Introduction
Cervical cancer is the second most common cancer in
women worldwide after breast cancer, and it is particularly
prevalent in developing countries (1), accounting for ~275,100 deaths annually
worldwide (2). Cervical cancer is
caused by persistent infection by high-risk types of the human
papillomavirus (HPV) (3).
Histologically, cervical cancer is almost entirely classified into
two types: Squamous cell carcinoma (SCC; ~80%) and adenocarcinoma
(~5–20%) (4). SCC develops from
cervical intraepithelial neoplasia (CIN), whereas adenocarcinoma
develops from intraepithelial adenocarcinoma and glandular
dysplasia. To reflect their relative risk of progression to
cervical cancer, CIN1 is considered low-grade CIN, whereas CIN2-3
is considered high-grade CIN. High-grade CIN may arise 2–3 years
after high-risk HPV infection, subsequently causing cervical cancer
after ≥10 years (5).
The most widely known tumor marker for cervical
cancer is SCC antigen, which is a tumor-associated antigen
identified by Kato et al in 1977 (6). The positive detection rate of SCC
antigen in each clinical stage of cervical cancer is 2.4% (stage
0), 22.2% (stage I), 56.7% (stage II), 76.4% (stage III), 76.9%
(stage IV) and 87% in recurrent cancer (7). However, the positive detection rate is
low in early stages.
MicroRNAs (miRNAs) are small (typically 19–25
nucleotides), non-coding, endogenous, single-stranded RNAs. miRNAs
were first described in 1993 by Lee et al in
Caenorhabditis elegans (8).
The majority of miRNAs negatively regulate target mRNAs in diverse
biological processes, including cell proliferation,
differentiation, development, metabolism and death. miRNAs are
often aberrantly expressed in a number of human malignancies, and
play key roles in tumor initiation and development. miRNAs suppress
the translation of target mRNAs, mainly by binding to their
3′-untranslated region (9). Moreover,
miRNAs significantly affect the expression of tumor oncogenes and
suppressor genes, being involved in both tumor promotion and tumor
suppression. Furthermore, epigenetic changes, such as DNA
methylation, are involved in the abnormal expression of certain
miRNAs (2,10). A single miRNA may have thousands of
targets and approximately one-third of human genes may be
controlled by miRNAs (11). The
mechanism of miRNA expression in cancer cells has not been fully
elucidated. However, altered expression of miRNAs has been detected
in various types of cancer and appears to play a major role in the
onset and progression of cancer.
It was recently discovered that miRNAs embedded in
the small granules of exosomes are secreted into extracellular
regions, including serum, urine and saliva (12). Serum miRNA detection may serve as a
diagnostic or prognostic biomarker in cancer patients. Accordingly,
our study was designed to analyze circulating miRNA levels in
subjects with cervical cancer, in order to assess the potential
value of miRNAs as diagnostic biomarkers.
Materials and methods
Study design and study population
A total of 131 subjects participated in the study,
which was conducted at the Tokyo Medical University Hospital from
April, 2010 to March, 2012. Of these subjects, 45 had cervical
cancer, 55 had CIN and 31 were healthy. The research protocol was
approved by our Institutional Review Board (approval no. 3268) and
informed consent was obtained from all the subjects. Of the 45
patients with cervical cancer, 7 were stage Ia, 16 were stage Ib,
10 were stage IIb, 3 were stage III and 2 were stage IV. Of the 55
subjects with CIN, 15 were CIN1, 16 were CIN2 and 24 were CIN3. The
patient backgrounds were obtained through interviews. Blood samples
were collected prior to chemotherapy and radiation therapy.
Cervical cancer and CIN were diagnosed on the basis of histological
examinations. The characteristics of the subjects are summarized in
Table I.
 | Table I.Characteristics of the subjects
participating in the study. |
Table I.
Characteristics of the subjects
participating in the study.
| Characteristics | Cervical cancer
(n=45) | CIN1 (n=15) | CIN2 (n=16) | CIN3 (n=24) | Normal (n=31) |
|---|
| Age, years |
|
| Median ±
SD | 49.0±14.1 | 34.0±6.7 | 35.0±9.2 | 34.0±4.9 | 39.0±11.2 |
|
Range | 28–88 | 27–50 | 19–55 | 26–43 | 24–69 |
| Smoking | 9 | 3 | 5 | 3 | 3 |
| Histological
type |
|
| SCC | 38 |
|
|
Adenocarcinoma | 9 |
|
| Clinical
stage |
|
| Ia | 7 |
|
| Ib | 16 |
|
| IIa | 7 |
|
| IIb | 10 |
|
|
IIIIV | 3 |
|
| IV | 2 |
|
| Serum SCC antigen,
ng/ml |
|
| Median ±
SD | 2.65±6.61 |
|
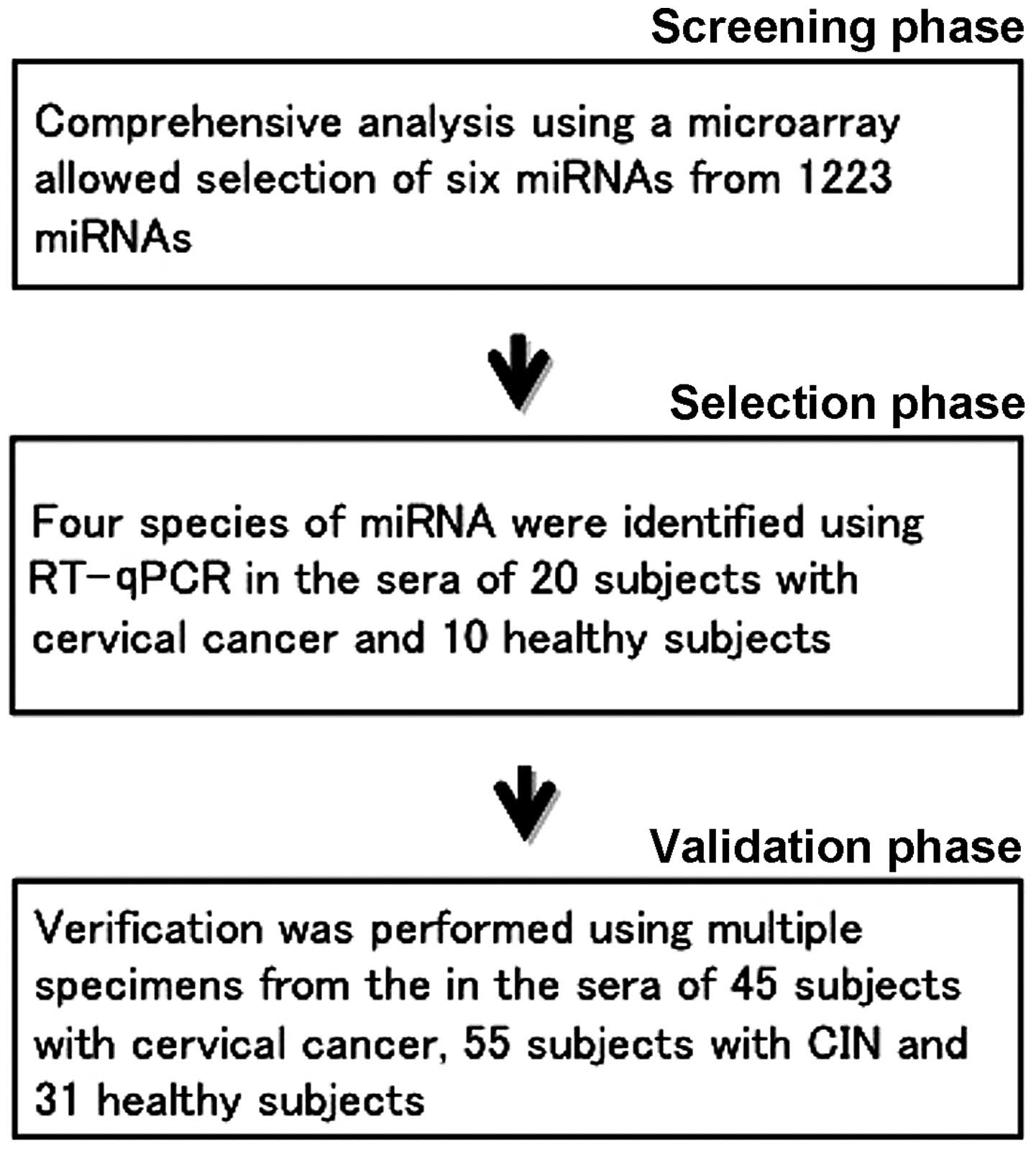
The present study was conducted in three phases
(Fig. 1). In the first phase
(screening phase), we performed comprehensive analysis of the serum
miRNA of the cervical cancer subjects using a microarray, and
selected 6 candidate miRNAs. We used 10 samples in total, 5 from
cervical cancer subjects and 5 from healthy subjects. In the second
phase (selection phase), the 6 candidate miRNAs were narrowed down
to 4 using reverse transcription-quantitative polymerase chain
reaction (RT-qPCR). For the RT-qPCR, we used serum miRNA from 20
subjects with cervical cancer and 10 healthy subjects. In the third
phase (validation phase), we performed RT-qPCR on the 4 miRNAs
identified during the selection phase. In order to validate these 4
miRNAs, we examined the serum miRNA of 45 subjects with cervical
cancer, 55 subjects with CIN and 31 healthy subjects, performed
statistical processing and investigated the potential of each miRNA
as a tumor marker.
Comprehensive analysis of serum miRNAs
by microarray
In the screening phase, total RNA was extracted
using ISOGEN-LS (Nippon Gene, Tokyo, Japan) and the serum of 5
subjects with cervical cancer and that of 5 healthy subjects. Total
RNAs were purified using a column. Each total RNA was labeled with
Hy3 or Hy5 using high-affinity locked nucleic acid (LNA) probes,
hybridized overnight and scanned to be digitized. We selected the
miRNAs exhibiting an expression level in the cervical cancer
subjects that was more than three times higher compared with that
in healthy subjects, as indicated by miRNA profiling, which left us
with 6 miRNA species out of the original 1,223.
Identification of 4 miRNA species by
qPCR
In the selection phase, total RNA was extracted from
the serum of 20 subjects with cervical cancer and 10 healthy
subjects. RT-qPCR was performed on the total RNA, and 4 miRNA
species were selected (miR-483-5p, miR-1246, miR-1275 and miR-1290)
from the 6 miRNA species. In the validation phase, multiple samples
were used to validate the miRNAs. Total RNA was extracted from the
serum of 45 subjects with cervical cancer, 55 subjects with CIN and
31 healthy subjects, and subjected to RT-qPCR.
Serum preparation and total RNA
extraction
Venous blood samples were collected from cervical
cancer patients and healthy controls. The samples were separated
into blood cells and serum by centrifugation, and stored at −5°C.
Total RNA in the serum was isolated using ISOGEN-LS, according to
the manufacturer's instructions. Briefly, 250 µl of serum was
homogenized in 750 µl of ISOGEN-LS and 200 µl of chloroform was
added to the sample, which was centrifuged. Following additional
chloroform extraction and precipitation with isopropanol, the RNA
sample was suspended in 20 µl of nuclease-free water (13).
Validation of miRNA expression by
qPCR
We quantified miRNAs using TaqMan MicroRNA assays
(Applied Biosystems, Foster City, CA, USA) with modifications and
miRNA-specific stem-loop primers (has-miR-1290; Applied
Biosystems). First, each miRNA was specifically reverse-transcribed
to cDNA according to the manufacturer's protocol, using TaqMan
miRNA RT-Kit with stem-loop RT-primer and the Applied Biosystems
9800 Fast Thermal Cycler. Second, qPCR was performed for each
specific miRNA using an RT-primer with Universal Master Mix on the
Applied Biosystems StepOnePlus™ Real-Time PCR system (Life
Technologies Corporation, Carlsbad, CA, USA). Sequence detection
was performed according to the manufacturer's protocol. The
reaction mixtures were incubated at 95°C for 2 min, followed by 50
cycles at 95°C for 15 sec and 60°C for 1 min. Each sample was
analyzed in duplicate. The cycle thresholds (Ct) for subjects with
cervical cancer, subjects with CIN and healthy subjects were
calculated and normalized to miR-16, which was found in the
literature to be the most widely-used endogenous control miRNA for
RT-qPCR (14). The expression levels
of miRNAs in subjects with cervical cancer and those with CIN
relative to healthy controls were calculated using the comparative
Ct method. The average Ct value of the control miR-16 for every
sample was subtracted from the Ct value for each respective mature
miRNA reaction, resulting in the ΔCt value. The -ΔΔCt value was
calculated by subtracting the -ΔCt value of a normal sample from
the respective -ΔCt values of the patient samples. Expression of
miR-1290 was normalized using the 2−ΔΔCt method
(13,14). The miR-1290 expression profile was
used to create a receiver operating characteristic (ROC) curve,
which is a graphical plot of the true-positive vs. the
false-positive rate. The area under the ROC curve represents the
discrimination accuracy.
Statistical analysis
Statistical analysis of the causal association
between the clinical background and expression level of the miRNAs
was performed using EZR software (15). A P-value of <0.05 was considered to
indicate statistically significant differences.
Results
miRNA expression status in cervical
cancer
The characteristics of the 131 subjects are
summarized in Table I. The samples
used in this study were obtained from subjects with CIN, subjects
with cervical cancer undergoing surgery or biopsy and healthy
volunteers, following informed consent. Table I shows the characteristics of the
patients with cervical cancer. Total RNA was extracted from the
serum. We first investigated the miRNA expression profiles using
miRCURY LNA microRNA array (Exiqon, Copenhagen, Denmark) in serum
samples from 6 patients with cervical cancer and 6 healthy subjects
to screen for candidate miRNAs associated with the development or
progression of cervical cancer. The purpose of this analysis was to
screen for a specific miRNA which may serve as a diagnostic or
prognostic biomarker for cervical cancer patients. Initially, we
compared miRNA expression in the serum of cervical cancer patients
to that in the serum of healthy subjects. Of the 1,223 miRNAs
compared in serum samples from cervical cancer patients and healthy
controls using a miRCURY LNA microRNA array, 6 were found to
exhibit a >3.0-fold change in their expression level
(P<0.01).
To perform a technical selection of the array
results, we analyzed 6 miRNAs in the serum of 20 patients with
cervical cancer and 10 healthy subjects by RT-qPCR using TaqMan
gene expression assays (Applied Biosystems). Among the 6 miRNAs
examined, miR-485-5p, miR-1246, miR-1275 and particularly miR-1290,
were found by RT-qPCR to be expressed at significantly higher
levels in cervical cancer patients compared with healthy controls.
As the difference in expression was particularly high for miR-1290,
we focused on this miRNA in the next step.
Elevated miR-1290 in the serum of
cervical cancer patients
We next determined whether miR-1290 could be
detected in the serum and if it was more abundant in subjects with
CIN and cervical cancer. We measured the miR-1290 concentration in
100 subjects with cervical neoplasia and 31 healthy controls. The
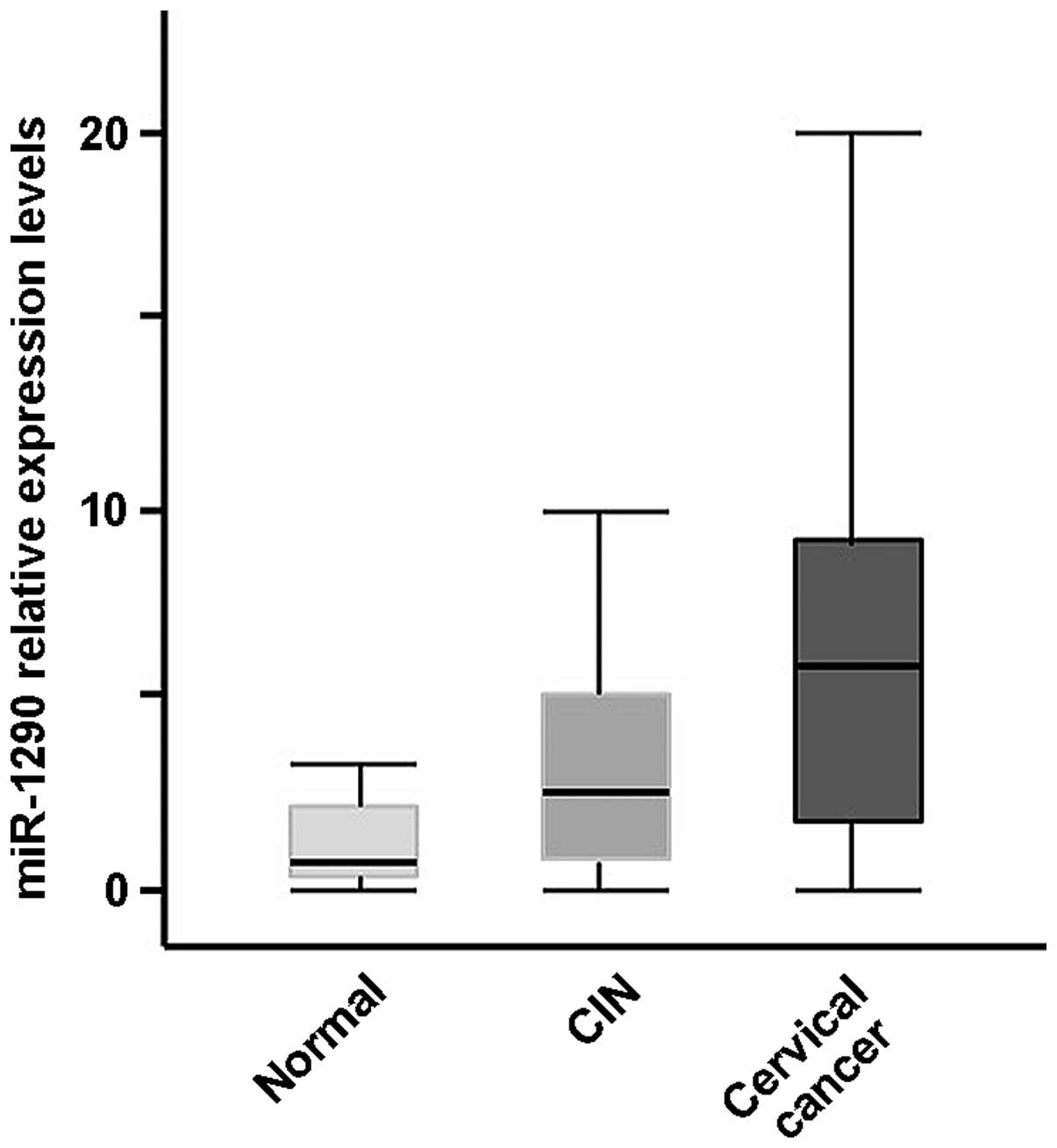
expression level of miR-1290 was significantly elevated in the
serum samples from subjects with cervical cancer, but not in those
from subjects with CIN, compared with healthy controls (P<0.001
and P>0.05, respectively, t-test; Fig.
2; miR-16 was used as a reference). The expression level of
miR-1290 was significantly elevated in the serum of subjects with
cervical cancer compared with that of CIN subjects (P<0.01,
t-test; Fig. 2; miR-16 was used as a
reference). The comprehensive analysis of the serum was performed
using the multi-analyte serum miR-1290, which was significantly
higher in subjects with cervical cancer compared with healthy
subjects. The expression level of serum miR-1290 was the lowest in
healthy subjects, higher in CIN subjects, and the highest in
cervical cancer patients. The median serum miRNA-1290 expression
level was 0.78 in healthy subjects, 2.46 in CIN subjects and 4.04
in cervical cancer patients (P<0.01, Kruskal Wallis test).
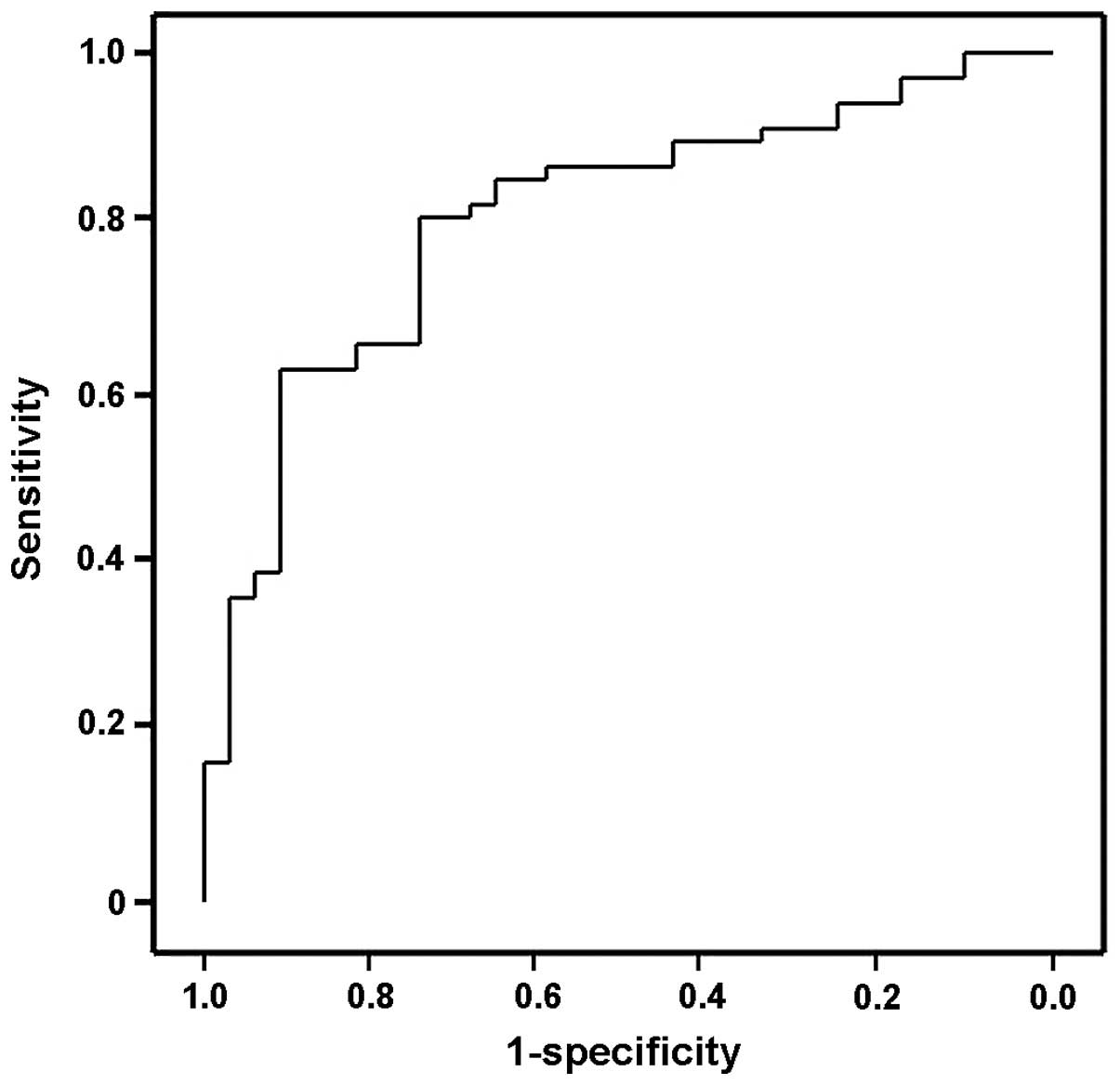
The ROC curve indicated that the serum levels of
miR-1290 may differentiate subjects with cervical cancer from
healthy controls, with ROC curve areas of 0.7957 (95% confidence
interval: 0.6937–0.8977) (Fig. 3).
With the cut-off at 3.959 (relative expression value), miR-1290 had
a 90.3% sensitivity for cervical cancer and a specificity of 62.2%,
compared with healthy controls.
Serum miR-1290 in subjects at each
stage of cervical cancer
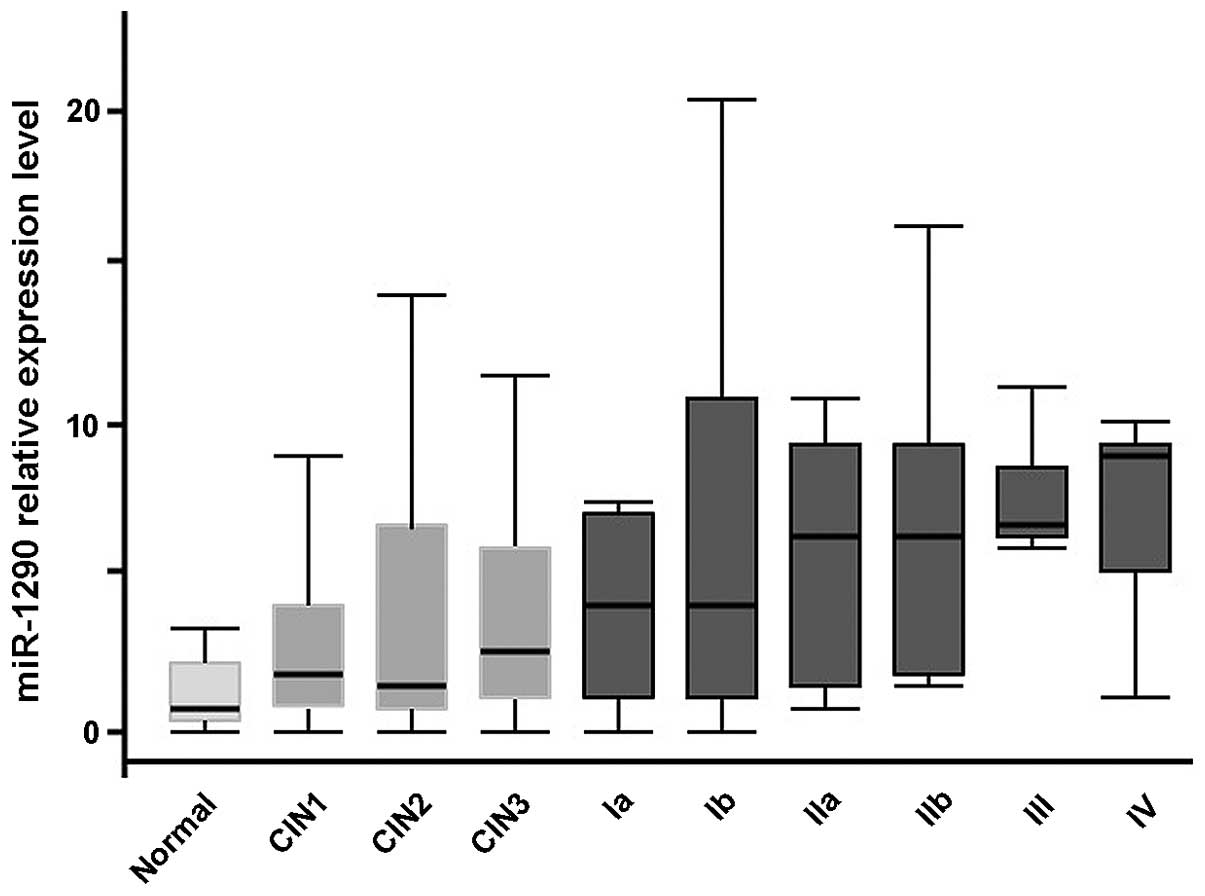
The median expression level of serum miR-1290
increased along with the stage of cervical cancer. The level in
each group was as follows: Healthy group, 0.78; CIN1-2 group, 1.81;
CIN3 group, 2.74; cervical cancer stage I group, 4.00; stage II
group, 5.66; and stage III–IV group, 5.59 (P<0.01,
Kruskal-Wallis test, Fig. 4). There
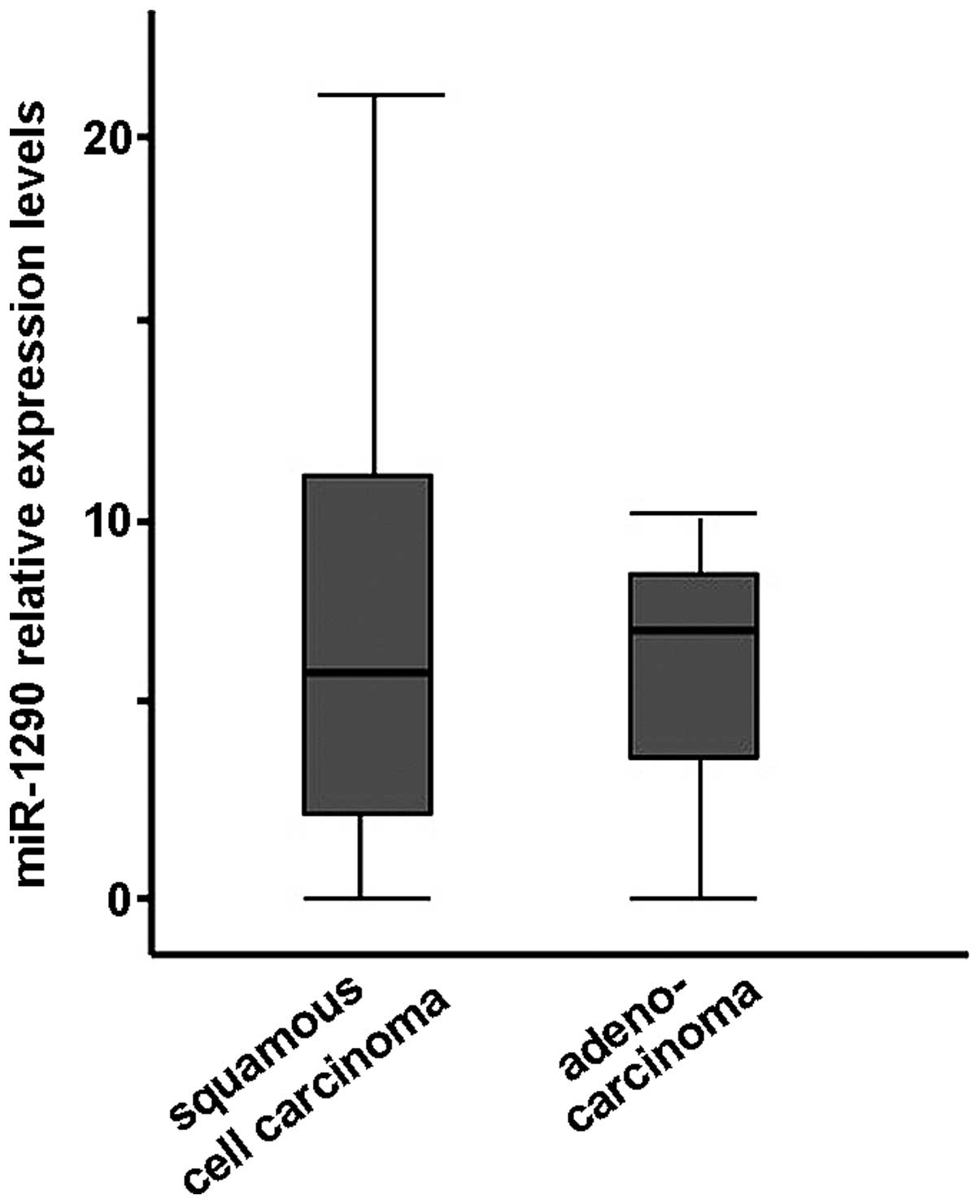
was no significant difference by histological type, i.e., SCC and
adenocarcinoma (Welch's t-test, Fig.
5). In addition, there was no correlation between clinical
data, such as smoking, SCC antigen level, carcinoembryonic antige
level and lymph node metastasis, and miRNA expression (Mann-Whitney
U test).
Discussion
As some biomarkers are able to detect cancer at
early stages, they may improve patient survival; thus, they are key
to the prognosis and diagnosis of cancer. SCC antigen is a tumor
marker in cervical cancer, but the level of this marker is not
increased during the early stages. Several clinicians have long
stressed the need for novel biomarkers for early detection of
cervical cancer. Abnormal expression of miRNAs has been reported in
a number of cervical cancer tissues and cell lines (16). These miRNAs are able to promote cell
proliferation and reduce apoptosis, affect cell invasion, and
eventually contribute to the initiation and progression of cervical
cancer (1). miRNAs are very stable
and permit easy detection of specific types of cancer (17).
The identification of circulating miRNAs is one of
the major scientific breakthroughs in recent years and it has
revolutionized cell biology and medical science. Growth factors,
such as cytokines, are the principal communication tools between
cells, but communication by exosomes between the peripheral cells
and cancer cells has also been noted to be significant (18). Circulating miRNAs function as
‘extracellular communication RNAs’ that play an important role in
cell proliferation and differentiation (12). Blood-based miRNA profiling is not as
reliable as tissue-based miRNA-profiling, but offers the potential
for early, non-invasive, sensitive and specific cervical cancer
detection and screening. Recently, several circulating miRNAs have
been identified as potential serum biomarkers in different cancer
types (19). These serum miRNAs may
be effective as predictive biomarkers in cancer. Chen et al
revealed that serum miRNA expression level is correlated with
specific types of cancer, such as lung and colorectal cancer
(17). Serum miRNAs are non-invasive
biomarkers that may permit early detection of cancer.
We investigated the expression of serum miRNAs in
healthy subjects, subjects with CIN and patients with cervical
cancer using an miRNA microarray and RT-qPCR. The expression of
serum miR-1290 was the lowest in healthy subjects, higher in CIN
subjects, and the highest in cervical cancer patients. Moreover,
the expression level of serum miR-1290 was higher in high-grade CIN
subjects compared with that in subjects with lower-grade CIN. The
expression level of miR-1290 tended to increase with advancing
clinical stage of cervical cancer. High-risk HPV infection plays a
central role in cervical carcinogenesis. These high-risk HPV
serotypes are found in CIN, even in CIN1. HPV infection may
increase miR-1290 expression directly or indirectly, since miR-1290
is already increased in CIN1. In order to normalize the miRNA
expression level, miR-16 was used as an internal control, as
previously reported (13,14,20).
The miR-1290 gene is present in chromosome
1:19,223,565–19,223,642 (http://www.mirbase.org/). Some of the functions of
miR-1290 have been previously reported. Wu et al suggested
that miR-1290 is significantly upregulated in colon cancer tissues
(21); moreover, they found that
upregulation of miR-1290 impairs cytokinesis and leads to the
formation of multinucleated cells in vitro and in
vivo, while also resulting in Akt and nuclear factor-κB
activation, which maintains cell proliferation (21). Endo et al suggested that
miR-1290 and its potential target genes, forkhead box protein A1
(FOXA1) and N-acetyltransferase (NAT) 1, may be associated with the
characteristics of estrogen receptor-positive breast cancer
(22). FOXA1 is a forkhead family
transcription factor, and arylamine NATs, known as drug- and
carcinogen-metabolizing enzymes, transfer an acetyl group from
acetyl coenzyme A to arylamines. They also suggested that B-cell
lymphoma 2 (BCL2) and microtubule-associated protein tau (MAPT) are
potential targets of miR-1290 according to in silico
analysis. BCL2 is an anti-apoptotic protein that exerts an
antiproliferative effect through affecting cell cycle entry. MAPT
binds to both the outer and the inner surfaces of microtubules,
leading to tubulin assembly and microtubule stabilization (22). It has been reported that 6 miRNAs,
including miR-1290, are upregulated in drug-sensitive cells
following Y-Box protein 1 inhibition, but no differences in miRNA
expression have been detected in multidrug-resistant gastric
carcinoma cells (23). It has also
been reported that 36 miRNAs, including miR-1290, circulate at
higher levels in subjects with renal cell carcinoma compared with
those in healthy controls (24).
We found that serum circulating miR-485-5p, miR-1246
and miR-1275, as well as miR-1290, were significantly higher in
cervical cancer subjects compared with healthy controls. The
expression of miR-485-5p and miR-1275 was significantly
downregulated in hepatocelllular carcinoma (HCC) tissues (25,26). In
particular, miR-485-5p expression was inversely correlated with TNM
stage and metastasis in HCC samples (25). Serum miR-1246 was significantly higher
in primary colorectal and esophageal cancer patients compared with
in healthy controls and significantly correlated with TNM stage
(27,28). Therefore, these three miRNAs
(miR-485-5p, miR-1246 and miR-1275), may be biomarkers for cervical
cancer with lower specificity.
In conclusion, serum miR-1290 appears to be a useful
biomarker for the early detection of cervical cancer. It may reduce
the need for invasive cervical biopsies and be useful in predicting
the prognosis of cervical cancer. Larger studies are required to
fully elucidate the role of miR-1290 in cervical cancer, and to
determine whether serum miR-1290 may serve as a diagnostic marker
or biomarker of treatment efficacy in this disease.
Acknowledgements
The authors would like to thank Ms. Chinatsu
Yoneyama (Department of Obstetrics and Gynecology, Tokyo Medical
University) and Dr Tomohiro Umezu (Department of Molecular
Oncology, Institute of Medical Science, Tokyo Medical University)
for providing invaluable experimental assistance. This study was
supported by the Private University Strategic Research Based
Support Project (grant no., S1311016) and Grants-in-Aids (grant
no., 24592533) from the Ministry of Education, Culture, Sports,
Science and Technology of Japan.
References
|
1
|
Li BH, Zhou JS, Ye F, Cheng XD, Zhou CY,
Lu WG and Xie X: Reduced miR-100 expression in cervical cancer and
precursors and its carcinogenic effect through targeting PLK1
protein. Eur J Cancer. 47:2166–2174. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yamamoto N, Kinoshita T, Nohata N, Itesako
T, Yoshino H, Enokida H, Nakagawa M, Shozu M and Seki N: Tumor
suppressive microRNA-218 inhibits cancer cell migration and
invasion by targeting focal adhesion pathways in cervical squamous
cell carcinoma. Int J Oncol. 42:1523–1532. 2013.PubMed/NCBI
|
|
3
|
Bosch FX, Lorincz A, Muñoz N, Meijer CJ
and Shah KV: The causal relation between human papillomavirus and
cervical cancer. J Clin Pathol. 55:244–265. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pisani P, Bray F and Parkin DM: Estimates
of the world-wide prevalence of cancer for 25 sites in the adult
population. Int J Cancer. 97:72–81. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
McCredie MR, Sharples KJ, Paul C, Baranyai
J, Medley G, Jones RW and Skegg DC: Natural history of cervical
neoplasia and risk of invasive cancer in women with cervical
intraepithelial neoplasia 3: A retrospective cohort study. Lancet
Oncol. 9:425–434. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kato H, Morioka H, Aramaki S and Torigoe
T: Radioimmunoassay for tumor-antigen of human cervical squamous
cell carcinoma. Cell Mol Biol Incl Cyto Enzymol. 25:51–56.
1979.PubMed/NCBI
|
|
7
|
Kato H: Squamous cell carcinoma antigen.
Serological cancer Markers. Sell S: Humana Press. (Totowa).
437–451. 1992. View Article : Google Scholar
|
|
8
|
Lee RC, Feinbaum RL and Ambros V: The
C. elegans heterochronic gene lin-4 encodes small RNAs with
antisense complementarity to lin-14. Cell. 75:843–854. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lytle JR, Yario TA and Steitz JA: Target
mRNAs are repressed as efficiently by microRNA-binding sites in the
5′UTR as in the 3′UTR. Proc Natl Acad Sci USA. 104:9667–9672. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Valadi H, Ekström K, Bossios A, Sjöstrand
M, Lee JJ and Lötvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ohyashiki K, Umezu T, Yoshizawa S, Ito Y,
Ohyashiki M, Kawashima H, Tanaka M, Kuroda M and Ohyashiki JH:
Clinical impact of down-regulated plasma miR-92a levels in
non-Hodgkin's lymphoma. PLoS One. 6:e164082011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Schrauder MG, Strick R, Schulz-Wendtland
R, Strissel PL, Kahmann L, Loehberg CR, Lux MP, Jud SM, Hartmann A,
Hein A, et al: Circulating micro-RNAs as potential blood-based
markers for early stage breast cancer detection. PLoS One.
7:e297702012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kanda Y: Investigation of the freely
available easy-to-use software ‘EZR’ for medical statistics. Bone
Marrow Transplant. 48:452–458. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Martinez I, Gardiner AS, Board KF, Monzon
FA, Edwards RP and Khan SA: Human papillomavirus type 16 reduces
the expression of microRNA-218 in cervical carcinoma cells.
Oncogene. 27:2575–2582. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K,
Guo J, Zhang Y, Chen J, Guo X, et al: Characterization of microRNAs
in serum: A novel class of biomarkers for diagnosis of cancer and
other diseases. Cell Res. 18:997–1006. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Al-Nedawi K, Meehan B, Micallef J, Lhotak
V, May L, Guha A and Rak J: Intercellular transfer of the oncogenic
receptor EGFRvIII by microvesicles derived from tumour cells. Nat
Cell Biol. 10:619–624. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Allegra A, Alonci A, Campo S, Penna G,
Petrungaro A, Gerace D and Musolino C: Circulating microRNAs: New
biomarkers in diagnosis, prognosis and treatment of cancer
(review). Int J Oncol. 41:1897–1912. 2012.PubMed/NCBI
|
|
20
|
Xu J, Cao Z, Liu W, You L, Zhou L, Wang C,
Lou W, Sun B, Miao Y, Liu X, et al: Plasma miRNAs effectively
distinguish patients with pancreatic cancer from controls: A
multicenter study. Ann Surg Jun. 25:(Epub ahead of print).
2015.
|
|
21
|
Wu J, Ji X, Zhu L, Jiang Q, Wen Z, Xu S,
Shao W, Cai J, Du Q, Zhu Y and Mao J: Up-regulation of
microRNA-1290 impairs cytokinesis and affects the reprogramming of
colon cancer cells. Cancer Lett. 329:155–163. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Endo Y, Toyama T, Takahashi S, Yoshimoto
N, Iwasa M, Asano T, Fujii Y and Yamashita H: miR-1290 and its
potential targets are associated with characteristics of estrogen
receptor α-positive breast cancer. Endocr Relat Cancer. 20:91–102.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Belian E, Kurucz R, Treue D and Lage H:
Effect of YB-1 on the regulation of microRNA expression in
drug-sensitive and drug-resistant gastric carcinoma cells.
Anticancer Res. 30:629–633. 2010.PubMed/NCBI
|
|
24
|
Wulfken LM, Moritz R, Ohlmann C,
Holdenrieder S, Jung V, Becker F, Herrmann E, Walgenbach-Brünagel
G, von Ruecker A, Müller SC and Ellinger J: MicroRNAs in renal cell
carcinoma: Diagnostic implications of serum miR-1233 levels. PLoS
One. 6:e257872011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sun X, Liu Y, Li M, Wang M and Wang Y:
Involvement of miR-485-5p in hepatocellular carcinoma progression
targeting EMMPRIN. Biomed Pharmacother. 72:58–65. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fawzy IO, Hamza MT, Hosny KA, Esmat G, El
Tayebi HM and Abdelaziz AI: miR-1275: A single microRNA that
targets the three IGF2-mRNA-binding proteins hindering tumor growth
in hepatocellular carcinoma. FEBS Lett. 589:2257–2265. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ogata-Kawata H, Izumiya M, Kurioka D,
Honma Y, Yamada Y, Furuta K, Gunji T, Ohta H, Okamoto H, Sonoda H,
et al: Circulating exosomal microRNAs as biomarkers of colon
cancer. PLoS One. 9:e929212014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Takeshita N, Hoshino I, Mori M, Akutsu Y,
Hanari N, Yoneyama Y, Ikeda N, Isozaki Y, Maruyama T, Akanuma N, et
al: Serum microRNA expression profile: miR-1246 as a novel
diagnostic and prognostic biomarker for oesophageal squamous cell
carcinoma. Br J Cancer. 108:644–652. 2013. View Article : Google Scholar : PubMed/NCBI
|