic acids and their hydroxamic acid analogs on vertebrate
development and neuronal chromatin. ACS Med Chem Lett. 2:39–42.
2010.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Gilbert KM, DeLoose A, Valentine JL and
Fifer EK: Structure-activity relationship between carboxylic acids
and T cell cycle blockade. Life Sci. 78:2159–2165. 2006.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Han R, Yang H, Li Y, Ling C and Lu L:
Valeric acid acts as a novel HDAC3 inhibitor against prostate
cancer. Medical Oncol. 39(213)2022.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Markham A: Bempedoic acid: First approval.
Drugs. 80:747–753. 2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Esperion Therapeutics Inc.: NEXLETOL
(bempedoic acid): tablets, for oral use. Initial U.S. Approval:
2020. https://www.accessdata.fda.gov/drugsatfda_docs/label/2020/211616s000lbl.pdf.
Accessed December 12, 2022.
|
|
26
|
Rose A, Rodriguez-Aguilera JR, Schicht G,
Lohrenz A, Tvardovskiy A, Büscher J, Hoffmann A, Damm G, Laufs U,
Seehofer D, et al: The impact of cholesterol lowering drugs on
metabolism and epigenetics. Atherosclerosis. 355:179–180. 2022.
|