Introduction
Gastric cancer is the most common gastrointestinal
cancer, with the highest morbidity and mortality rates in China,
ranking second in the incidence of malignant tumors worldwide.
Therefore, novel, more effective therapeutic options are required
for the prevention and treatment of gastric cancer.
The occurrence and development of gastric cancer is
a complicated, multistage and multifactorial process. An imbalance
between cell apoptosis and proliferation is a key reason resulting
in excessive proliferation of tumor cells. Survivin, which was
screened and separated from the human genomic library by Ambrosini
et al (1) by means of
effector cell protease receptor-1 cDNA, is a novel member of the
inhibitor of apoptosis (IAP) family and a bifunctional protein that
may inhibit cell apoptosis and regulate cell division. Survivin is
highly expressed in embryonic and developing fetal tissues, but it
is almost absent from the terminally differentiated tissues of
normal adults, whereas it is selectively expressed in tumors.
Survivin expression was previously detected in colon, gastric,
esophageal and non-small-cell lung cancers (2–5) and
a previous study indicated that survivin is highly expressed in
gastric cancer tissues, but is almost absent from the normal
tissues surrounding gastric cancer (6). However, Yao et al (7) detected survivin expression in gastric
adenocarcinoma using the immunohistochemical
streptavidin-peroxidase method and the results demonstrated that
the positive expression rate of survivin in the primary focus of
gastric cancer, metastatic cancer cells of lymph nodes and basal
germinal layer cells of normal glands were 49.2, 64.0, and 17.5%,
respectively. Therefore, further investigation into the expression
of survivin in tumor cells is required to elucidate the mechanisms
underlying the occurrence and development of gastric cancer.
The majority of gastric cancers are middle- to
advanced-stage at diagnosis and exhibit a low remission rate with
chemotherapy. The anticancer role of traditional Chinese medicine
(TCM) is under ongoing investigation in the medical field. In TCM,
gastric cancer belongs to the classification of ‘sick’,
‘dysphagia’, ‘stomachache’ and ‘accumulation’ disease. Qizhu, an
empirical formula for gastrointestinal tumors, has been subjected
to numerous years of clinical research. Our earlier study (8) demonstrated that the Qizhu formula was
able to inhibit the expression of telomerase and its related genes
in MGC-803 gastric cancer cells.
In the present study, the effect of Qizhu on the
downregulation of survivin protein and mRNA expression in MGC-803
human gastric adenocarcinoma cells was investigated by western blot
analysis and reverse transcription-polymerase chain reaction
(RT-PCR), with the aim to further demonstrate the induction of
gastric cancer cell apoptosis by Qizhu and provide reliable
experimental evidence regarding multitargeted treatment and
application of TCM compounds in the treatment of gastric
cancer.
Materials and methods
Tumor strain
The MGC-803 human gastric cancer cells were
purchased from Nanjing Kaiteng Technological Corporation (Nanjing,
China) and preserved by subculturing.
Drugs
Qizhu formula crude extracts (referred to as Qizhu
formula hereafter) were purchased from Nanjing Zhongshan
Pharmaceutical Factory (Nanjing, China). Qizhu formula consists of
Zhihuangqi (Radix Astragali Praeparata cum Melle), Yuzhu
(Rhizoma Polygonati Odorati), Fabanxia (Rhizoma Pinelliae
Praeparatum), Shengyiyiren (Semen Coicis, raw),
Xianhecao (Herba Agrimoniae), Ezhu (Rhizoma Curcumae)
and Baihuasheshecao (Herba Hedyotis Diffusae). All the
compounds in the formula were identified by TCM Identification
Teaching and Research Section of Nanjing University of Chinese
Medicine and conformed to the national pharmacopoeia criterion.
Steam distillation was applied to extract zedoary oil, with 11.25 g
extracted from 500 g of crude Ezhu, yielding 11.25 ml, for a yield
of 2.25%, namely 1 g=1 ml. The extracted residual Ezhu liquid was
mixed with other crude drugs and the crude drugs were then
extracted by poaching and alcohol precipitation. A total of 480.5 g
crude extracts (dry extracts, 496.25 g + zedoary oil, 11.25 g) was
collected from 4,500 g of crude drugs, for a yield of 10.68%
(480.5/4,500).
Reagents
Western blotting
Methanol, 200 ml 10X blotting buffer, 1,400 ml
deionized water and Coomassie brilliant blue G250 were
collected for the western blotting procedure. First, 20 mg of
Coomassie brilliant blue G250 were dissolved into 10 ml
of 95% alcohol, followed by the addition of 20 ml
H3PO4 and deionized water to a final volume
of 200 ml. The mixture was filtered using filter paper and stored
at 4°C. Subsequently, 0.05 g bovine serum albumin (BSA) were
dissolved in 100 ml of double-distilled water (0.5 mg/ml) and
stored at 4°C. Finally, 0.01742 g phenylmethanesulfonyl fluoride
(PMSF) were dissolved in 1 ml of isopropanol (100 mM), subpackaged
and stored at −20°C. Acrylamide and ammonium persulfate were
purchased from Longxi Chemical Company (Nanjing, China),
tetramethylethylenediamine was obtained from American Amresco Co.
(Solon, OH, USA) and horseradish peroxide solutions A and B were
purchased from Beijing ZSGB-Biotechnology Co., Ltd. (Beijing,
China).
RT-PCR
TRIzol (DP-405) was purchased from Beijing Tianwei
Time Technology Co., Ltd. (Beijing, China), general RT-PCR kits (20
reactions) were obtained from Beijing Dingguo Biotechnology Co.,
Ltd. (Beijing, China) and synthetic primers of survivin and β-actin
genes were obtained from Shanghai Sangon Biotech Service Co., Ltd.
(Shanghai, China). The TRIzol kits were provided by Tianyao Time
Company (Tianjin, China) and diethyl pyrocarbonate was obtained
from American Amresco Co. Pure chloroform, isopropanol and ethanol
were all analyzed by the Longxi Chemical Company and agarose was
obtained from Oxoid Limited (Hampshire, UK).
Main experimental apparatus
A laminar flow hood was obtained from Suzhou
Purification Equipment Factory (Suzhou, China); a Forman 3111
CO2 incubator was purchased from (Thermoscientific, West
Palm Beach, FL, USA); a 550 enzyme-linked immunoassay detector was
obtained from Bio-Rad Laboratories (Hercules, CA, USA); an LDZ5-2
medical centrifuge was purchased from Beijing Medical Centrifuge
Factory (Beijing, China); a UV-2450 ultra-violet spectrophotometer
was obtained from Shimadzu Co. (Milton Keynes, UK); a PHS-3C
Precision pH meter was purchased from Shanghai Torpedo-Magnetic
Instrument Plant (Shanghai, China); an FAZ104 electronic balance
was obtained from Meigele Hardware Co., Ltd. (Guangzhou, China); a
101AS-3 stainless steel digital electrothermal air dry oven was
purchased from Shanghai Shengxin Scientific Instrument Co., Ltd.
(Shanghai, China); a YXQ-CS-30L double-layer stainless vertical
sterilizer was obtained from the Medical Equipment Factory of
Shanghai Boxun Industrial Co., Ltd. (Shanghai, China); a WD-940153
horizontal shaker was purchased from Beijing Liuyi Instrument Plant
(Beijing, China); an XSZ-O2 inverted metallographic microscope was
obtained from Chongqing Optical Instrument Plant (Chongqing,
China); a PTC-100 PCR amplifier was purchased from MJ Reserach Inc.
(Waltham, MA, USA); a SHO 3014 tabletop high-speed refrigerated
centrifuge was obtained from Eppendorf (Hamburg, Germany); a
Mini-PROTEAN vertical electrotransfer chamber was purchased from
Bio-Rad Laboratories; and an FR-980 biological electrophoresis
image analysis system was obtained from Shanghai Furi Technology
Co., Ltd (Shanghai, China).
Assessment of Qizhu formula effect on
survivin protein expression in MGC-803 cells by western blot
analysis Grouping
A negative control group, a dimethylsulfoxide (DMSO)
solvent control group and two medication groups with different
concentrations of Qizhu formula (250 and 500 μg/ml) were set
up.
Methods
The MGC-803 cells were first cultured to a certain
concentration. Four 10-ml culture flasks were then inoculated with
2×106 cells and placed in a 5% CO2 incubator for 24 h at
37°C. Subsequently, a negative control group, a 0.1% DMSO solvent
control group and two medication groups with different
concentrations of Qizhu formula (250 and 500 μg/ml) were set
up. Finally, 10 ml of the cultured cells were added to each of the
four groups and placed in a 5% CO2 incubator at 37°C for
24 h.
Cell collection
Cell scraping paper was used to collect 10 ml of
cells in each group. The cells were centrifuged for 5 min at 352×g,
followed by the addition of 1 ml PBS to transfer the centrifuged
cells to Eppendorf tubes and the mixture was centrifuged at 1,570×g
for 1 min. The supernatant was then removed, the cells were washed
again with PBS, beakers were prepared and tap water was added and
heated to 95°C. Furthermore, 500 μl of lysis buffer were
freshly prepared (497 μl lysis buffer, 2.5 μl
dithiothreitol and 0.5 μl PMSF).
Cell lysis
Lysis buffer (110 μl) was added to each group
to create a cell suspension and placed on ice for 30 min. A
separation gel and a stacking gel were then prepared. Following
lysis, the cells were centrifuged at 3,070×g for 10 min, the
supernatant was absorbed and another 1.5-ml Eppendorf tube was
prepared to measure the protein concentration and conduct protein
quantification. Finally, equal amounts of protein were collected
from each sample, 10 μl lysis buffer and 20 μl 2X
sample buffer were added and the solution was incubated at 95°C for
5 min. The electrophoresis 10X running buffer was diluted to 1X,
the samples were placed on ice for 5 min, centrifuged at 1,570×g
for 1 min and the supernatant was absorbed to prevent trailing. The
samples were placed in loading buffer and subjected to sodium
dodecyl sulfate-polyacrylamide gel electrophoresis at 80 V for 2
h.
Electrotransfer membranes
In total, 500 ml 1X blotting buffer (100 ml
methanol, 50 ml 10X blotting buffer and 350 ml water) were freshly
prepared. The gel was cut into 5.5×5 cm pieces, placed in 1X
blotting buffer and marked; two pieces of the PVDF mebrane were
sheared into 6.5×6 cm pieces and activated in methanol over 5 min.
Eight filter papers with two pieces on one side were then placed
into 1X blotting buffer to siphon, the PVDF was removed from the
methanol, washed once with distilled water and placed into 1X
blotting buffer with the gel. In total, four sponges were prepared
and soaked in 1X blotting buffer; subsequently, the sponges, two
flat pieces of filter paper, the gel (peripheral
absorption-desorption 1X blotting buffer) and the flat membranes
(moistened with 1X blotting buffer) were placed on a wet-rotating
device in turn to compress the filter papers and sponges, followed
by placement in a 100 V vertical electrophoresis chamber for 1 h.
The chamber was filled with 1X blotting buffer and an ice box was
placed along its side.
Protein detection
The capsular membranes were placed into two small
containers filled with blocking buffer at 4°C for 8 h to block the
non-reactive sites on the membranes in order to inhibit the
non-specific absorption of antibodies. The primary goat polyclonal
β-actin (1:500) and rabbit polyclonal survivin (1:500) antibodies
(Santa Cruz Biotechnology Inc., Santa Cruz, CA) were then diluted
with blocking buffer, the opening was sealed with film and the
table was agitated horizontally for 1 h to combine the antigens and
antibodies. The membranes were washed three times with
Tris-buffered saline (TBS) for 15 min each time, the secondary
donkey anti-rabbit antibodies (1:5,000; Santa Cruz Biotechnology
Inc.) with horseradish peroxidase were added and the mixture was
diluted with blocking buffer. The opening was sealed with film and
the table was agitated horizontally for 45 min. Finally, the
samples were washed three times with TBS for 15 min per wash and
developed in a dark room by X-ray radiography.
Assessment of Qizhu formula effect on
survivin protein expression in MGC-803 cells using RT-PCR
Grouping
A 0.1% DMSO group and three medication groups with
different concentrations of Qizhu formula (125, 250 and 500
μg/ml) were set up.
Methods
Cell collection
First, the MGC-803 cells were collected at the
logarithmic phase and added to four 30-cm2 Petri dishes,
with 4.5×106 cells per dish. The cells were placed in a 5%
CO2 incubator at 37°C for 24 h, followed by the addition
of 6 ml 0.1% DMSO, 125 250, or 500 μg/ml Qizhu formula and
placement in a 5% CO2 incubator at 37°C for 24 h.
Total MGC-803 cell RNA extraction
TRIzol one-step extraction was performed according
to the manufacturer's instructions as follows: First, the
supernatant was removed, 3 ml TRIzol (1 ml/10 cm2) were
added to each Petri dish, the homogenate samples were repeatedly
absorbed and blown 20 times with a 1-ml spearhead and the sample
was left to rest for 5 min at 15°C. Second, the supernatant was
obtained after a 10-min centrifugation at 10,000 × g at 4°C; the
sample was then vigorously agitated for 15 sec following the
addition of 0.6 ml trichloromethane (0.2 ml CHCl3/1 ml
TRIzol) and was kept for 3 min at room temperature. Third, the
sample was centrifuged for 15 min at 10,000 × g at 4°C, the upper
layer of fluid was transferred to another dry Eppendorf tube, 1.5
ml of isopropanol (0.5 ml isopropanol/1 ml TRIzol) were added, the
sample was kept for 10 min at room temperature after blending and
centrifuged for 10 min at 10,000 × g at 4°C. Fourth, the
supernatant was discarded, the sediment was removed, 3 ml 75%
alcohol (1 ml 75% alcohol/1 ml TRIzol) were added and the solution
was blended. The supernatant was then discarded by absorption with
a spearhead following centrifugation for 5 min at <7,500 × g at
4°C and the sample was dried for 5–10 min at room temperature.
Fifth, 20 μl water without RNA enzymes were added and the
pellet was dissolved using the blow and beat of the spearhead. The
optical density (OD) at 260 and 280 nm was measured using an
ultraviolet spectrophotometer following addition of water without
RNA enzymes to 2 μl RNA to a final volume of 1 ml. Finally,
the RNA concentration was calculated according to the formula
[(OD260nm - ODH O) × H2O
(ml)/template (μl)] × 40 μg/μl and the RNA
purity to OD260/OD280. The ranges of
extracted RNA OD260/OD280 of 1.78–2.0 were
diluted to 1 μg/μl and kept at −72°C.
RT-PCR was performed according to the instructions
of a general RT-PCR kit. Based on the primer design of the gene
sequences provided by the gene pools, the primer sequences were as
follows through Blast comparison: β-actin gene primer sequence:
upstream, 5′-ATCATGTTTGAGACCTTCAACA-3′ and downstream,
5′-CATCTCTTGCTCGAAGTCCA-3′ with a product length of 318 bp;
survivin gene primer sequence: upstream,
5′-CACCGCATCTCTACATTCAAG-3′ (97,118) and downstream,
5′-GAAGCAGCCACTGTTACCAG-3′ (681,700) with an amplified fragment
length of 612 bp.
RT reaction
Total RNA (5 μg) was transferred to a 0.2-ml
centrifuge tube, maintained at 65°C for 10 min and placed in an ice
bath following centrifugation for several seconds. We then prepared
a 20 μl reactive system of table preparation in the
centrifugal tube: 1 μl solution A [random primer (RP)], 1
μl solution B (dNTPs), 4 μl solution D (5X RT
buffer), 1 μl solution E (enzymatic mixed solution), 5
μl RNA and mRNA samples, followed by the addition of 8
μl solution G to the final volume of 20 μl. The
sample was left to rest for 10 min at room temperature and the
temperature was maintained for 30 min at 37°C, 5 min at 95°C and
4°C for 5 min, respectively. The samples were immediately used or
stored at −20°C.
PCR reaction
The 50-μl PCR sample was prepared as follows:
4 μl RT reactive products, 4.5 μl solution F (10X PCR
buffer), 1 μl solution B (dNTPs), 50 pmol upstream primers,
50 pmol downstream primers, 1 μl solution C (Taq
enzyme) and solution G to a volume of 50 μl. A PCR amplifier
was then added following centrifugation for several seconds and the
result was finally observed using agarose gel electrophoresis. The
amplification parameters were as follows: denaturation for 2 min at
94°C (denaturation at 94°C for 45 sec; renaturation at 55°C for 45
sec; extension at 72°C for 45 sec), amplification for 30 cycles,
extension for 5 min at 72°C and storage at 4°C.
Semi-qualitative agarose
electrophoresis and analysis
First, ethidium bromide was added to 10 μl of
the PCR products and electrophoresis was conducted on a 2% agarose
gel (1 h at 80 V). Images of the electrophoretogram were captured
using an FR-980 biological electrophoresis image analysis system
(Shanghai Furi Technology Co., Ltd., Shanghai, China) and the
results were analyzed using SmartView analysis software (Everett,
WA, USA). The survivin/β-actin ratio of each sample was calculated
using β-actin as an internal standard. A smaller ratio indicated a
lower level of survivin mRNA expression.
Results
Experimental results of the Qizhu
formula on survivin protein expression in MGC-803 cells with
western blotting
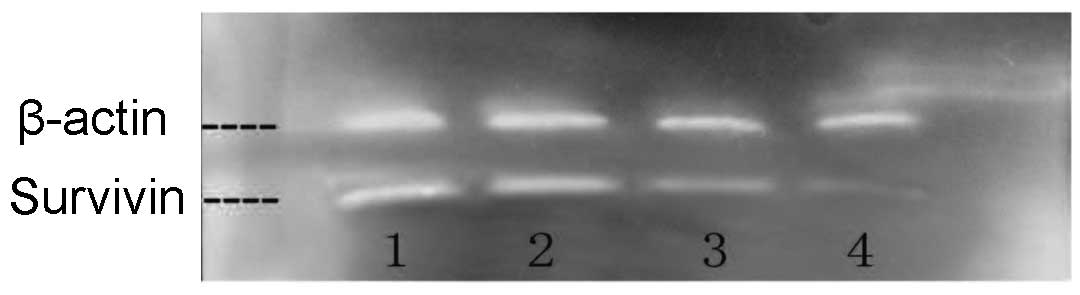
A gray qualitative analysis of β-actin and survivin
indicated that the Qizhu formula exerted no explicit effects on the
protein expression of the housekeeping β-actin gene; however, it
exerted a significant inhibitory effect on the protein expression
of the apoptosis-related survivin gene at concentrations of 250
μg/ml and, particularly, 500 μg/ml (Fig. 1).
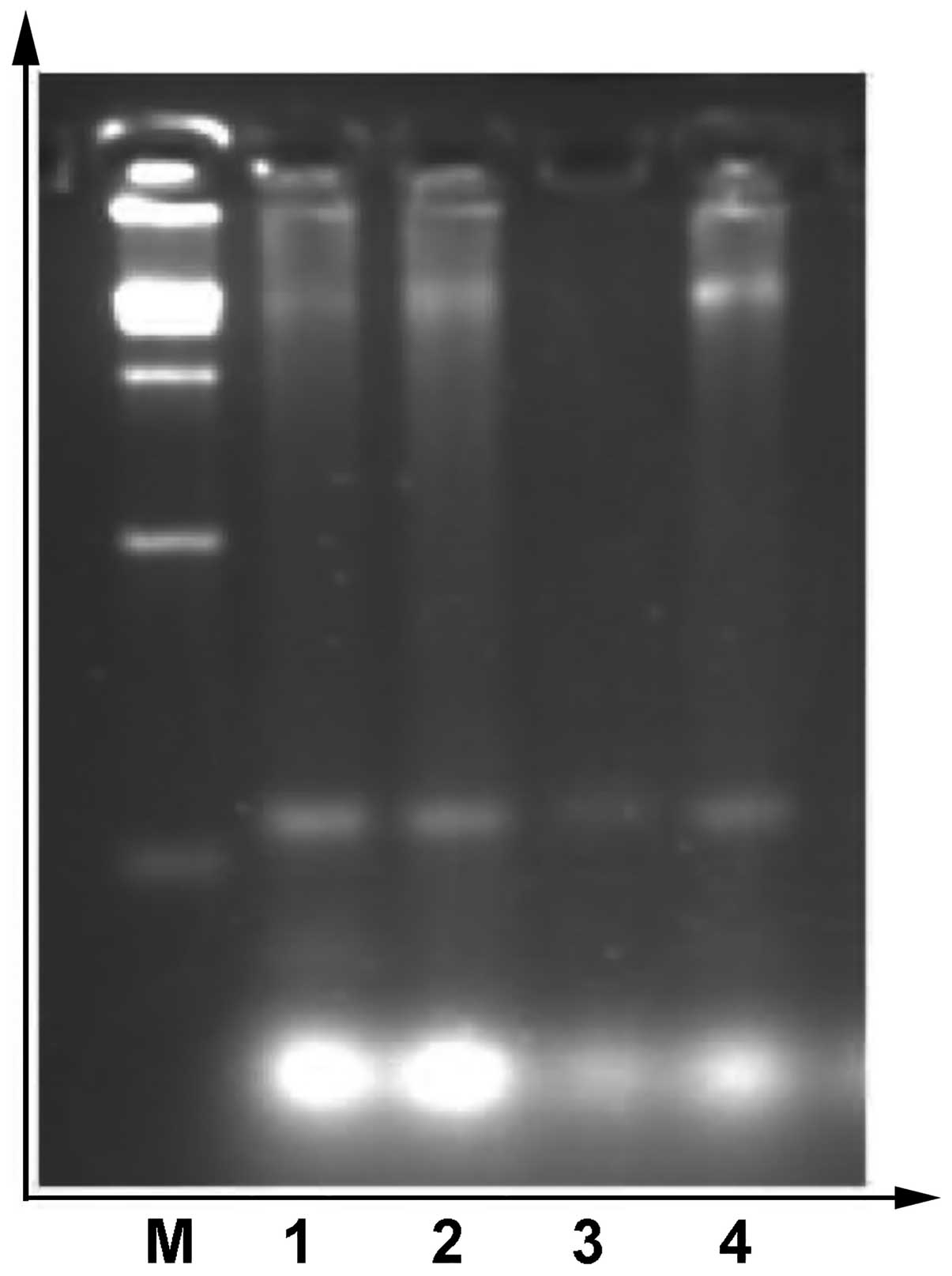
Experimental results of survivin mRNA
expression in MGC-803 cells with RT-PCR
The effect of Qizhu formula on survivin mRNA
expression in MGC-803 human gastric adenocarcinoma cells is shown
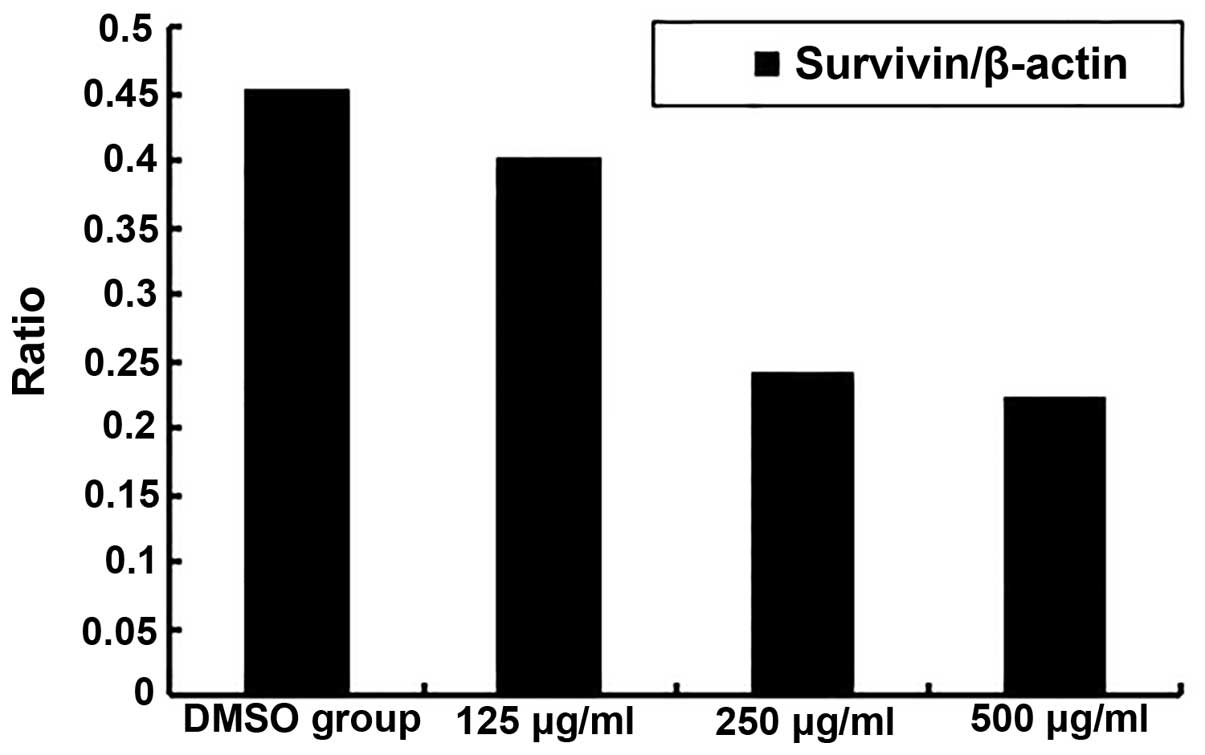
in Fig. 2. The ratios of
survivin/β-actin in the 0.1% DMSO group and in each of the Qizhu
formula groups (125, 250 and 500 μg/ml) were 0.4543, 0.4025,
0.2415, and 0.2235, respectively (Fig.
3).
The Qizhu formula was shown to modulate the protein
expression of survivin in MGC-803 human gastric adenocarcinoma
cells, indicating that this formula acts by downregulating the
inhibition of apoptosis of tumor cells. Additionally, Qizhu formula
exerted a significant inhibitory effect on survivin mRNA expression
in MGC-803 human gastric adenocarcinoma cells in a dose-dependent
manner. These results further indicate that Qizhu formula may
induce apoptosis of gastric cancer cells by downregulating IAP
survivin mRNA expression.
Discussion
Cell proliferation and apoptosis are essential in
maintaining body homeostasis. Tumor cells accelerate cell cycles
and downregulate apoptosis-related genes using complex molecular
mechanisms, consequently contributing to the malignant
proliferation characteristics of tumors. Apoptosis is a key event
in the process of tumor growth and tumor cells frequently exhibit
defective apoptotic signal transduction pathways.
Previous studies indicated that the high expression
of survivin, the most potent IAP currently identified and the
smallest of the IAP family, is able to inhibit apoptosis induced by
various apoptosis-stimulating factors, such as caspases. Wen et
al (9) demonstrated that
survivin expression in gastric cancer tissue was negatively
associated with caspase-3 expression, whereas the changes in the
expression levels of caspase-3 in gastric cancer tissues were
correlated with the occurrence and development of gastric cancer,
indicating that survivin exerts its inhibitory effect on cell
apoptosis by inhibiting caspase-3, resulting in tumorigenesis and
cancer progression.
Our earlier study (8) also indicated that Qizhu formula not
only decreases telomerase activity in MGC-803 human gastric
adenocarcinoma cells and protein expression of the
telomerase-related hTERT gene, but it can also induce caspase-3, an
apoptosis-related protease in its core position.
The present study demonstrated the ability of Qizhu
formula to modulate protein and mRNA expression of the survivin
gene in MGC-803 cells. The gray qualitative analysis of β-actin and
survivin revealed that this formula exerted no explicit effects on
the protein expression of the β-actin housekeeping gene, whereas it
exerted a significant inhibitory effect on the protein expression
of the apoptosis-related survivin gene at concentrations of 250
μg/ml and, particularly, 500 μg/ml. RT-PCR was used
to detect the effect of Qizhu formula on survivin mRNA expression
in MGC-803 human gastric adenocarcinoma cells. The ratios of
survivin/β-actin in the 0.1% DMSO group and in each of the Qizhu
formula groups (125, 250 and 500 μg/ml) were 0.4543, 0.4025,
0.2415 and 0.2235, respectively, indicating that Qizhu formula
exerted a significant inhibitory effect on MGC-803 survivin mRNA
expression in a dose-dependent manner. These results demonstrated
that the Chinese medicinal Qizhu formula decreases protein and mRNA
expression of the IAP survivin gene, which may represent an
additional target pathway via which Qizhu formula intervenes in the
process of tumor apoptosis and exerts its antitumor effects.
It was previously demonstrated that the Qizhu
formula inhibits the telomerase proliferation of MGC-803 gastric
cancer cells and induces caspase-3, an apoptosis-related protease,
in the core position and decreases the protein and mRNA expression
of the IAP survivin gene (8).
Therefore, it may be deduced that Qizhu formula intervenes in
gastric cancer through multiple pathways, multiple targets and
bidirectional regulation, which further provides objective evidence
of the anticancer efficacy of this Chinese medicinal compound.
References
|
1.
|
Ambrosini G, Adida C and Altieri DC: A
novel anti-apoptosis gene, survivin, expressed in cancer and
lymphoma. Nat Med. 3:917–921. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Liang QL, Wang BR and Li GH: DcR3 and
survivin are highly expressed in colorectal carcinoma and closely
correlated to its clinicopathologic parameters. J Zhejiang Univ Sci
B. 10:675–682. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Hu ZD, Zhong MZ and Zhou HJ: Expression
and significance of MMP-7 and survivin in gastric cancer. Cancer
Res Prev Treat. 39:1349–1352. 2012.
|
|
4.
|
Hoffmann AC, Vallböhmer D, Grimminger P,
et al: Preoperative survivin mRNA detection in peripheral blood is
an independent predictor of outcome in esophageal carcinoma.
Pharmacogenomics. 11:341–347. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Krepela E, Dankova P, Moravcikova E, et
al: Increased expression of inhibitor of apoptosis proteins,
survivin and XIAP, in non-small cell lung carcinoma. Int J Oncol.
35:1449–1462. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Meng H, Lu C, Mabuchi H and Tanigawa N:
Prognostic significance and different properties of survivin
splicing variants in gastric cancer. Cancer Lett. 216:147–155.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Yao XQ, Liu FK and Qi XP: Research on the
correlation between survivin gene expression in gastric
adenocarcinoma tissue and cell proliferation as well as apoptosis.
Chin J Surg. 42:145–148. 2004.(In Chinese).
|
|
8.
|
Wang XH and Shan ZW: Effects of Qizhu
Formula on caspase-3 and telomerase proliferation and apoptosis in
human gastric adenocarcinoma cells MGC-803. Chin J Integr Trad West
Med Dig. 14:90–92. 2006.(In Chinese).
|
|
9.
|
Wen YY and Liu BH: Functions of survivin
gene and caspase-3 in the genesis of gastric cancer. Dig Surg.
3:415–418. 2004.
|