1. Introduction
Since Dr. Stephen Paget proposed the ‘seed and soil’
theory of metastasis in 1889(1),
the mechanism of cancer metastasis has been an area of interest
(2). Cancer metastasis has been
shown to be a complex process involving multiple factors including
sustaining proliferative signalling and evading growth suppressors,
resisting cell death, enabling replicative immortality, inducing
angiogenesis, activating invasion, migration and drug resistance
(3,4). Increasing numbers of oncogenes, such
as ErbB2, PI3KCA, MYC, CCND1 and tumour suppressor genes, such as
p53 and Rb have been identified as being related to tumour
metastasis (5). Additionally, many
more signalling pathways have been found to play an essential role
in the development and progress of cancer including TGF-β (6), MAPK (7), Wnt (8), NOTCH (9), Fak (10), PI3K/Akt (11,12).
Among all these oncogenes, signal-induced proliferation-associated
protein 1 (SIPA1), a mitogen-inducible gene encoding a
GTPase-activating protein for Rap1 and Rap2, was identified as an
important moderator participating in several cancer related
signalling pathways (13). In this
paper, we focus on SIPA1 - the structures of SIPA1 gene and
protein, the relationships between polymorphisms of SIPA1 and
tumour susceptibility, the interaction between SIPA1 and other
molecules and the functions of SIPA1 in solid tumours.
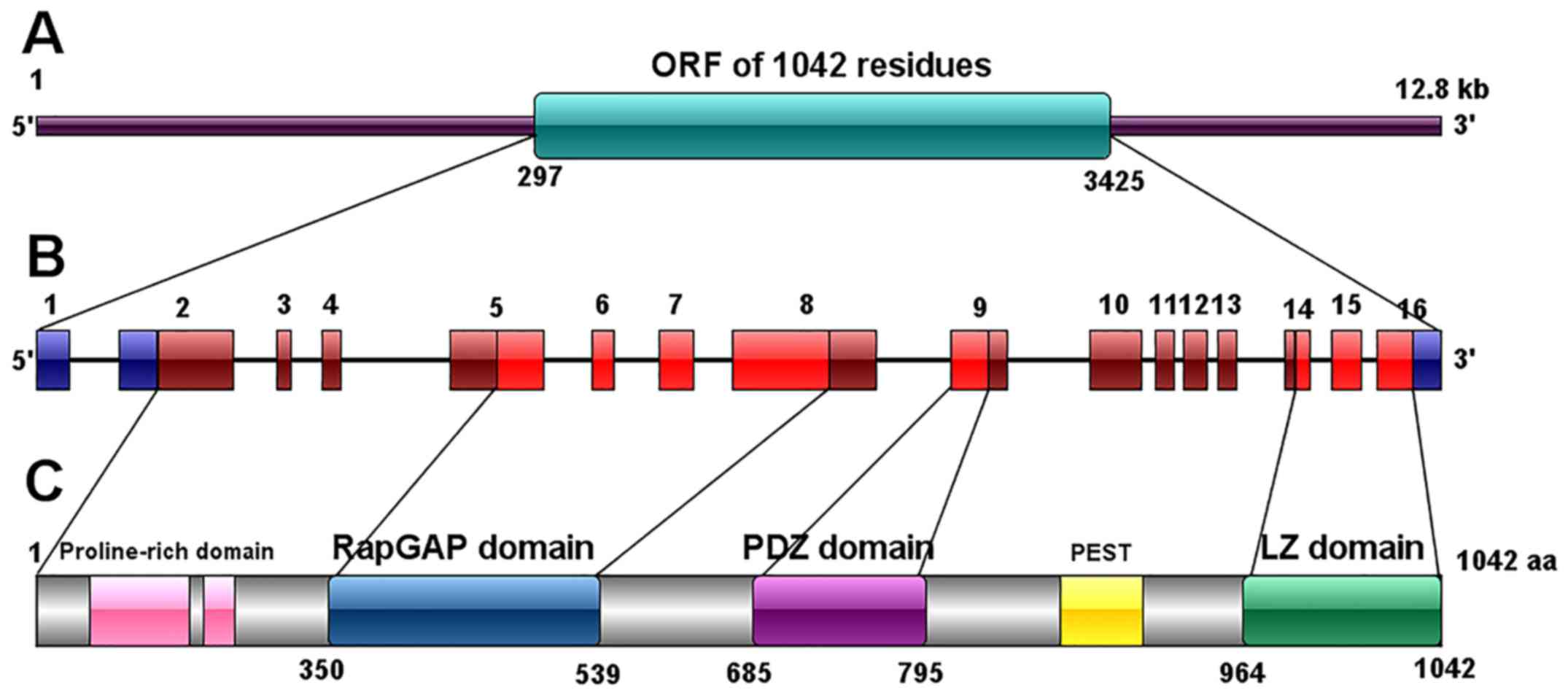
2. Cloning and identification of the SIPA1
gene
The SIPA1 gene, also known as SPA1 (suppressor of
phyA-105), was first cloned in 1995 from a murine lymphoid cell
line, LFD 14, after IL-2 stimulation (14). SIPA1 in mice was originally
described as 3,518 bp long with a long open reading frame (ORF)
(from position 1,199 to 3,280) and several short ORFs at the 5'-end
(14). In 1997, the same research
team cloned the human SIPA1 cDNA from human peripheral blood
lymphocytes (PBL) after stimulation with phytohemagglutinin and TPA
(15). The human SIPA1 gene was
mapped to chromosome 11q13.1, spanning 12.8 kb. Human SIPA1 gene is
highly homologous to the murine gene, containing 16 exons, amongst
which exon 1, 91 bp of exon 2 (considered as 5'-UTR) and the
3'-region 205 bp of exon 16 are untranslated (13). The human SIPA1 genome contains a
much longer ORF of 1,042 residues (from position 297 to 3,425)
(15). In the 5' terminal region of
SIPA1 gene, residues from position 192 to 539 (containing part of
the exon 5, exon 6, 7 and part of exon 8 in human) were found to be
highly homologous to human Rap1-GAP (Gap related domain, GRD).
Upstream of GRD there is a proline-rich domain with the potential
ability to bind SH3 and downstream of GRD there is the PDZ domain
which consists of part of exon 9. The 3' region, part of exon 14,
exon 15 and translated part of exon 16 encode a leucine zipper
(LZ)-like domain, which was also found to be conserved in both
humans and mice (13,15).
3. The structure and expression of SIPA1
protein
In mice, the SIPA1 gene encodes a 68 KDa protein
(P68) with 693 amino acids, mostly located in the nuclei (14), whilst the human SIPA1 protein
contains 1,042 amino acids with a molecular mass of 130 KDa
(15). Three domains were
identified in the SIPA1 protein: i) RapGTPase-activating protein
(GAP) related domain (GRD) (350-539); ii) PDZ domain (685-759);
iii) Leucine zipper like (LZ) domain (964-1042) which is similar to
myosin tail (13).
There is a proline-rich domain including possible
SH3-binding motifs located in the N terminal to the GRD and
upstream the LZ domain has a probable PEST sequence (Fig. 1) (15). Expression levels and localization of
SIPA1 protein vary in different human tissues and cells. SIPA1
protein is most highly expressed in the lymphohematopoietic system
such as spleen, bone marrow and thymus. SIPA1 may be located in the
cytoskeleton, plasma membranes and nuclei depending on the type of
cell and SIPA1's interaction with other proteins (13).
4. Germline polymorphisms in SIPA1
There are thousands of single-nucleotide
polymorphisms (SNPs) in the SIPA1 gene, of which seven SNPs located
in the promoter or encoding regions of SIPA1 have been identified
as being associated with tumourigenesis, metastasis and prognosis
in previous studies (16-20).
Amongst them, most research has focused on three SNPs: i) rs931127,
a G>A SNP located in the promoter region; ii) rs3741378, a
C>T or C>G SNP which encodes for the replacement of a serine
(Ser) to phenylalanine (Phe) amino acid in exon 3; iii) rs746429, a
G>A SNP which encodes for a synonymous amino acid (Alanine, Ala)
transformation in exon14.
These three SNPs are considered to be related to the
development, metastasis and prognosis of breast, lung and cervical
cancer (16-25).
Other SNPs have been shown to be involved in breast
cancer: rs2306364, a G>A SNP rs2448490, a G>A, G>C or
G>T SNP, both of which encode for a synonymous amino acid
(Alanine, Ala) transformation (16,19,25) In
addition, rs75894763, a G>A SNP also encoding for a synonymous
amino acid (Valine, Val) transformation (16) and rs75894763 have been shown to be
related to lung cancer (25). The
G>T SNP, rs3741379, encoding for the replacement of an Ala to
Ser amino acid has been reported to be involved in the process of
lung cancer metastasis (25).
5. SIPA1 family and Rap1-GTPase activating
proteins
The initial function of SIPA1 was believed to be
specific GAP activity for Ras-related mediating proteins, Rap1,
Rap2, Rsr1 and nuclear Ran (15),
although recent research suggests that SIPA1 cannot work as a GAP
for Ran or other small GTPases (15). SIPA1 overexpression induces rounding
and eventual detachment of inherently adherent cells from the
extracellular matrix by inhibiting endogenous Rap1 activation,
indicating that Rap1 signals are involved in the regulation of cell
adhesion and SIPA1 functions as a negative regulator of cell
adhesion (14).
Human RapGTPase activating protein mainly consists
of two subfamilies: i) SIPA1 family; ii) rapGAP family.
In addition to SIPA1, the SIPA1 family also includes
SIPA1-like1 (SIPAL1, also called E6TP1 or SPAR), SIPAL2, and
SIPAL3; and the rapGAP family contains rapGAP1 and rapGAP2(13). All the RapGAPs share a homologous
catalytic GRD domain in addition to an analogous PDZ domain
(13,15).
6. SIPA1 interacting molecules
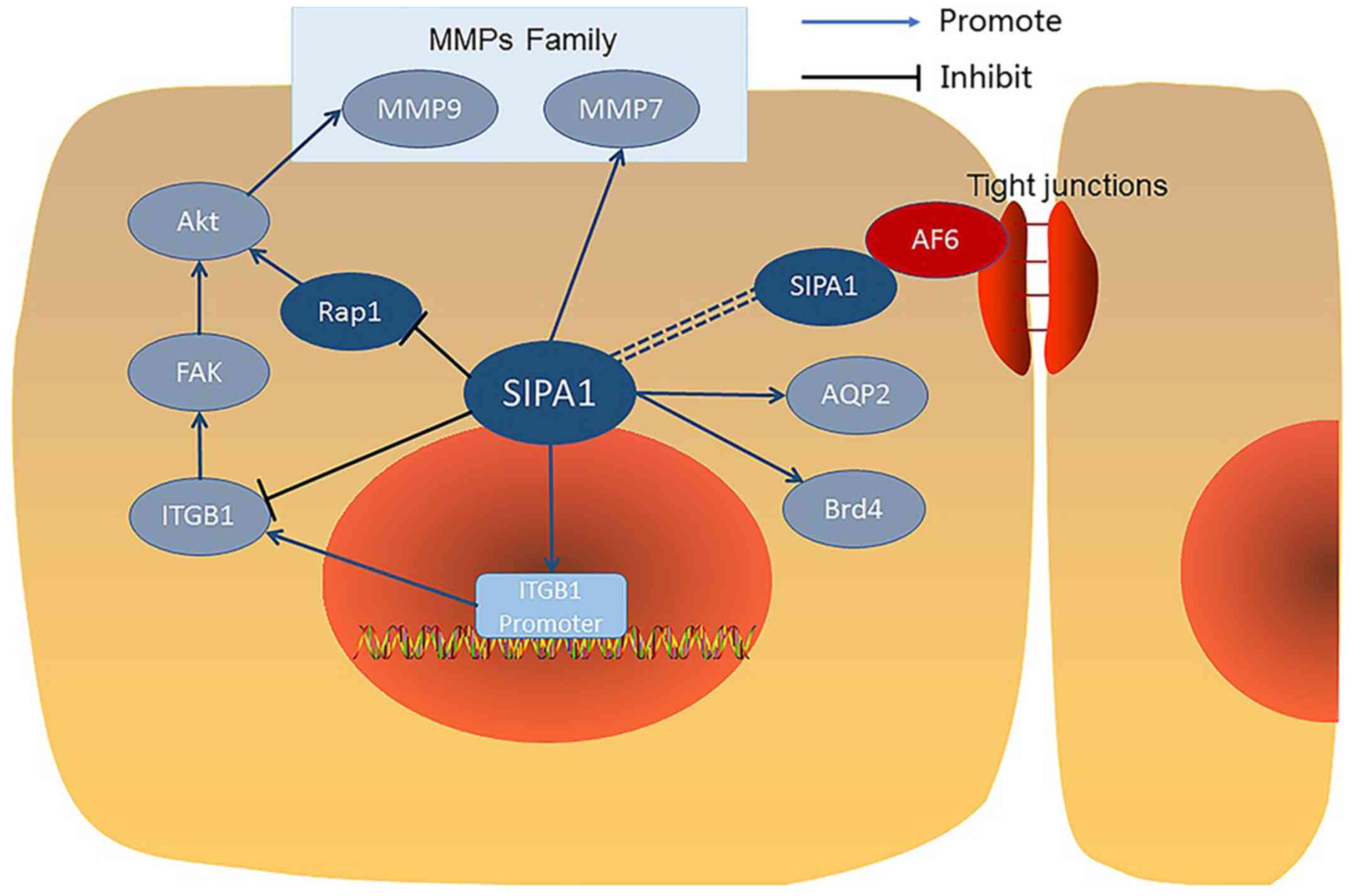
Nuclear SIPA1 and integrin β1.
Nuclear SIPA1 has been shown to interact with a
number of other proteins (Fig. 2)
and activate the integrin β1 promoter in breast cancer cells
(26). Nuclear SIPA1 has been shown
to interact with and activate the integrin β1 promoter in breast
cancer cells (26). After SIPA1
knock down in MDA-MB-231 cells, mRNA levels of SIPA1 were
significantly downregulated. Chromatin immunoprecipitation
experiments revealed that nuclear SIPA1 could interact with the
ITGB1 promoter and increase its transcription activity, causing
phosphorylation changes in the integrin-mediated FAK/Akt signalling
pathway, thereby affecting the adhesion and invasion capability of
cancer cells (26).
Association of SIPA1 and AF6.
Co-immunoprecipitation experiments demonstrated that
SIPA1 bonded specifically with AF6 (afadin) in 293T cells
transfected with both SIPA1 and AF6. Further studies revealed that
the binding occurred between the GRD domain of SIPA1 and the PDZ
domain of AF6. AF6 was reported to localize at cell adhesion sites
and have an association with the tight junction (TJ) protein ZO-1
(Zonula Occludens 1), SIPA1 also co-localized with AF6 in similar
cell to cell adhesion sites, so it was suggested that SIPA1 may
also regulate TJs via AF6, although there is only scant evidence
for this (27).
SIPA1 colocalizes with AQP2 in renal
collecting ducts.
Studies have shown that in renal collecting ducts,
SIPA1 directly binds to aquaporin-2 (AQP2) and is involved in the
regulation of trafficking AQP2 to the apical membrane. AQP2 has the
ability to bind to molecules containing a PDZ domain, so SIPA1 with
a PDZ domain can combine with AQP2 and play a role in the
intracellular transport of AQP2. Studies also demonstrated that
Rap1 signalling pathway was involved in the AQP2 intracellular
transport system. As a regulation factor of Rap1, SIPA1 can control
the transport process by inhibiting Rap1. Therefore the interaction
between SIPA1 and AQP2 may be both direct and indirect (28).
Brd4 interacts with SIPA1.
In HeLa cells, the bromodomain protein Brd4 was
found to bind to the GRD domain of SIPA1 directly and this
interaction is mainly located in the cell nucleus especially at or
near the inner aspect of the nuclear membrane and enhanced the
RapGTPase activity of SIPA1 for Rap1 and Rap2 (29,30).
The combination of SIPA1 and Brd4 promotes the cell cycle through M
to G1 phases, which may also regulate cancer development (30).
SIPA1 is a metastasis efficiency
modifier Mtes1.
Genetic mapping revealed thar SIPA1 a candidate
modifier for the Mtes1 (Metastasis efficiency suppressor gene 1),
which locates on the mouse chromosome 19 and contains a sequence of
human's metastasis suppressor gene Brms1(31). In mice, SIPA1 works as a candidate
for underlying Mtes1. The mice kidney cancer cell line cos-7 cells
with the SIPA1 protein which had alanine (A) (SIPA1/741A) at the
amino acid position 741 showed higher metastatic potential than
those which had threonine (T) (SIPA1/741T) at the Position 741 in
the PDZ domain (31). The higher
metastatic potential could be induced through the mechanism that
the Rap1GAP activating level was higher in the cells with
SIPA1/741A SNP than those with SIPA1/741T SNP (31).
7. The multiple roles of SIPA1 in different
cancer types
Breast cancer.
Most research on the role of SIPA1 in cancer and
cancer metastasis has focused on breast cancer. Immunohistochemical
staining has shown that in breast cancer patients, SIPA1 localized
to the nuclear region. Its expression level may also be a
predictive factor for lymph node metastatic status. Similarly, in
breast cancer cell lines, SIPA1 is mainly localized to the nucleus
in the aggressive breast cancer cell line MDA-MB-231 cells after
transduction to overexpress SIPA1(26). In vitro cell function
experiments demonstrated that knockdown of SIPA1 reduced adhesion,
migration and invasion in the MDA-MB-231 breast cancer cell line,
but promoted the cells to proliferate (26). These changes may be due to the fact
that nuclear SIPA1 interacts with and activates the integrin β1
promoter (ITGB1), thereby regulating the adhesion and invasion of
breast cancer cells. Knockdown of SIPA1 in the MDA-MB-231 cell line
suppressed the integrin mediated FAK/Akt-MMP9 signal pathway by
markedly decreasing the phosphorylation levels of FAK and Akt and
extracellular secretion of MMP9(26). Thus the mechanism of SIPA1 promoting
breast cancer cell adhesion, invasion and metastasis may occur by
regulating the integrin β1/FAK/Akt-MMP9 signalling pathway.
Prostate cancer.
In patients with prostate cancer (CaP), high
expression of SIPA1 was associated with poor prognosis and tumour
metastasis (32). Similarly in the
human CaP cell lines, LNCaP with low metastatic capacity inoculated
in SCID mice was accompanied by undetectable levels of SIPA1, while
PC3 cells with high metastatic capacity in SCID mice was
accompanied by high levels of SIPA1 expression (32). After transduction of SIPA1, the low
metastatic LNCaP cells inoculated into the testis of SCID mice
exhibited a propensity to metastasize to the abdominal lymph nodes.
Following knockdown of SIPA1, the highly metastatic PC3 cells
exhibited reduced metastatic potential and there was no significant
change in the size of primary tumour in both groups (32). Thus the effect of SIPA1 on prostate
metastasis is greater than the effect on proliferation. In
vitro, after transduction of SIPA1 in LNCaP cells, expression
of SIPA1 resulted in decreased adhesion of CaP cells to the
extracellular matrix (ECM). Nuclear Brd4 and ECM-related gene
expression were down-regulated, which was regulated by Rap1
activation (32). A meta-analysis
of human gene expression data (Oncomine website: https://www.oncomine.org/resource/login.html) from
prostate, lung and a variety of solid tumours showed overexpression
of SIPA1 in human primary prostate cancer tissues which was related
to cancer metastasis progress.
Oral squamous cell carcinoma.
Expression of SIPA1 in both human oral squamous cell
carcinoma (OSCC) and OSCC cells is higher than in normal tissue,
and is related to regional lymph node metastasis in OSCC patients.
In the OSCC cell line HSC-3 and HSC-4, the knockdown of SIPA1
reduced the ability of cells to invade and migrate, but increased
the adhesion of cells and had no significant effect on
proliferation compared with the control group. In the same cell
line, knockdown of SIPA1 down-regulated the cytosolic expression of
BRD4, but abundant BRD4 protein was still expressed in the nucleus
(33). The interaction between
SIPA1 and BRD4 may promote OSCC metastasis. Expression of ITGB1, an
integrin which is known to be an important marker in cell invasion
and adhesion, was significantly higher in SIPA1 knock down cells
than control cells. However, expression of MMP7 (membrane
metalloproteinase 7) which has an essential role in tumour
invasion, growth and metastasis, was markedly reduced after
knockdown of SIPA1(33). Therefore,
SIPA1 and its interaction with BRD4 may impact the development and
metastatic progression of OSCCs by regulating ITGB1 and MMP7.
Colorectal cancer.
In human colorectal adenocarcinoma patients, there
was increased expression of SIPA1 in tumours, especially in
well-differentiated and moderately differentiated tumours compared
to poorly differentiated tumours. Patients with a higher expression
level of SIPA1 had a poor prognosis. In vitro experiments
demonstrated that after knockdown of SIPA1 in HT115 and Caco-2
colorectal cancer cell lines, the potential of cancer cells to
invade, adhere and migrate was increased compared to the control
group, while the ability to proliferate was decreased (34). This suggests that SIPA1 may have an
active role during progression of colorectal adenocarcinoma.
Cervical cancer.
Two SNPs in SIPA1 (rs931127, 313G>A and rs746429,
2760G>A) are potentially related to an increased risk of nodal
metastasis in cervical cancer. The G allele at both rs931127 and
rs746429 in SIPA1 was associated with nodal disease in cases and
controls. In terms of tumour size, patients with smaller stage IB1
tumours having the G allele showed an increased risk of nodal
metastases at SIPA1 rs746429 and at rs931127, but the correlated
trend between polymorphisms in SIPA1 and nodal metastasis were not
significant in Stage IB2 tumours (which are larger lesions). The G
allele in SIPA1 at both rs746429 and rs931127 was significantly
related to nodal disease in patients without lymphovascular
invasion (LVI), which was considered as an independent poor
prognostic factor in cervical cancer patients. The GG genotype was
associated with a markedly higher risk of nodal disease in both
SNPs of SIPA1 in patients without LVI. Histologically, SNPs in
SIPA1 rs746429 and rs931127 were not related to histology types
(adenocarcinoma or squamous cancer). Moreover, SNPs of SIPA1 made
no difference in the overall survival of cervical cancer patients
(20).
Lung cancer.
Previous studies have focused on the polymorphism of
SIPA1 in lung cancer development and metastasis. Three SNPs were
found to be involved in lung cancer, rs931127 A>G, rs2448490
G>A and rs3741379 G>T (24,25).
In a southern Chinese population, a high frequency of the G allele
at rs931127 was significantly correlated with the risk of lung
cancer, but no significant connection was observed for the other
two SNPs. From the perspective of tumour staging and grading, the
fusion of the G allele in rs931127 was also associated with worse
clinical stage, nodal metastasis and distal metastasis. The SNP
rs931127 A>G in the promoter of SIPA1 was highly associated with
tumourogenesis and metastasis of lung cancer (25). Another study suggested that G allele
fusion in SNP rs931127 A>G was significantly correlated with
more serious progression free survival (PFS) in the patients with
non-small cell lung cancer (NSCLC) (24). Therefore the SIPA1 SNP rs931127
A>G may be identified as an independent prognostic predictive
factor for progression free survival (PFS) in NSCLC patients
(24,25).
Gastric cancer.
There has been little previous research regarding
the role of SIPA1 in gastric cancer. A single study demonstrated
both mRNA level (detect by qPCR) and protein expression (detected
by western blotting) of SIPA1 in gastric tumour tissues was lower
than in adjacent normal tissues. However, on the contrary, IHC that
positive staining of SIPA1 was significantly higher in gastric
cancer tissue than in adjacent normal tissue (35). Positive staining of SIPA1 in the
tumour from gastric cancer patients was markedly associated with
the degree of differentiation, lymph node metastases and clinical
grading (35). The expression and
function of SIPA1 in gastric cancer and its mechanisms of action
need to be further explored.
Melanoma.
Limited research regarding the role of SIPA1 in
melanoma has been conducted. One study indicated that in
fast-growing melanoma there was significant overexpression of
SIPA1, resulting in the inactivation of Rap1 and aggressive
melanoma cell models. When knockdown down of SIPA1 in these
melanoma models was carried out, adhesion capability was enhanced
but clonogenic potential and migration ability were reduced
(36). These data suggest that
SIPA1 interacts with Rap1 and may have a complex role in the
regulation of melanoma development and metastasis.
8. Conclusion
Although it has been nearly 30 years since the
discovery of SIPA1, there has been a scarcity of research as
regards the role of SIPA1 in cancer development or prognosis. In
almost all types of cancers studied, SIPA1 was associated with
lymph node metastasis, which suggests that SIPA1 might be a marker
of, or at least associated with, cancer development and hence
patients' prognosis. Whether it could prove to be a reliable
biomarker for prognosis remains to be seen and further work is
necessary.
Numerous studies have demonstrated that interactions
between SIPA1 and several other molecules, known to be involved in
control of cell growth and division, could offer a potential model
for studying the process of cancer metastasis. In relation to this,
the role of SIPA1 in Rap1 signalling pathway may also shed light on
the regulation of this pathway in metastatic disease and provide a
novel target in the future.
We can conclude that SIPA1 may be a new target for
cancer therapy in those patients with tumours which have resistance
to current chemotherapy agents. But so far there were no ongoing or
potential clinical trials for targeting SIPA1. Potential inhibitors
to SIPA1 could have a direct bearing on its putative role as a
therapeutic target, the importance of which has yet to be
determined.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
CL, TAM, RH and WGJ made substantial contributions
to conception and design of the review. CL, TAM, RH and WGJ were
involved in drafting, revising and intellectual content. All
authors were given final approval of the version to be
published.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Paget S: The distribution of secondary
growths in cancer of the breast. Lancet. 133:571–573.
1889.PubMed/NCBI
|
|
2
|
Fidler IJ: The pathogenesis of cancer
metastasis: The ‘seed and soil’ hypothesis revisited. Nat Rev
Cancer. 3:453–458. 2003.PubMed/NCBI View
Article : Google Scholar
|
|
3
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Valastyan S and Weinberg RA: Tumor
metastasis: Molecular insights and evolving paradigms. Cell.
147:275–292. 2011.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Lee EY and Muller WJ: Oncogenes and tumor
suppressor genes. Cold Spring Harb Perspect Biol.
2(a003236)2010.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Seoane J and Gomis RR: TGF-β family
signaling in tumor suppression and cancer progression. Cold Spring
Harb Perspect Biol. 9(a022277)2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Dhillon AS, Hagan S, Rath O and Kolch W:
MAP kinase signalling pathways in cancer. Oncogene. 26:3279–3290.
2007.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Logan CY and Nusse R: The Wnt signaling
pathway in development and disease. Annu Rev Cell Dev Biol.
20:781–810. 2004.
|
|
9
|
Ranganathan P, Weaver KL and Capobianco
AJ: Notch signalling in solid tumours: A little bit of everything
but not all the time. Nat Rev Cancer. 11:338–351. 2011.PubMed/NCBI View
Article : Google Scholar
|
|
10
|
Sulzmaier FJ, Jean C and Schlaepfer DD:
FAK in cancer: Mechanistic findings and clinical applications. Nat
Rev Cancer. 14:598–610. 2014.PubMed/NCBI View
Article : Google Scholar
|
|
11
|
Vanhaesebroeck B, Guillermet-Guibert J,
Graupera M and Bilanges B: The emerging mechanisms of
isoform-specific PI3K signalling. Nat Rev Mol Cell Biol.
11:329–341. 2010.PubMed/NCBI View
Article : Google Scholar
|
|
12
|
Goncalves MD, Hopkins BD and Cantley LC:
Phosphatidylinositol 3-kinase, growth disorders, and cancer. N Engl
J Med. 379:2052–2062. 2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Hattori M: SIPA1 (signal-induced
proliferation-associated 1). Atlas of Genetics and Cytogenetics in
Oncology and Haematology, 2011.
|
|
14
|
Hattori M, Tsukamoto N, Nur-e-Kamal MS,
Rubinfeld B, Iwai K, Kubota H, Maruta H and Minato N: Molecular
cloning of a novel mitogen-inducible nuclear protein with a Ran
GTPase-activating domain that affects cell cycle progression. Mol
Cell Biol. 15:552–560. 1995.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Kurachi H, Wada Y, Tsukamoto N, Maeda M,
Kubota H, Hattori M, Iwai K and Minato N: Human SPA-1 gene product
selectively expressed in lymphoid tissues is a specific
GTPase-activating protein for Rap1 and Rap2. Segregate expression
profiles from a rap1GAP gene product. J Biol Chem. 272:28081–28088.
1997.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Roberts MR, Hong CC, Edge SB, Yao S,
Bshara W, Higgins MJ, Freudenheim JL and Ambrosone CB: Case-only
analyses of the associations between polymorphisms in the
metastasis-modifying genes BRMS1 and SIPA1 and breast tumor
characteristics, lymph node metastasis, and survival. Breast Cancer
Res Treat. 139:873–885. 2013.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Crawford NP, Ziogas A, Peel DJ, Hess J,
Anton-Culver H and Hunter KW: Germline polymorphisms in SIPA1 are
associated with metastasis and other indicators of poor prognosis
in breast cancer. Breast Cancer Res. 8(R16)2006.PubMed/NCBI View
Article : Google Scholar
|
|
18
|
Gaudet MM, Hunter K, Pharoah P, Dunning
AM, Driver K, Lissowska J, Sherman M, Peplonska B, Brinton LA,
Chanock S and Garcia-Closas M: Genetic variation in SIPA1 in
relation to breast cancer risk and survival after breast cancer
diagnosis. Int J Cancer. 124:1716–1720. 2009.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Hsieh SM, Look MP, Sieuwerts AM, Foekens
JA and Hunter KW: Distinct inherited metastasis susceptibility
exists for different breast cancer subtypes: A prognosis study.
Breast Cancer Res. 11(R75)2009.PubMed/NCBI View
Article : Google Scholar
|
|
20
|
Brooks R, Kizer N, Nguyen L, Jaishuen A,
Wanat K, Nugent E, Grigsby P, Allsworth JE and Rader JS:
Polymorphisms in MMP9 and SIPA1 are associated with increased risk
of nodal metastases in early-stage cervical cancer. Gynecol Oncol.
116:539–543. 2010.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Pei R, Xu Y, Wei Y, Ouyang T, Li J, Wang
T, Fan Z, Fan T, Lin B and Xie Y: Association of SIPA1 545 C>T
polymorphism with survival in Chinese women with metastatic breast
cancer. Front Med. 7:138–142. 2013.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Ugenskienė R, Myrzaliyeva D, Jankauskaitė
R, Gedminaitė J, Jančiauskienė R, Šepetauskienė E and Juozaitytė E:
The contribution of SIPA1 and RRP1B germline polymorphisms to
breast cancer phenotype, lymph node status and survival in a group
of Lithuanian young breast cancer patients. Biomarkers. 21:363–370.
2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Hsieh SM, Smith RA, Lintell NA, Hunter KW
and Griffiths LR: Polymorphisms of the SIPA1 gene and sporadic
breast cancer susceptibility. BMC Cancer. 9(331)2009.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Gdowicz-Kłosok A, Giglok M, Drosik A,
Suwiński R and Butkiewicz D: The SIPA1-313A>G polymorphism is
associated with prognosis in inoperable non-small cell lung cancer.
Tumour Biol. 36:1273–1278. 2015.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Xie C, Yang L, Yang X, Yang R, Li Y, Qiu
F, Chen M, Fang W, Bin X, Deng J, et al: Sipa1 promoter
polymorphism predicts risk and metastasis of lung cancer in
Chinese. Mol Carcinog. 52 (Suppl 1):E110–E117. 2013.PubMed/NCBI View
Article : Google Scholar
|
|
26
|
Zhang Y, Gong Y, Hu D, Zhu P, Wang N,
Zhang Q, Wang M, Aldeewan A, Xia H, Qu X, et al: Nuclear SIPA1
activates integrin beta1 promoter and promotes invasion of breast
cancer cells. Oncogene. 34:1451–1462. 2015.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Su L, Hattori M, Moriyama M, Murata N,
Harazaki M, Kaibuchi K and Minato N: AF-6 controls
integrin-mediated cell adhesion by regulating Rap1 activation
through the specific recruitment of Rap1GTP and SPA-1. J Biol Chem.
278:15232–15238. 2003.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Noda Y, Horikawa S, Furukawa T, Hirai K,
Katayama Y, Asai T, Kuwahara M, Katagiri K, Kinashi T, Hattori M,
et al: Aquaporin-2 trafficking is regulated by PDZ-domain
containing protein SPA-1. FEBS Lett. 568:139–145. 2004.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Alsarraj J, Faraji F, Geiger TR, Mattaini
KR, Williams M, Wu J, Ha NH, Merlino T, Walker RC, Bosley AD, et
al: BRD4 short isoform interacts with RRP1B, SIPA1 and components
of the LINC complex at the inner face of the nuclear membrane. PLoS
One. 8(e80746)2013.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Farina A, Hattori M, Qin J, Nakatani Y,
Minato N and Ozato K: Bromodomain protein Brd4 binds to
GTPase-activating SPA-1, modulating its activity and subcellular
localization. Mol Cell Biol. 24:9059–9069. 2004.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Park YG, Zhao X, Lesueur F, Lowy DR,
Lancaster M, Pharoah P, Qian X and Hunter KW: Sipa1 is a candidate
for underlying the metastasis efficiency modifier locus Mtes1. Nat
Genet. 37:1055–1062. 2005.PubMed/NCBI View
Article : Google Scholar
|
|
32
|
Shimizu Y, Hamazaki Y, Hattori M, Doi K,
Terada N, Kobayashi T, Toda Y, Yamasaki T, Inoue T, Kajita Y, et
al: SPA-1 controls the invasion and metastasis of human prostate
cancer. Cancer Sci. 102:828–836. 2011.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Takahara T, Kasamatsu A, Yamatoji M, Iyoda
M, Kasama H, Saito T, Takeuchi S, Endo-Sakamoto Y, Shiiba M,
Tanzawa H and Uzawa K: SIPA1 promotes invasion and migration in
human oral squamous cell carcinoma by ITGB1 and MMP7. Exp Cell Res.
352:357–363. 2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Ji K, Ye L, Toms AM, Hargest R, Martin TA,
Ruge F, Ji J and Jiang WG: Expression of signal-induced
proliferation-associated gene 1 (SIPA1), a RapGTPase-activating
protein, is increased in colorectal cancer and has diverse effects
on functions of colorectal cancer cells. Cancer Genomics
Proteomics. 9:321–327. 2012.PubMed/NCBI
|
|
35
|
Li JY, Wang JB, Liu CB, Ma DL and Ma JH:
Dynamic relationship between SIPA1 gene and protein expression and
the development of gastric cancer. Genet Mol Res.
16:2017.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Mathieu V, Pirker C, Schmidt WM,
Spiegl-Kreinecker S, Lötsch D, Heffeter P, Hegedus B, Grusch M,
Kiss R and Berger W: Aggressiveness of human melanoma xenograft
models is promoted by aneuploidy-driven gene expression
deregulation. Oncotarget. 3:399–413. 2012.PubMed/NCBI View Article : Google Scholar
|