Introduction
Chronic myeloid leukemia (CML) is one of the most
common hematologic malignancies and accounts for 15% of all cases
of leukemia in adults (1). The
incidence of CML is approximately 1.6/100,000. The cytogenetic
marker of the disease is the presence of a distinctive Philadelphia
chromosome (Ph) in more than 95% of the patients (2). It is a reciprocal translocation between
the long arms of chromosomes 9 and 22. The translocation involves
the transfer of the Abelson or ABL1 gene on chromosome 9 to
the breakpoint cluster region, BCR, of chromosome 22,
resulting in a fusion BCR/ABL gene. The fusion gene produces
BCR/ABL, a tyrosine kinase with deregulated activity that plays a
key role in the development of CML.
Our case is an 83-year old woman who is directed to
the genetic laboratory for a cytogenetic and molecular-genetic
analysis suspected to be Ph positive [(+)]. Her initial diagnosis
is primary aplastic anemia with additional diagnosis of primary
(essential) hypertonia. The anamnesis is taken both from medical
documentation and the statements of patient's relatives. She is
accepted for the first time in the clinic having leukocytosis with
neutrophilia, moderate-to-severe anemic syndrome-microcytic,
hypochromic anemia; the thrombocytes are in the reference ranges.
She has a consuming syndrome accompanied with a preserved and/or
increased appetite and sore throat. Fever and feverish night sweats
are lacking, there is no bleeding.
High levels of leucocytes have been diagnosed
February, 2018. The leucocyte numbers increase in the following
several months up to 85G/l. The levels from March, 2005 are
documented as follows: St 12%; Sg 17%; Мо 2%; Ly 10%; Eo 5%; Bl +
ProM 25%; M 21%; Meta M 9%.
Upon her acceptance in the hematological ward she is
adequate and orientated. Her skin and mucosal membranes are pale
pink with isolated suffusions and hematomas; no icterus is present.
Peripheral lymph nodes are not enlarged on palpation; breathing is
clear vesicular, double-sided, without wheezing. Cardiovascular
system-there is arrhythmic cardiac activity, clear tones, systolic
noise at Ao and cardiac apex. Stomach-soft, painless; liver-1-2 cm
below the ribs' arch; spleen-non-enlarged. There are no swellings
of the limbs.
Materials and methods
At the Clinic of Hematology the patient is subjected
to routine diagnostic procedures: Whole blood count test and
biochemical analysis (Table I),
morphological analysis of the blood cells, restriction analysis for
V617F in JAK2 gene (Fig. 1),
quantitative molecular-biological analysis (Real-time PCR) at the
time of her first visit and three months after treatment (Fig. 2), and karyotyping (Fig. 3). For all procedures and tests the
patient has provided a written informed consent.
 | Table I.WBC and biochemical analysis result of
the patient at the time of her acceptance in the ward. |
Table I.
WBC and biochemical analysis result of
the patient at the time of her acceptance in the ward.
| Biochemical
characteristic | Result | Units | Reference values | Parameter | Result | Units | Referent values |
|---|
| WBC | 69.77 |
x109/l | 3.50–10.50 | Gluc | 5.58 | mmol/l | 3.50–6.10 |
| RBC | 4.34 |
x1012/l | 3.70–5.30 | Creatinine | 161.00 | µmol/l | up to 96.00 |
| HGB | 87 | g/l | 120-160 | TBil | 8.50 | µmol/l | up to 21.00 |
| HTC | 0.295 | l/l | 0.360–0.480 | ASAT | 27.60 | U/l | up to 32.00 |
| MCV | 67.8 | fl | 80.0–96.0 | ALAT | 7.00 | U/l | up to 33.00 |
| MCH | 19.90 | pg | 27.0–33.0 | LDH | 2783.00 | U/l | up to 460 |
| MCHC | 294 | g/l | 300-360 | Na | 143.00 | mmol/l | 135-151 |
| Platelets | 182 |
x109/l | 130-360 | K | 3.20 | mmol/l | 3.5–5.6 |
| % lymphocytes | 3.60 | % | 20.0–48.0 | Cl | 103.00 | mmol/l | 93-112 |
| % monocytes | 6.30 | % | 1.0–11.0 | Fe | 15.80 | µmol/l | 6.60–28.00 |
| % eosinophils | 0.80 | % | Up to 6.5 | TIBC | 50.30 | µmol/l | 42.00–78.00 |
| % basophils | 7.40 | % | Up to 2.0 | UIBC | 34.50 | µmol/l | 27.8–63.6 |
| % neutrophils | 88.00 | % | 40.0–70.0 | CRP | 0.20 | mg/l | <6 |
| No lymphocytes | 2.54 |
x109/l | 1.00–4.00 |
|
|
|
|
| No monocytes | 4.36 |
x109/l | Up to 0.80 |
|
|
|
|
| No eosinophils | 0.59 |
x109/l | Up to 0.50 |
|
|
|
|
| No basophils | 5.14 |
x109/l | Up to 0.14 |
|
|
|
|
| No neutrophils | 61.39 |
x109/l | 2.00–7.00 |
|
|
|
|
| MPV | 9.40 | fl | 6.3–12.5 |
|
|
|
|
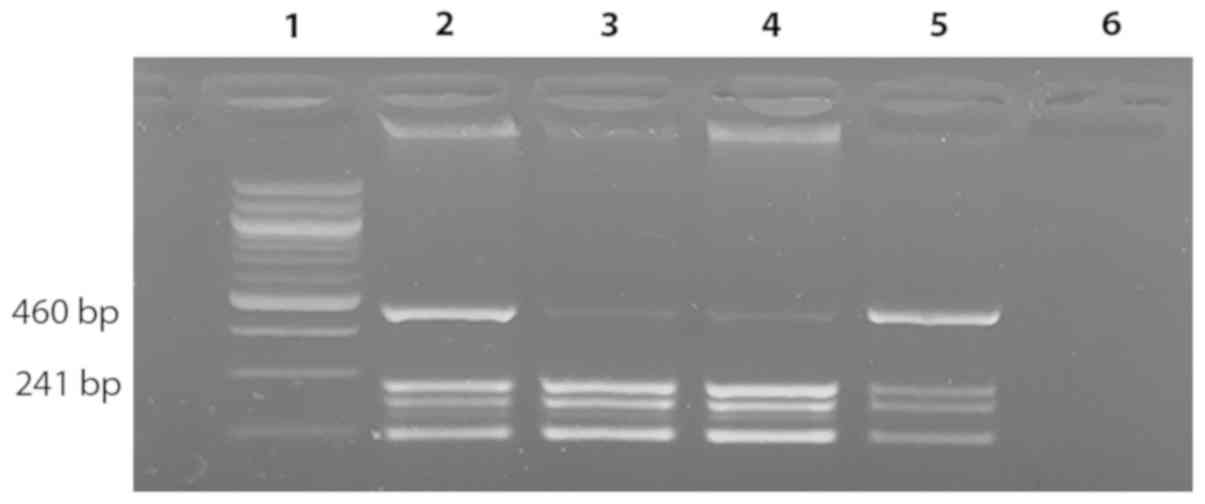
PCR amplification and restriction
analysis for JAK2 V716F mutation detection
DNA is extracted from 200 µl venous blood taken in
K2EDTA tube using QIAamp DNA Mini kit (Qiagen).
JAK2 mutation has been checked as a part of the routine
diagnostic procedure of the Clinic of Haematology when chronic
myeloproliferative process is suspected. DNA is amplified using
JAK2 codon 617 mutation specific primers (V617F)
(JAK2F 5′-GGGTTTCCTCAGAACGTT-3′ and JAK2R
5′-TCATTGCTTTCCTTTTTC-3′) for 32 cycles using Taq polymerase
(Qiagen), annealing temperature of 60°C, and standard amplification
conditions as described previously by Baxter et al (3). The amplified 460-bp fragment is
enzymatically digested using BsaXI restriction enzyme
(BioLabs™). Fragments of three different sizes (241, 189 and 30 bp)
are received after digestion of the wild type allele, while the
mutant allele remains undigested (Fig.
1). Digested fragments are separated in 2% agarose gel.
Visualization of the restriction fragments is achieved by ethidium
bromide (Fig. 1).
RT-qPCR analysis
It is carried out using the Cepheid
GeneXpert® platform. RNA has been automatically
extracted from 4 ml venous blood and converted to cDNA. Xpert
BCR-ABL kit is used for quantitative detection of BCR-ABL
chromosomal translocation mRNA transcripts and the ABL endogenous
control mRNA transcripts in peripheral blood specimen from patients
with CML. The Xpert BCR-ABL Ultra quantifies the BCR-ABL mRNA level
on the IS and is calibrated to the first World Health
Organization (WHO) international genetic reference panel for
quantitation of BCR-ABL mRNA. The GeneXpert software calculates the
%BCR-ABL/ABL (IS) using the following equation where the
Delta Ct (∆Ct) value is obtained from ABL Ct minus BCR-ABL Ct:
%BCR-ABL/ABL IS=E∆Ct(∆Ct) × 100
× Scaling Factor (SF) (4). The
efficiency value is embedded in the barcode of the Xpert BCR-ABL
Ultra cartridge; the SF is lot-specific (Xpert® BCR-ABL
Ultra Handbook) (Fig. 2).
Cytogenetic analysis
The bone marrow sample of the patient is taken by
trepanobiopsy and sent to the cytogenetic laboratory of the
Military Medical Academy (Sofia, Bulgaria). The cells are cultured
in RPMI culture medium supplied with antibiotics and growth factors
at 37°C for 24 h. 70 µl of colcemid is added to the sample and
incubated in 37°C for 20 min. After centrifugation at 1,500 rpm,
0.075 M KCl is added and the sample is incubated at 37°C for 10
min. After two times fixation and centrifugation, several
microscopic slides are prepared. They undergo standard Giemsa
staining and are analyzed under microscope (Fig. 3).
Results
The morphological analysis of cells' populations
shows hypercellular bone marrow with an altered ratio between the
neutrophil, granulocyte and erythroblast clones up to 12.1:1
(granulocytes 0.90; erythroblasts 0.07); hyperplasia of the
neutrophil population with sustained maturation, increase of
myelocytes and promyelocytes. Index of maturation 1.3 (reference
values 0.5–0.8). Cytological features of gigantism in a part of the
metamyelocytes. Substantially reduced erythroblast population with
single representatives of all maturation forms. Well-presented
megakaryocyte apparatus with a dominating population of polyploidic
granulated megakaryocytes. The conclusion of the morphological
analysis is ‘Chronic myeloproliferative process’. The ratio of
different cell populations in the bone marrow is as follows: 1.5%
lymphocytes, 87.9% granulocytes, 0.1% myeloblasts, 6.2% mature
monocytes, 4.2% promonocytes. The whole blood count test of the
patient and the biochemical analysis result at the time of her
acceptance in the haematological ward of ‘St. Ivan Rilski’ Hospital
is presented in Table I.
Based on the myelography the patient is directed for
molecular-biological analysis for JAK2 mutation and Ph
chromosome. The result of the restriction analysis is presented in
Fig. 1 showing the genotype status
of the patient (homozygous carrier of two wild type alleles for
V617F JAK2 gene mutation).
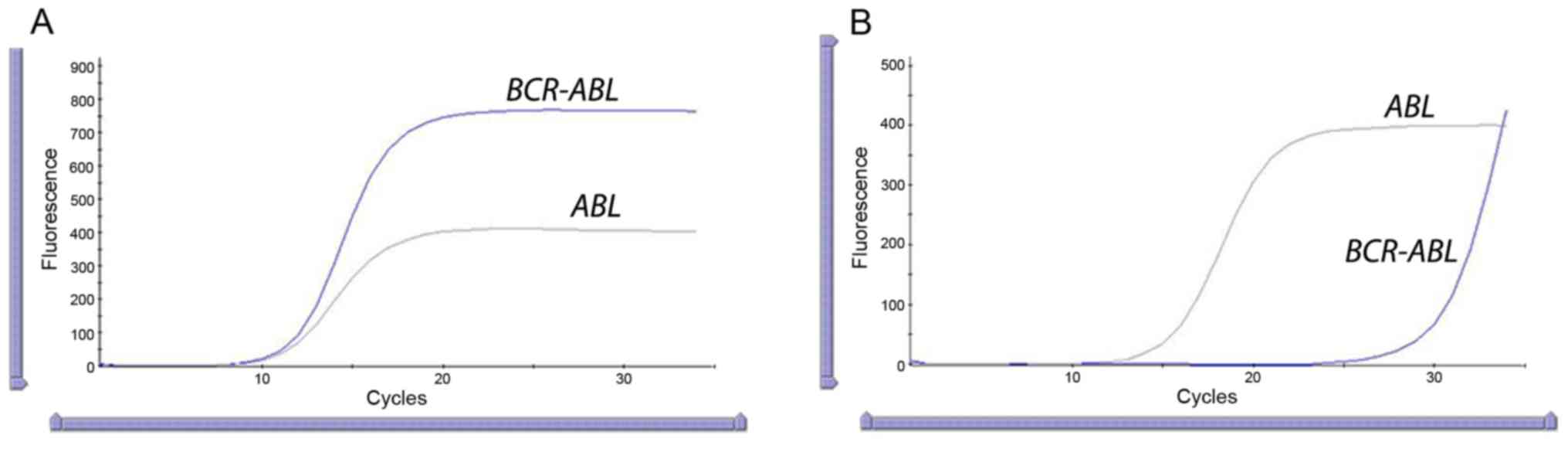
Quantitative Real-time PCR analysis of the patient
is performed (May, 2018) to confirm the presence of the molecular
marker of CML-fusion gene BCR/ABL. The graph clearly shows
the positive result of the amplification for both ABL
(endogenous control) and BCR-ABL genes. The software
estimates automatically the level of the fusion gene of 120%
(IS) on the International Standardized Scale (Fig. 2A). Real-time analysis is performed
second time (September, 2018) after four months' treatment with
Tasigna (Nilotinitib) to monitor the molecular improvement of the
patient. The result demonstrates 0,0019% (IS) (Fig. 2B) level which is much less compared
to the patient's earlier data.
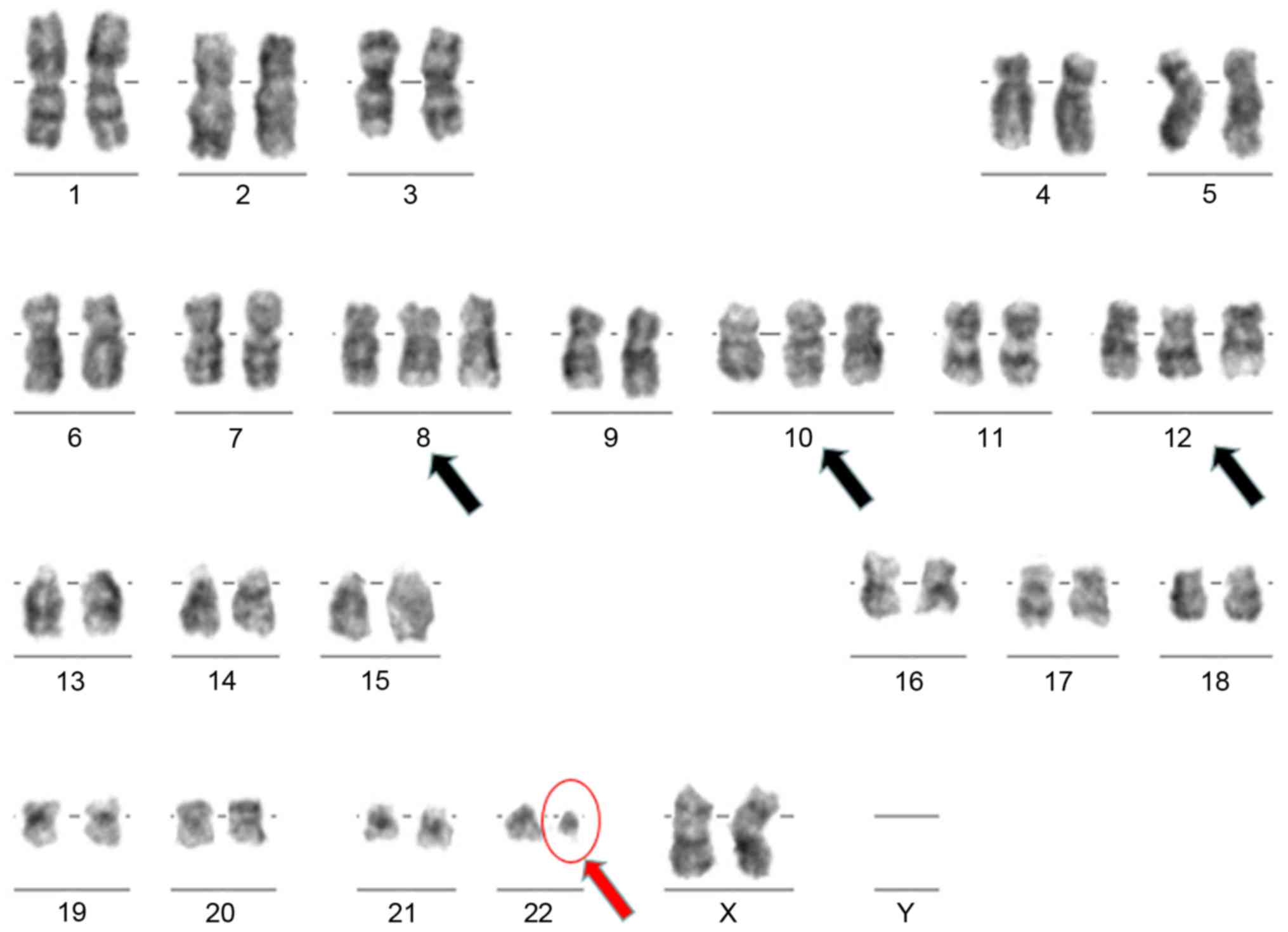
Karyotyping is performed after 24-h cultivation time
of stimulated bone marrow specimen. Chromosomes are obtained,
stained following Giemsa standard protocol and subsequently
analyzed under light microscope (5).
The cytogenetic analysis shows two different clones of cells:
Hyperdiploid with additional chromosomes 8, 10 and 12 and Ph
chromosome; and second clone which is Ph(+) with no hyperdiploidy
(Fig. 3).
Discussion
Hyperdiploidy is a phenomenon of having additional
chromosomes rather than the diploid chromosome number in the
karyotype. According to Onodera et al, hyperdiploid
karyotype arises by a simultaneous gain of multiple chromosomes
from a diploid karyotype during a single abnormal cell division
(6). Generally hyperdiploidy is not
a common event in CML patients (7).
Other chromosome changes, such as chromosomal translocations, are
more common, especially at the time of blast transformation
(8). Hyperdiploidy could be a bad
prognostic factor in CML (7,9). Further investigation on the topic shows
that the existing statements about hyperdiploidy are ambiguous.
A case report (10)
presents a Ph(+) CML case with hyperdiploidic karyotype and an
additional T315I kinase domain (KD) mutation in the BCR-ABL
gene. The patient responded well after therapy with Nilotinib.
Though T315I mutation remained after the treatment, targeted drug
eliminated the hyperdiploid clone (10).
In a study on acute lymphoblastic leukemia (ALL) in
children high hyperdiploidy is characterized by a favourable
prognosis (11). A paper studying
chromosomal aberrations in pediatric and adult ALL defines high
hyperdiploidy as present in 25–30% of children with favorable
prognosis and 7–8% in adults with favorable/intermediate prognostic
value (12). In multiple myeloma
patients hypоdiploidy but not hyperdiploidy, is a poor indicator
for the disease's development (13).
According to some authors (14–16)
hypodiploid DNA content is related to a poor response to
chemotherapy and a very short survival time; according to others
(17) hyperdiploid karyotype is
associated with a better prognosis. The chromosomes which usually
appear in addition in ALL karyotypes are 21, X, 14, 6, 18, 4, 17
and 10 which tend to be gained in blasts (18). The acquisition of chromosome 21 is
seen in 95% of hyperdiploid cases.
There are only few reports about hyperdiploid
karyotypes in CML cases. Rojas et al analyzes 63 cases with
CML and not even one additional chromosome is found (19). There is only one case with i(17q);
and one case with 21q deletion (19). Roland and Blahey reported a
case of a patient with breast cancer who developed hyperdiploidy at
the accelerated phase of the disease, near blast crisis, presenting
with seven additional chromosomes besides Ph chromosome (+6, +8,
+11, +18, +19, +20, +21) (8). Gains
of chromosomes 6 and 19 are common in CML hyperdiploic karyotypes
(8). Other commonly reported
chromosomes are 8 and 19 in Ph(+) cases of CML (20). In a study of 256 patients with CML
only one is with 51 chromosomes and trisomy +6, +10, +13 and +19
(21). Chromosomes +7, +8, +9, +10,
+12, +15, +19 are reported in a case with near triploid karyotype
in pre-imatinib mesylate CML patient with T315I mutation in BCR-ABL
kinase domain (10).
The reported case in this study has been diagnosed
as CML as for of an absence of a blast infiltration in the bone
marrow. The myelogram and the immunophenotyping characteristics of
the cells reveal the percentage of myeloblasts in the bone marrow
estimated at 0.1%. Besides being Ph(+), the patient shows a
hyperdiploid karyotype with additional chromosomes 8, 10 and 12
(see ‘Results’). To our knowledge, this is the only report of
chromosome 12 in a hyperdiploid CML patient.
The underlying mechanism of hyperdiploidy and how
this phenomenon can potentially influence the expression profile of
the genes is still unknown (22).
The influence of hyperdiploidy in the course of CML remains a
disputable question in literature. From clinical point of view, the
chromosomal gain is associated with increased risk for blast crisis
(23,24). To our knowledge, there is only one
more report which presents higher percentage level of BCR-ABL
transcript detected quantitatively and expressed in % (IS)
(25). The detected level of the
BCR-ABL transcript was measured to be 187% (IS) in bone
marrow and 152% (IS) in peripheral blood of a 28-year-old
patient.
As a whole the cytogenetic complexities play major
role in the prognostic evaluation of CML. Along with the Ph
chromosome, various chromosomal aberrations can be associated with
CML. 5–10% of these cases showing complex translocation involving
another chromosome in addition to the Ph chromosome. Our case is
Ph(+) with an additional hyperdiploid clone with trisomy 8, 10, 12.
Cases like this could sometimes present with higher results of
BCR-ABL/ABL transcripts detected quantitatively.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during the present
case study are included in this article.
Authors' contributions
DN performed the restriction analysis, Real-time PCR
test, performed the literature review and wrote the manuscript, VD
assisted in the laboratory data interpretation, VH and MM
interpreted the clinical data of the patient and monitored the
treatment, LM and AA performed the cytogenetic analysis, AR and DT
acquired relevant data and revised the manuscript critically. All
authors have read and approved the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Written informed consent was obtained from the
patient for the publication of the obtained data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Tamascar I and Ramanarayanan J: Targeted
treatment of chronic myeloid leukemia: Role of imatinib. Onco
Targets Ther. 2:63–71. 2009.PubMed/NCBI
|
|
2
|
Jabbour E and Kantarjian H: Chronic
myeloid leukemia: 2016 update on diagnosis, therapy, and
monitoring. Am J Hematol. 91:252–265. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Baxter EJ, Scott LM, Campbell PJ, East C,
Fourouclas N, Swanton S, Vassiliou GS, Bench AJ, Boyd EM, Curtin N,
et al: Acquired mutation of the tyrosine kinase JAK2 in human
myeloproliferative disorders. Lancet. 365:1054–1061. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Safaei A, Shokripour M and Omidifar N:
Bone marrow and karyotype findings of patients with pancytopenia in
southern iran. Iran J Med Sci. 39:333–340. 2014.PubMed/NCBI
|
|
6
|
Onodera N, McCabe NR and Rubin CM: Rubin:
Formation of a hyperdiploid karyotype in childhood acute
lymphoblastic leukemia. Blood. 80:203–208. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Belurkar S, Manohar C and Kurien A:
Chronic myeloid leukemia with hyperdiploidy: A case report with
review of literature. Indian J Med Sci. 67:188–192. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Roland B and Blahey WB: A case of
near-triploidy in chronic myelogenous leukemia. Cancer Genet
Cytogenet. 121:96–98. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yehuda O, Abeliovich D, Ben-Neriah S,
Sverdlin I, Cohen R, Varadi G, Orr R, Ashkenazi YJ, Heyd J, Lugassy
G and Ben Yehuda D: Clinical implications of fluorescence in situ
hybridization analysis in 13 chronic myeloid leukemia cases:
Ph-negative and variant Ph-positive. Cancer Genet Cytogenet.
114:100–107. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Al-Achkar W, Moassass F, Ikhtiar A, Liehr
T, Othman MA and Wafa A: Hyperdiploidy associated with T315I
mutation in BCR-ABL kinase domain in an accelerated phase-chronic
myeloid leukemia case. Mol Cytogenet. 7:892014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Paulsson K and Johansson B: High
hyperdiploid childhood acute lymphoblastic leukemia. Genes
Chromosomes Cancer. 48:637–660. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mrózek K, Harper DP and Aplan PD:
Cytogenetics and molecular genetics of acute lymphoblastic
leukemia. Hematol Oncol Clin North Am. 23991–1010. (v)2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Smadja NV, Bastard C, Brigaudeau C, Leroux
D and Fruchart C; Groupe Français de Cytogénétique Hématologique, :
Hypodiploidy is a major prognostic factor in multiple myeloma.
Blood. 98:2229–2238. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Barlogie B, Alexanian R, Dixon D, Smith L,
Smallwood L and Delasalle K: Prognostic implications of tumor cell
DNA and RNA content in multiple myeloma. Blood. 66:338–341. 1985.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Latreille J, Barlogie B, Dosik G, Johnston
DA, Drewinko B and Alexanian R: Cellular DNA content as a marker of
human multiple myeloma. Blood. 55:403–408. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Morgan RJ Jr, Gonchoroff NJ, Katzmann JA,
Witzig TE, Kyle RA and Greipp PR: Detection of hypodiploidy using
multi-parameter flow cytometric analysis: A prognostic indicator in
multiple myeloma. Am J Hematol. 30:195–200. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Garcia-Sanz R, Orfão A, González M, Moro
MJ, Hernández JM, Ortega F, Borrego D, Carnero M, Casanova F,
Jiménez R, et al: Prognostic implications of DNA aneuploidy in 156
untreated multiple myeloma patients. Castelano-Leones (Spain)
cooperative group for the study of monoclonal gammopathies. Br J
Haematol. 90:106–112. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Heerema NA, Raimondi SC, Anderson JR,
Biegel J, Camitta BM, Cooley LD, Gaynon PS, Hirsch B, Magenis RE,
McGavran L, et al: Specific extra chromosomes occur in a modal
number dependent pattern in pediatric acute lymphoblastic leukemia.
Genes Chromosomes Cancer. 46:684–693. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rojas A, Pineda L, González S, Soto M,
Avila E, Urdaneta B, Prieto-Carrasquero M and González R:
Chromosomal abnormalities in malignant hematologic diseases. Acta
Cient Venez. 51:109–114. 2000.(In Spanish). PubMed/NCBI
|
|
20
|
Werner M, Kaloutsi V, Buhr T, Delventhal
S, Vykoupil KF and Georgii A: Cytogenetics of chronic myelogenous
leukemia (CML) correlated to the histopathology of bone marrow
biopsies. Ann Hematol. 63:201–205. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Meng CY: Cytogenetics and molecular
studies in chronic myeloid leukemiaClements University; 2001
|
|
22
|
Stagno F, Vigneri P, Consoli ML, Cupri A,
Stella S, Tambè L, Massimino M, Manzella L and Di Raimondo F:
Hyperdiploidy associated with a high BCR-ABL transcript level may
identify patients at risk of progression in chronic myeloid
leukemia. Acta Haematol. 127:7–9. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
AM B: Cancer cytogeneticsThe principles of
clinical cytogenetics. Totowa, New Jersey: Humana Press Inc;
1999
|
|
24
|
Godley LA and Le Beau MM: Williams
Hematology8. Cytogenetics and molecular abnormalities. McGraw Hill;
2008
|
|
25
|
Wang Z, Zen W, Meng F, Xin X, Luo L, Sun
H, Zhou J and Huang L: Chronic myeloid leukemia with variation of
translocation at (Ph) [ins (22;9) (q11;q21q34)]: A case report. Int
J Clin Exp Pathol. 8:13707–13710. 2015.PubMed/NCBI
|