Introduction
Lung cancer remains one of the most prevalent and
deadly types of cancer globally (1), with the majority of patients with
non-small cell lung cancer (NSCLC) first presenting with advanced
or metastatic disease. Treatment planning in these patients is
typically dependent upon tumor genomic profiling in order to
identify the therapies to which a given patient is most likely to
respond. Asian patients with NSCLC commonly harbor certain
epidermal growth factor receptor (EGFR)-sensitizing mutations, such
as L858R or exon 19 deletions (2,3), and
the treatment of these patients with EGFR tyrosine kinase
inhibitors (EGFR-TKIs) has been shown to achieve significant
improvements in progression-free survival (PFS) compared with
first-line chemotherapy (4,5). As such, EGFR-TKI monotherapy or
combination treatment is a primary approach to the management of
NSCLC patients with EGFR mutations (6). However, these patients generally
develop acquired resistance to first-generation EGFR-TKIs over a
median period of 10-14 months (7,8).
Patients exhibiting progressive disease (PD)
consistent with acquired resistance to first-generation EGFR-TKIs
must typically undergo re-biopsy in order to establish the
mechanism of resistance. The EGFR T790M mutation is the most common
cause of such resistance and it is present in 50-60% of the
patients, with others exhibiting resistance as a result of either
histological/phenotypic transformation or the activation of
alternative pathways (9-11).
There are several approaches to detecting gene
mutations in patient tissue or plasma samples. For patients with
primary NSCLC, detecting gene mutations in tumor tissue samples
using an amplification refractory mutation system assay is the
clinical gold standard (12). This
assay is based upon a quantitative PCR approach, and utilizes
specific probes to detect mutations in tissue samples containing as
little as 1% mutated DNA on a background of 99% normal DNA
(13). Importantly, this assay has
been approved for clinical use in China by the China Federal Drug
Administration. However, this approach may be inadequate for
re-biopsy tissue samples due to their limited DNA content. In order
to reduce the incidence of false-negative results, more sensitive
gene detection methods have been developed. Next-generation
sequencing (NGS), which is based on the massively parallel
sequencing of millions of different DNA molecules, allows for the
detection of multiple mutations across genes. By using focused gene
panels and by narrowing the coverage on target genes, each read is
sequenced thousands of times, ensuring a high degree of sensitivity
(14). The NGS approach facilitates
the detection of rare circulating tumor DNA (ctDNA) sequences in
the blood or other bodily fluids, and represents a new gold
standard approach to ctDNA analysis (15). Roche Cobas z480 (Cobas) is an
allele-specific polymerase chain reaction assay that enables the
detection of known common EGFR mutations. The validity of this
approach has been confirmed using negative and positive controls,
revealing a sensitivity of 0.1-0.5% (16).
In some cases, conducting tissue biopsy can be
dangerous due to the condition of the patient or the location of
the tumor. Liquid biopsy may serve as an alternative option in
these patients, as EGFR mutation status can be assessed using
ctDNA. In certain clinical trials, droplet digital PCR (ddPCR) has
been adopted to compare the performance of EGFR testing in ctDNA
vs. tumor tissue samples, achieving high sensitivity and
specificity (17,18). However, as this approach is
targeted, it limits the potential for the simultaneous analysis of
additional emergent mutations. The super amplification refractory
mutation system (SuperARMS) assay is a novel modification of the
ARMS assay wherein primers and reaction conditions have been
optimized to improve sensitivity, achieving a detection limit of
0.2% (6).
Osimertinib is a third-generation EGFR-TKI that has
been shown to be effective against tumors harboring T790M mutations
and to achieve adequate central nervous system (CNS)
concentrations. The AURA3 study demonstrated that osimertinib
treatment achieved a significantly longer median PFS duration (10.1
vs. 4.4 months; P<0.001) and a better objective response rate
(71 vs. 31%; P<0.001) relative to platinum-based chemotherapy
combined with pemetrexed (19).
Patients with acquired T790M mutation may be treated with
osimertinib. The treatment of patients with other drug-resistant
mechanisms, including MET rearrangement, HER-2 mutation and
pathological transformation, may be changed to other targeted drugs
or chemotherapy. Consequently, re-biopsy is necessary. Acquired
T790M mutation represents 50-60% of all resistance mechanisms, and
patients with acquired T790M mutation show a good therapeutic
response after receiving osimertinib as subsequent therapy
(19). However, in the real-world
clinical setting, the number of patients who receive osimertinib is
lower compared with the rate of acquired T790M mutations reported
in the literature, which may be attributed to the gene detection
technologies or the patients' financial status (20). A number of different approaches have
been used to assess tumor tissue and plasma samples in order to
establish the presence of the T790M mutation, but the relative
sensitivity and reliability of these different methods remain to be
firmly established. In addition, the data from real-world clinical
settings regarding the frequency of acquired T790M positivity in
patients and their subsequent treatment are limited. Furthermore,
tissue re-biopsy is not feasible for all patients; therefore, the
present study was focused on plasma re-biopsy. The aim of the
present study was to assess the relative sensitivity and
reliability of different technologies for the detection of T790M
mutations in plasma samples from patients with NSCLC and to assess
re-biopsy rates and subsequent treatment strategies in a real-world
clinical setting.
Materials and methods
Patients
Relevant data for all patients eligible for
inclusion in this study were retrospectively gathered from the
electronic records of the Department of Respiratory Medicine of the
First Hospital Affiliated to Wenzhou Medical University between
December 2014 and July 2018. All the patients were diagnosed with
advanced NSCLC as confirmed by both radiological and
cytological/pathological evidence. In addition, all patients
harbored classical EGFR mutations (exon 19 deletion or the L858R
mutation in exon 21), and all patients exhibited PD while under
treatment with first-generation EGFR-TKIs.
Acquired resistance to first-generation EGFR-TKIs
was defined based on a study conducted by Jackman et al
(20) as follows: Patients
exhibited significant and sustained benefits upon initial treatment
with first-generation EGFR-TKIs (>6 months), but experienced PD
within the past 30 days despite sustained treatment. Patients were
excluded from the present study if they underwent additional
treatment prior to re-biopsy, if they had additional cancers, or if
they harbored de novo T790M mutations. The baseline disease
status in each patient was evaluated within 28 days of EGFR-TKI
treatment initiation, and patients were re-evaluated every 8 weeks
via chest high-resolution computed tomography (CT) and every 2-3
months via brain, adrenal gland, lymph node, bone and abdominal
imaging.
The Response Evaluation Criteria in Solid Tumors
(version 1.1) (22) were used to
define tumor progression. PFS-1 was defined as the time from
initiation of first-generation EGFR-TKI treatment to the first
documentation of PD, while PFS-2 was defined as the time from the
initiation of sequential therapy until the date of objective PD or
death.
All patient demographic and clinical information was
retrieved retrospectively and based on real-world evidence,
including patient age, sex, smoking history, tumor histological
findings, mutation detection strategy, EGFR-activating mutation
status, type of EGFR-TKI treatment, CNS metastasis status and type
of PD.
This study was approved by the Ethics of Committees
of the First Hospital Affiliated to Wenzhou Medical University, and
was conducted in accordance with the 1964 Helsinki Declaration and
its later amendments or comparable ethical standards. Informed
patient consent was waived due to the retrospective study
design.
Re-biopsy procedure
Tissue re-biopsy was conducted in patients via
CT-guided needle biopsy, transbronchial biopsy, malignant lymph
node biopsy, thoracoscopy biopsy and cytology from malignant
effusions, as appropriate. All patients provided informed consent
before re-biopsy was conducted. When tissue re-biopsy failed or
when the obtained sample was insufficient for gene sequencing,
plasma samples were collected for ctDNA analyses with patient
consent. A peripheral whole blood sample (10 ml) from each patient
was collected into EDTA tubes and was used for subsequent gene
detection analyses.
Assessment of EGFR T790M mutation
status
Tissue samples from all patients were subjected to
histological review, and when sufficient tumor cells were present,
DNA was extracted from formalin-fixed paraffin-embedded (FFPE)
samples for molecular analyses. Five to ten 5-µm sections per tumor
tissue sample were used for DNA extraction.
The ARMS gene detection strategy utilized a QIAamp
DNA FFPE Tissue Kit (Qiagen GmbH) according to the manufacturer's
instructions. The Illumina Hiseq NovaSeq6000 S2 (Geneseeq
Technology Inc.) platform was used for NGS analysis, while a
multiplex Taqman real-time PCR by Cobas z480 (Roche Molecular
Systems, Inc.) was used for Cobas. These systems were used to
detect the presence of the T790M mutation and for other genomic
analyses assessing ERBB2, ALK, RET fusion and ROS1 fusion
status.
When plasma re-biopsy was required, ctDNA was
extracted from the blood by centrifuging samples within 1 h of
collection at room temperature (2,000 x g for 10 min), followed by
an additional spin at 8,000 x g for 10 min to isolate the plasma,
which was then stored at -80˚C until ctDNA extraction. Plasma ctDNA
was extracted from 4 ml of plasma from each patient. The samples
were analyzed using an ADx-SuperARMS EGFR mutation detection kit
(Amoy Diagnostics Co., Ltd.), Bio-Rad QX200 Droplet Digital PCR
(Bio-Rad Laboratories, Inc.) and an NGS platform (Illumina Hiseq
NovaSeq6000 S2, Geneseeq Technology Inc.). Eluted DNA was
immediately used to detect EGFR mutations. All kits and techniques
were used based on the manufacturer's instructions.
When one or more tissue gene detection analyses
indicated T790M positivity, the patient was considered to harbor
the T790M mutation in the analyzed tissue; the same was also true
for plasma ctDNA detection results.
Statistical analysis
SPSS v24.0 (IBM Corp.) was used for all statistical
testing. Sensitivity was measured by assessing the frequency of
concordant sample positivity as a fraction of total tissue sample
positivity, whereas specificity was determined based on the
frequency of concordant negative samples as a fraction of total
negative tissue samples. Concordance rates were determined based
upon the sum of positives and negatives in both sample types
divided by the total number of matched samples. The concordance of
T790M detection in tissue and plasma samples was evaluated via the
McNemar χ2 test and using κ values. All time-to-event
outcomes were estimated using the Kaplan-Meier method and compared
across groups with the log-rank test or the Cox proportional
hazards model. A two-sided P<0.05 was considered to indicate
statistically significant differences.
Results
Patient characteristics
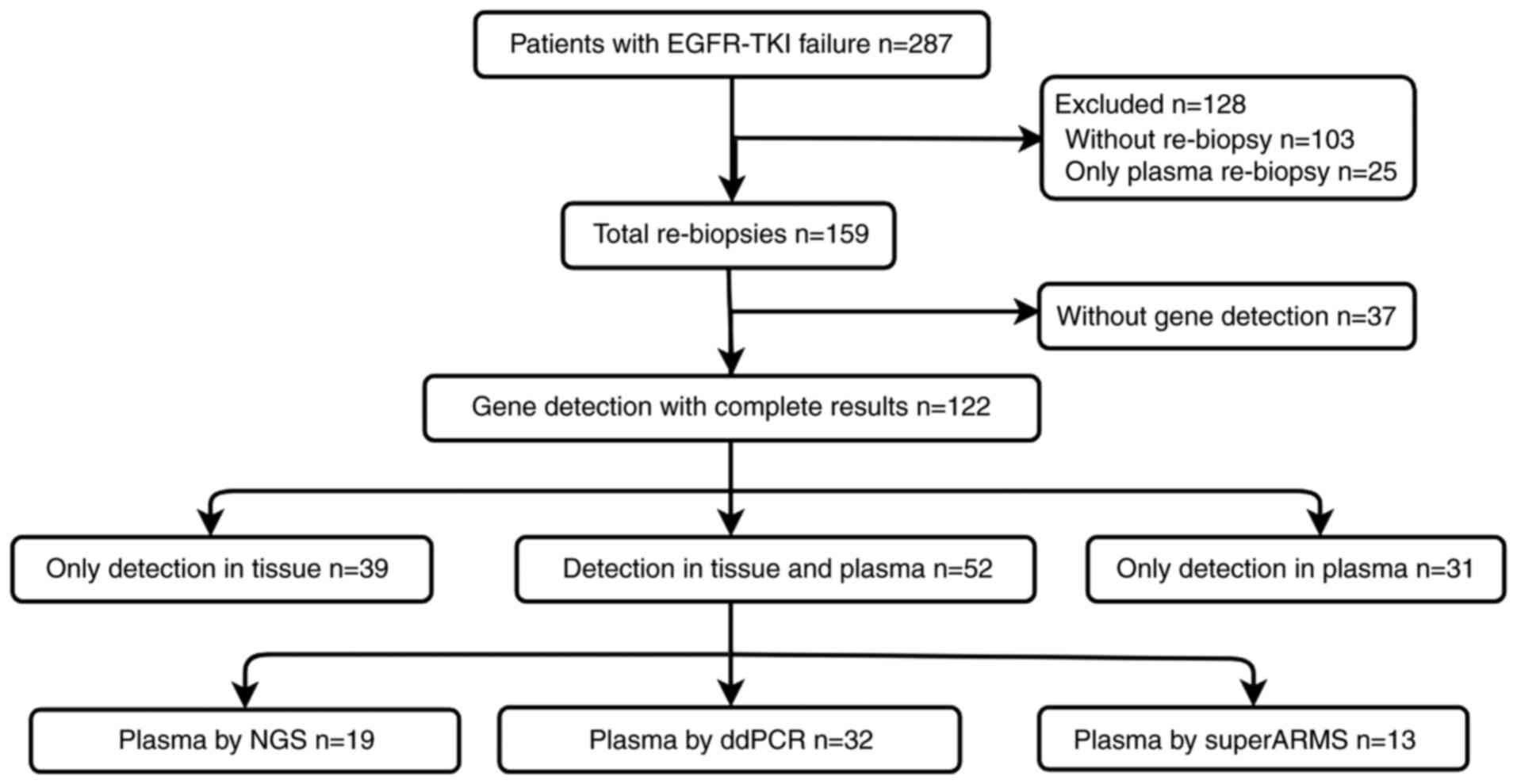
In total, 287 patients were eligible for inclusion
in the present study, of whom 103 did not undergo re-biopsy, and 25
only underwent plasma-based ctDNA analysis. The 159 patients who
underwent tissue re-biopsy were included in our final patient
cohort. These patients had a median age of 63 years (range, 32-85
years), 63.5% were female, 78.6% were never-smokers and the
remaining 21.4% were current or former smokers. In-frame exon 19
EGFR deletions were identified in 56% of included patients, with
the remaining 44% exhibiting L858R mutations in EGFR exon 21.
Pathological analyses confirmed that the type of tumor was
adenocarcinoma in all the patients. The first-generation EGFR-TKIs
used to treat these patients included gefitinib (n=74), icotinib
(n=82) and erlotinib (n=3). The median PFS of the patients was 10.7
months. In total, 194 specimens were obtained upon re-biopsy of
these 159 patients. All patient characteristics are listed in
Table I, while the primary biopsy
and re-biopsy methods are shown in Table II. A flowchart of the study design
is shown in Fig. 1.
 | Table ICharacteristics of enrolled
patients. |
Table I
Characteristics of enrolled
patients.
|
Characteristics | N (%) |
|---|
| Age (years), median
(range) | 66 (40-81) |
| Sex | |
|
Male | 58 (36.5) |
|
Female | 101 (63.5) |
| Smoking status | |
|
Never
smokers | 125 (78.6) |
|
Former and
current smokers | 34 (21.4) |
| Pathology at
diagnosis | |
|
Adenocarcinoma | 157 (98.7) |
|
Poorly
differentiated carcinoma | 1 (0.6) |
|
Sarcomatous
degeneration | 1 (0.6) |
| Baseline EGFR
mutation | |
|
Exon19
deletions | 89 (56.0) |
|
Exon 21
L858R | 70 (44.0) |
| Initial EGFR-TKI
regimen | |
|
Gefitinib | 74 (46.5) |
|
Icotinib | 82 (51.6) |
|
Erlotinib | 3 (1.9) |
| PFS-1 (months) | 10.4 |
| PFS-2 (months) | 8.7 |
| Disease stage | |
|
<IIIB | 20 |
|
IIIB or
IV | 139 |
 | Table IIPrimary biopsy and re-biopsy
methods. |
Table II
Primary biopsy and re-biopsy
methods.
| Procedures | Primary biopsy, n
(%) (n=159) | Re-biopsy, n (%)
(n=194) |
|---|
| Computed
tomography-guided lung needle biopsies | 53 (33.3) | 70 (36.1) |
| Transbronchial
biopsy | 42 (26.4) | 26 (13.4) |
| Thoracentesis | 33 (20.8) | 76 (39.2) |
| Thoracoscopic
biopsy | 13 (8.2) | 0 |
| Lung resection | 11 (6.9) | 1 (0.5) |
| Lymph node
biopsy | 6 (3.8) | 11 (5.7) |
| Metastatic lesion
surgery | 1 (0.6) | 1 (0.5) |
|
Pericardiocentesis | 0 | 3 (1.5) |
|
Abdominocentesis | 0 | 4 (2.1) |
| Lumbar
puncture | 0 | 1 (0.5) |
Prevalence of the T790M detection
Tissue re-biopsy was conducted in 159 patients, of
whom 26 did not undergo any genetic analyses due to financial
limitations. In addition, sufficient tissue was not obtained for
genetic analyses from 23 patients, while 19 patients reportedly
underwent re-biopsy and genetic analyses, but had no available gene
reports and were lost to follow-up. In addition, 37 of these
patients declined to provide blood samples; finally, only 122
patients underwent gene detection analyses based upon tissue,
blood, or tissue and blood samples. The details on gene detection
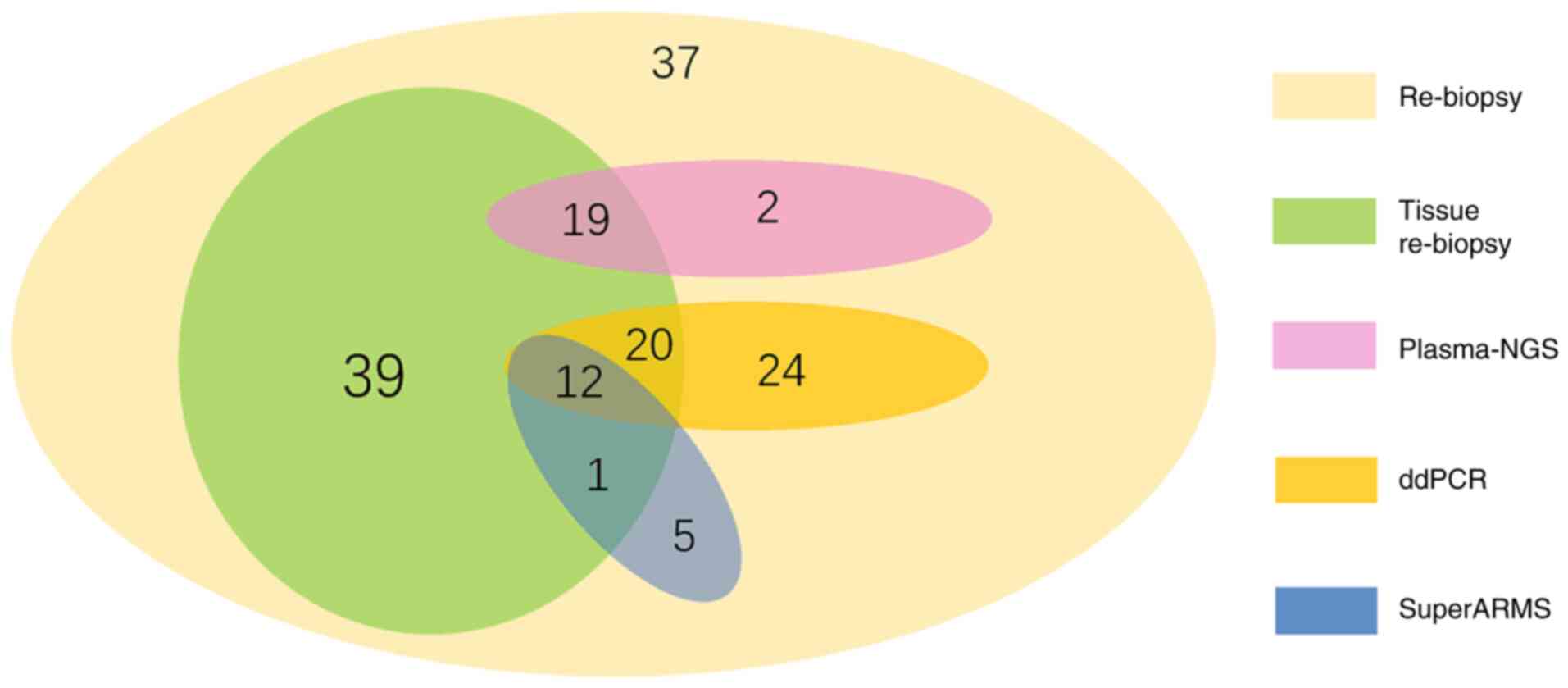
methods are shown in Fig. 4.
The presence of the T790M mutation was detected upon
tissue re-biopsy in 53.8% (49/91) of the patients, and upon plasma
biopsy analysis in 56.6% (47/83) of the patients. The rates of
T790M mutation positivity in tissue specimens were 57.8% (26/45),
60.0% (12/20) and 38.1% (16/42) when detection was conducted using
NGS, Cobas and ARMS assays, respectively. For plasma specimens, the
rates of T790M mutation positivity were 60.0% (12/20), 59.3%
(32/54) and 60.0% (9/15) when detection was conducted using NGS,
ddPCR and SuperARMS assays, respectively.
In the 52 patients who underwent paired tissue and
plasma sample analyses, the rates of T790M mutation positivity were
61.5 and 65.4%, respectively (P=0.065). The sensitivity and
specificity of the three methods for plasma T790M status
determination were next evaluated by comparing the rates of
detection of this mutation in paired plasma and tissue re-biopsy
samples, with tissue samples serving as a reference. The T790M
status in tissue and plasma is shown in Table III.
 | Table IIIT790M status in tissue and
plasma. |
Table III
T790M status in tissue and
plasma.
| | Plasma T790M
status, n (%) |
|---|
| | By NGS | By ddPCR | By SuperARMS |
|---|
| Tissue T790M
status | + | - | Total | + | - | Total | + | - | Total |
|---|
| + | 8 | 2 | 10 (52.6) | 15 | 7 | 22 (68.8) | 6 | 4 | 10 (76.9) |
| - | 4 | 5 | 9 (47.4) | 4 | 6 | 10 (31.2) | 2 | 1 | 3 (23.1) |
| Total | 12 (63.2) | 7 (36.8) | 19 (100.0) | 19 (59.4) | 13 (40.6) | 32 (100.0) | 8 (61.5) | 5 (38.5) | 13 (100.0) |
An NGS approach was employed to analyze plasma ctDNA
samples from 19 patients. Of these, 8 patients were found to be
T790M-positive in both tissue and plasma analyses, while 5 were
negative in both analyses. Of the remaining patients, 2 were found
to be T790M-positive in tissue samples but not in plasma samples,
whereas 4 were found to be T790M-positive in plasma samples but not
in tissue samples. By comparing these results, it was determined
that the NGS-based detection of the T790M mutation in patient
plasma had sensitivity and specificity values of 80% (8/10) and
55.6% (5/9), respectively.
A ddPCR-based approach was used to evaluate plasma
ctDNA samples from 32 patients. Of these, 15 were found to be
positive for T790M mutations in both tissue and plasma samples,
whereas 6 were negative in both types of samples. By contrast, 7
patients were found to be positive for T790M mutations in tissue
samples but not in plasma samples, while the remaining 4 patients
were positive for T790M mutations in plasma samples but not in
tissue samples. By comparing these results, it was determined that
the ddPCR-based detection of T790M mutations in patient plasma had
sensitivity and specificity values of 68.2% (15/22) and 60.0%
(6/10), respectively.
A SuperARMS-based approach was used to evaluate the
plasma ctDNA samples of 13 patients. Of these, 6 were found to be
positive for T790M mutations in both tissue and plasma samples,
whereas 1 was found to be negative in both sample types. By
contrast, 4 patients were found to be positive for T790M mutations
in tissue samples but not in plasma samples, while the remaining 2
patients were positive for T790M mutations in plasma samples but
not in tissue samples. By comparing these results, it was
determined that the SuperARMS-based detection of the T790M
mutations in patient plasma had sensitivity and specificity values
of 60.0% (6/10) and 33.3% (1/3), respectively.
In conclusion, the rates of T790M positivity
associated with these three different approaches in tissue and
plasma were 52.6% (10/19) vs. 63.2% (12/19; P=0.687), 68.8% (22/32)
vs. 59.4% (19/32; P=0.549) and 76.9% (10/13) vs. 61.5% (8/13;
P=0.687), respectively. Further details may be found in Table IV. The rates of T790M detection did
not differ significantly between tissue and plasma samples
(P>0.05), with a low concordance rate between the two
(κ<0.4), possibly due to the insufficient sample size included
in this study.
 | Table IVSensitivity and specificity of
different detection methods. |
Table IV
Sensitivity and specificity of
different detection methods.
| | Detection rate
(%) | |
|---|
| Detection
method | Tissue | Plasma | P-value | Sensitivity
(%) | Specificity
(%) | κ |
|---|
| Plasma NGS | 52.6 | 63.2 | 0.687 | 80.0 | 55.6 | 0.360 |
| Plasma ddPCR | 68.8 | 60.0 | 0.549 | 68.2 | 60.0 | 0.261 |
| Plasma
SuperARMS | 76.9 | 61.5 | 0.687 | 60.0 | 33.3 | -0.054 |
Prevalence of other detected
mutations
In total, 122 patients underwent T790M genotyping,
of whom 72 were found to be positive for this mutation. Of these
patients, 2 exhibited dual MET and EGFR T790M mutations, while 1
harbored dual ALK and EGFR T790M mutations.
The remaining 50 patients were T790M-negative. Of
these patients, 4 had rare mutations, including ALK fusions as well
as HER-2, EGFR G719A and EGFR G719X mutations.
Subsequent treatment
A total of 76.7% (122/159) of the patients in the
present study underwent re-biopsy and/or plasma-based genotyping
following tumor progression. Of these patients, 72 (59%) were found
to be T790M-positive, yet only 32 patients (26.2%, 32/122) were
treated with osimertinib. Of the remaining patients, 19 continued
to receive first-generation EGFR-TKIs, 13 switched to a
chemotherapy regimen, 1 switched to anlotinib, 4 died from
pericardial or brain metastases, and 3 were lost to
follow-up.
Association between the T790M mutation
and survival in patients receiving osimertinib
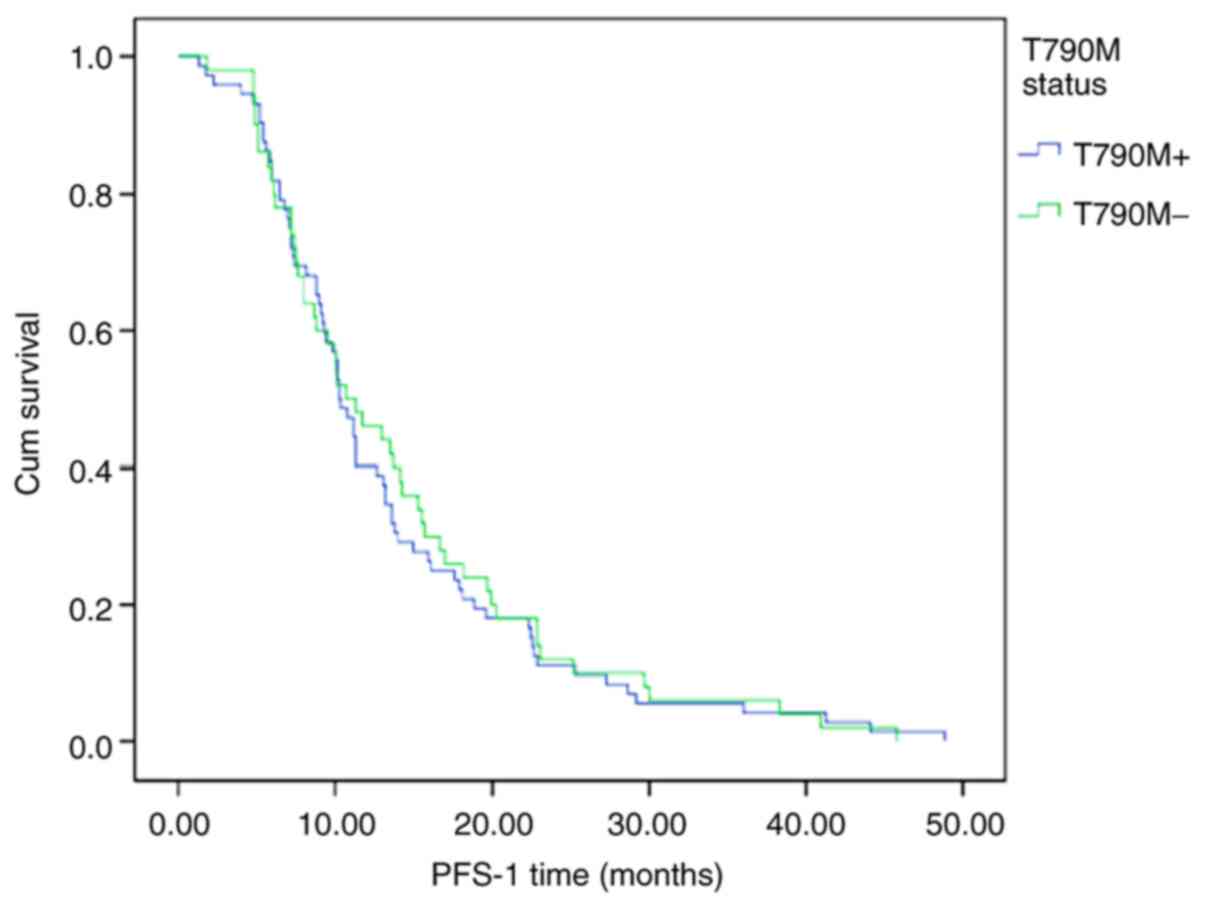
PFS-1 was assessed in all 122 patients based on
their tissue or plasma T790M status. In all patients, the median
PFS-1 was 10.4 months. The median PFS-1 in the T790M-positive group
was shorter compared with that in the T790M-negative group (10.3
vs. 10.7 months, respectively; P=0.752; Fig. 2).
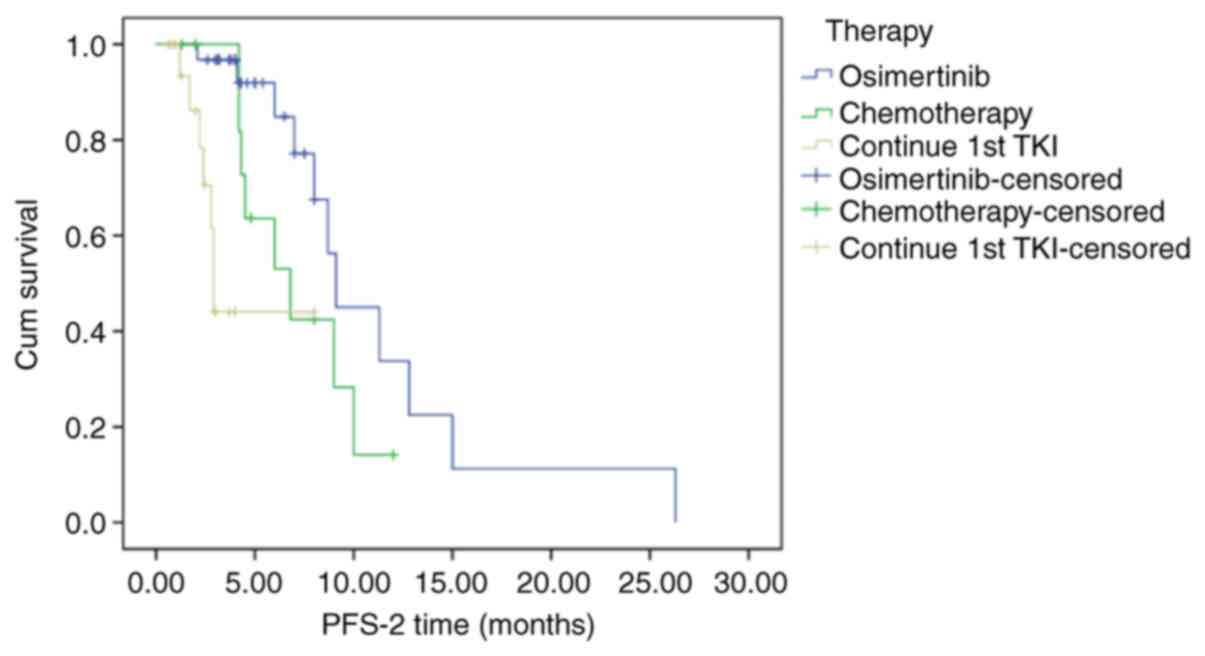
The survival of 64 patients with T790M mutations who
underwent different sequential treatments, including osimertinib,
chemotherapy and sustained first-generation TKI therapy, was also
investigated (Fig. 3). The median
PFS-2 in these patients was 8.7 months (95% CI: 6.338-11.026
months). Of these patients, the median PFS-2 of 32 patients
receiving osimertinib was 9.1 months (95% CI: 8.012-10.118), the
median PFS-2 of the 13 patients who switched to chemotherapy was
6.8 months (95% CI: 3.484-10.116), and the median PFS-2 of the 19
patients who received continuous TKI treatment was 2.9 months (95%
CI: 2.740-3.060). Of these three groups, the PFS-2 of patients
receiving osimertinib was significantly longer compared with that
of patients who underwent themotherapy or sustained
first-generation TKI treatment (P<0.001).
Discussion
EGFR mutations are among the most common driver
mutations observed in Asian patients with NSCLC. First-generation
EGFR-TKIs, such as gefitinib or erlotinib, or second-generation
EGFR-TKIs, such as afatinib, remain the standard of care for
patients with advanced NSCLC harboring EGFR-sensitizing mutations.
While these EGFR-TKIs are associated with good efficacy and
improved patient quality of life, drug resistance commonly develops
and remains an obstacle to positive long-term treatment
outcomes.
In patients exhibiting PD, re-biopsy is required in
order to understand the genetic basis for resistance and to guide
further treatment selection. Tissue re-biopsy samples exhibit
higher rates of T790M mutation and are more stable compared with
plasma samples. In addition, tissue samples enable detection of
histological transformation. Therefore, tissue specimens are used
for reference purposes upon re-biopsy (21). However, as tissue re-biopsy is an
invasive procedure, it may not be well-tolerated by certain
patients. In addition, it may be difficult to obtain such biopsy
samples when patients exhibit stable lung disease but experience
progression in other sites, such as the brain, lung, bone, or
peritoneum. Liquid biopsy, by contrast, is a low-risk and
relatively non-invasive procedure that can offer more comprehensive
information regarding tumor mutation status without the spatial
restrictions imposed by tissue re-biopsy. In theory, liquid biopsy
can offer a comprehensive overview of patient tumor burden and
mutational status, although in practice such biopsies generally
only reflect a small fraction of total tumor DNA present within
cancer patients (22). Indeed, in
some cases, plasma ctDNA analyses may fail to detect mutations that
are identified upon analysis of tumor tissue specimens. Therefore,
negative plasma ctDNA results should be confirmed by tumor tissue
sample analyses (23-25).
Recent work by Usui et al (26) utilized a Cobas approach to detect
the T790M mutation in plasma ctDNA re-biopsy samples, and
determined that this approach had a sensitivity of 2/9 (22.2%) and
a specificity of 15/2 2 (68.2%), with a T790M detection concordance
rate of 54.8% between plasma and tissue samples. Zugazagoitia et
al (27) concluded that
NGS-based analyses of ctDNA may serve as an alternative to tumor
tissue sample analyses in their small study of 53 patients. A study
of 95 patients conducted by Cui et al (28) found that SuperARMS and ARMS methods
exhibited high concordance in their detection of EGFR mutations.
With respect to T790M mutation status, 9 cases were identified via
SuperARMS, whereas only 1 of these cases was detected using an ARMS
assay. However, at present, there is not sufficiently strong
evidence to support the use of ctDNA-based analyses for the
identification of the EGFR T790M mutation in patients with
NSCLC.
In the present study, 52 patients who had undergone
simultaneous analyses of tissue and plasma re-biopsy samples were
identified, yielding respective rates of T790M positivity of 61.5
and 65.4% (P=0.065). In contrast to prior studies, the rate of
T790M detection herein was slightly lower in tissue samples. This
may be attributable to the different methodological approaches used
to analyze these samples, or it may be a consequence of tumor
tissue heterogeneity and the presence of T790M subclones in plasma
ctDNA samples (29,30).
In the present study, the T790M detection rates in
tissue samples were 57.8 and 60.0% by NGS and Cobas, respectively,
in line with the detection rates of 49-65% reported by prior
studies (19,31,32). A
number of retrospective analyses have explored ARMS-based
genotyping and found it to be modestly sensitive when detecting
T790M mutations, with a recent study having recorded a rate of
T790M positivity of 39.6% using an ARMS approach (33), similar to our findings. Relative to
other tested genotyping strategies, ARMS is not as sensitive,
particularly when detecting low-abundance mutations. While the
detection of T790M and other EGFR mutations via ARMS has been
increasingly common in the clinical setting, the relatively poor
sensitivity of this assay bears the risk of erroneously low rates
of accurate detection for this mutation in tissue samples.
To further evaluate the sensitivity and specificity
of different ctDNA-based T790M mutation detection approaches, these
52 patients were next subdivided based on whether they were
analyzed via NGS, ddPCR or SuperARMS. It was observed that NGS was
the most sensitive of these approaches and exhibited moderate
specificity, as well as the highest concordance with tissue testing
among these three strategies. All three methods had sensitivity
values of 60-80%, in line with the ranges reported in prior studies
(40-78%). These three methods had specificities of 55.6-60% (with
the exception of SuperARMS, which had a specificity of just 33.3%
due to an insufficient sample size and was thus omitted), with
these values being well below those reported by prior studies
(80-100%) (22,34-36).
This may be the result of the insufficient number of patients
undergoing simultaneous blood- and tissue-based genotyping in the
present study.
As the availability of osimertinib to patients
within the Chinese healthcare system has increased in recent years,
the proportion of patients undergoing re-biopsy has also increased.
Specimen types and analysis methods have also become more
diversified in recent years, leading to the more widespread use of
third-generation EGFR-TKIs. However, our results suggest that only
a limited subset of patients are actually treated with osimertinib
as second-line therapy after developing resistance to
first-generation EGFR-TKIs.
Of the 287 patients included in the present study,
128 did not undergo re-biopsy. For the majority of these patients,
the decision not to perform re-biopsy was associated with either an
intolerance to the necessary biopsy procedures, difficulty in
accessing the biopsy site, or financial limitations. In total, 159
patients underwent tissue re-biopsy, but the sequencing of tissue
samples from 68 of these patients failed due to insufficient tumor
content (n=25) or negative pathology (n=12). In total, 122 (76.7%,
122/159) patients underwent tissue and/or alternative plasma-based
genotyping of T790M. Of these, 72 patients (59%,72/122) were found
to be T790M-positive. Ultimately, only 32 patients (26.2%, 32/122)
were subsequently treated with osimertinib. Decisions regarding the
use of osimertinib were primarily related to the general patient
condition and financial factors.
These results clearly indicated that the percentage
of patients treated with third-generation EGFR-TKIs was markedly
lower than the rate at which this mutation was detected in this
real-world clinical setting. A number of different factors
ultimately determine whether patients are able to undergo re-biopsy
and to be treated with osimertinib if appropriate, including
limited financial means, difficulties in conducting re-biopsy due
to patient intolerance or tumor location, and failed re-biopsy. To
date, two real-world studies have confirmed that <25% of
patients exhibiting acquired resistance to first-generation
EGFR-TKIs were able to undergo treatment with osimertinib (37,38).
The FLAURA study found that the use of osimertinib as first-line
therapy in NSCLC patients with EGFR mutations was associated with a
significant increase in median PFS relative to first-line use of
first-generation EGFR-TKIs (18.9 vs. 10.2 months, respectively;
P<0.0001) (39). Therefore, the
use of osimertinib as first-line therapy may be beneficial for
patients with EGFR-mutated NSCLC.
Brain metastases are often detected in patients with
advanced NSCLC, with >25% of patients exhibiting such metastases
upon initial diagnosis and with their prevalence increasing with
disease progression. EGFR mutation positivity and EGFR-TKI
treatment are associated with a higher incidence of brain
metastases in patients with advanced NSCLC (40). The mechanism underlying this
phenotype may be distinct from that governing extracranial
progression. Specifically, while extracranial progression is often
associated with acquired resistance, intracranial progression is
generally associated with the limited ability of EGFR-TKIs to
penetrate the blood-brain barrier (BBB). Indeed, gefitinib and
erlotinib have just 1.1 and 2.8-5.1% BBB penetration rates,
respectively (41,42). A total of 97 patients in the present
study were evaluated for brain metastases during treatment. Of
these, 20 patients exhibited brain metastases at baseline, with 11
exhibiting intracranial progression during treatment and 8
exhibiting intracranial stability (8.2%, 8/97); 1 patient with
baseline lesions exhibited a partial response (1%, 1/97). Of the 77
patients without baseline brain metastases prior to first-line
EGFR-TKI treatment, 6 exhibited new intracranial metastases during
follow-up. In summary, 17 patients in the present study exhibited
intracranial progression, with 8 patients (47.1%, 8/17) having been
treated with osimertinib, 3 (17.6%, 3/17) having been treated via
chemotherapy, and 4 patients (23.5%, 4/17) under continued
treatment using first-generation EGFR-TKIs. In addition, 2 patients
exhibited rapid intracranial progression and succumbed to the
disease within 2 weeks.
A total of 2 patients in our overall study
population (1.3%, 2/159) who underwent re-biopsy exhibited a rare
histological transformation from adenocarcinoma to small cell lung
cancer (SCLC). As chemotherapy is the primary treatment used for
SCLC, 1 of these patients received chemotherapy, although it was
found to be ineffective after 2 treatment cycles. However, plasma
ctDNA analyses for this patient revealed weak T790M positivity,
suggesting that a combination of chemotherapy and targeted
third-generation EGFR-TKI treatment may have been more effective,
although further research is necessary to fully determine the
efficacy of this strategy. The other one of these 2 patients did
not receive treatment, and succumbed to the disease due to rapid
progression. At present, there is no standard treatment course for
transformed SCLC. Given that this condition is serious and has a
poor prognosis, it is crucial to develop novel treatments for
affected patients (43,44).
There were several limitations to the present study.
First, our data were derived from electronic health records, which
were not collected or organized with the goal of supporting
research, and as such their accuracy and reliability are unknown.
To compensate for this weakness, we have tried to collect original
data, including radiographic images and gene detection reports
where possible. In addition, as this was a single-center study of a
small population, these results may not necessarily apply to larger
populations. Furthermore, patients exhibiting gradual disease
progression are more suitable candidates for re-biopsy owing to
their overall good condition, but they are also able to continue
using first-generation EGFR-TKIs. Although these patients exhibited
T790M mutation positivity, they were not switched to osimertinib.
Due to the relatively short follow-up period of the preent study,
these patients were considered to have not changed their
therapeutic regimen, although they may have done so after the end
of our follow-up period.
In summary, the results of the present study suggest
that NGS-based approaches are the most sensitive for detecting EGFR
T790M mutations in plasma ctDNA samples from patients with NSCLC,
with this approach yielding the highest concordance with tissue
samples. In this study of a real-world clinical setting, it was
also found that fewer patients than expected underwent re-biopsy,
and that only a relatively limited subset of patients found to
harbor the acquired T790M mutations underwent third-generation
EGFT-TKI follow-up treatment. Furthermore, less than half of
patients suffering from intracranial progression received
osimertinib. Finally, it was also found that transformed SCLC with
acquired T790M mutation positivity may be associated with poor
prognosis.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Wenzhou
Municipal Science and Technology Bureau (grant. no. ZH2017001).
Availability of data and materials
The datasets generated and/or analyzed during the
present study are available from the corresponding author on
reasonable request.
Authors' contributions
TH, JZ and HX collected and organized the data, SS
were responsible for formal analysis, JY proposed the methodology,
and YL, TH and JZ wrote the manuscript. YL and TH revised the
manuscript and approved the final version. All the authors have
read and approved the final manuscript. TH and YL confirm the
authenticity of the raw data.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Review Board of the First Affiliated Hospital of Wenzhou Medical
University (approval. no. 2017161), and in accordance with the 1964
Helsinki Declaration and its later amendments or comparable ethical
standards.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Wei WE, Mao NQ, Ning SF, Li JL, Liu HZ,
Xie T, Zhong JH, Feng Y, Wei CH and Zhang LT: An analysis of EGFR
mutations among 1506 cases of non-small cell lung cancer patients
in Guangxi, China. PLoS One. 11(e0168795)2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Seo JS, Ju YS, Lee WC, Shin JY, Lee JK,
Bleazard T, Lee J, Jung YJ, Kim JO, Shin JY, et al: The
transcriptional landscape and mutational profile of lung
adenocarcinoma. Genome Res. 22:2109–2119. 2012.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Mok TS, Wu YL, Thongprasert S, Yang CH,
Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, et
al: Gefitinib or carboplatin-paclitaxel in pulmonary
adenocarcinoma. N Engl J Med. 361:947–957. 2009.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Rosell R, Carcereny E, Gervais R,
Vergnenegre A, Massuti B, Felip E, Palmero R, Garcia-Gomez R,
Pallares C, Sanchez JM, et al: Erlotinib versus standard
chemotherapy as first-line treatment for European patients with
advanced EGFR mutation-positive non-small-cell lung cancer
(EURTAC): A multicentre, open-label, randomised phase 3 trial.
Lancet Oncol. 13:239–246. 2012.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Tan CS, Gilligan D and Pacey S: Treatment
approaches for EGFR-inhibitor-resistant patients with
non-small-cell lung cancer. Lancet Oncol. 16:e447–e459.
2015.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Kobayashi S, Boggon TJ, Dayaram T, Jänne
PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG and Halmos
B: EGFR mutation and resistance of non-small-cell lung cancer to
gefitinib. N Engl J Med. 356:786–792. 2005.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Kosaka T, Yatabe Y, Endoh H, Yoshida K,
Hida T, Tsuboi M, Tada H, Kuwano H and Mitsudomi T: Analysis of
epidermal growth factor receptor gene mutation in patients with
non-small cell lung cancer and acquired resistance to gefitinib.
Clin Cancer Res. 12:5764–5770. 2006.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Camidge DR, Pao W and Sequist LV: Acquired
resistance to TKIs in solid tumours: Learning from lung cancer. Nat
Rev Clin Oncol. 11:473–481. 2014.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Sequist LV, Waltman BA, Dias-Santagata D,
Digumarthy S, Turke AB, Fidias P, Bergethon K, Shaw AT, Gettinger
S, Cosper AK, et al: Genotypic and histological evolution of lung
cancers acquiring resistance to EGFR inhibitors. Sci Transl Med.
3(75ra26)2011.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Zhao J, Shao J, Zhao R, Li R, Yu K, Zhu L
and Zhang J: Histological evolution from primary lung
adenocarcinoma harboring EGFR mutation to high-grade neuroendocrine
carcinoma. Thorac Cancer. 9:129–135. 2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Handorf EA, McElligott S, Vachani A,
Langer CJ, Demeter MB, Armstrong K and Asch DA: Cost effectiveness
of personalized therapy for first-line treatment of stage IV and
recurrent incurable adenocarcinoma of the lung. J Oncol Pract.
8:267–274. 2012.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Kimura H, Kasahara K, Kawaishi M, Kunitoh
H, Tamura T, Holloway B and Nishio K: Detection of epidermal growth
factor receptor mutations in serum as a predictor of the response
to gefitinib in patients with non-small-cell lung cancer. Clin
Cancer Res. 12:3915–3921. 2006.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Paweletz CP, Sacher AG, Raymond CK, Alden
RS, O'Connell A, Mach SL, Kuang Y, Gandhi L, Kirschmeier P, English
JM, et al: Bias-corrected targeted next-generation sequencing for
rapid, multiplexed detection of actionable alterations in cell-free
DNA from advanced lung cancer patients. Clin Cancer Res.
22:915–922. 2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Feng WN, Gu WQ, Zhao N, Pan YM, Luo W,
Zhang H, Liang JM, Yang J and Deng YM: Comparison of the superARMS
and droplet digital PCR for detecting EGFR mutation in ctDNA from
NSCLC patients. Transl Oncol. 11:542–545. 2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Li C, Jia R, Liu H, Zhang B and Wang C:
EGFR T790M detection and osimertinib treatment response evaluation
by liquid biopsy in lung adenocarcinoma patients with acquired
resistance to first generation EGFR tyrosine kinase inhibitors.
Diagn Pathol. 13(49)2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Zhu G, Ye X, Dong Z, Lu YC, Sun Y, Liu Y,
McCormack R, Gu Y and Liu X: Highly sensitive droplet digital PCR
method for detection of EGFR-activating mutations in plasma
cell-free DNA from patients with advanced non-small cell lung
cancer. J Mol Diagn. 17:265–272. 2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Vendrell JA, Mazieres J, Senal R,
Rouquette I, Quantin X, Pujol JL, Roch B, Bouidioua A, Godreuil S
and Coyaud E: Ultra-sensitive EGFR (T790M) detection as an
independent prognostic marker for lung cancer patients harboring
EGFR (del19) mutations and treated with first-generation TKIs. Clin
Cancer Res. 25:4280–4289. 2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Mok TS, Wu YL, Ahn MJ, Garassino MC, Kim
HR, Ramalingam SS, Shepherd FA, He Y, Akamatsu H, Theelen WS, et
al: Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung
cancer. N Engl J Med. 376:629–640. 2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Seto T, Nogami N, Yamamoto N, Atagi S,
Tashiro N, Yoshimura Y, Yabuki Y and Saka H: Real-World EGFR T790M
Testing in advanced nonsmall-cell lung cancer: A prospective
observational study in Japan. Oncol Ther. 6:203–215.
2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Jackman D, Pao W, Riely GJ, Engelman JA,
Kris MG, Janne PA, Lynch T, Johnson BE and Miller VA: Clinical
definition of acquired resistance to epidermal growth factor
receptor tyrosine kinase inhibitors in non-small-cell lung cancer.
J Clin Oncol. 28:357–360. 2010.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Eisenhauer EA, Therasse P, Bogaerts J,
Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S,
Mooney M, et al: New response evaluation criteria in solid tumours:
Revised RECIST guideline (version 1.1). Eur J Cancer. 45:228–247.
2009.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Lindeman NI, Cagle PT, Aisner DL, Arcila
ME, Beasley MB, Bernicker EH, Colasacco C, Dacic S, Hirsch FR, Kerr
K, et al: Updated molecular testing guideline for the selection of
lung cancer patients for treatment with targeted tyrosine kinase
inhibitors: Guideline from the college of American pathologists,
the international association for the study of lung cancer, and the
association for molecular pathology. Arch Pathol Lab Med.
142:321–346. 2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Wei Z, Shah N, Deng C, Xiao X, Zhong T and
Li X: Circulating DNA addresses cancer monitoring in non small cell
lung cancer patients for detection and capturing the dynamic
changes of the disease. Springerplus. 26(531)2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Diehl F, Schmidt K, Choti MA, Romans K,
Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, et al:
Circulating mutant DNA to assess tumor dynamics. Nat Med.
14:985–990. 2008.PubMed/NCBI View
Article : Google Scholar
|
|
26
|
Dawson SJ, Tsui DW, Murtaza M, Biggs H,
Rueda OM, Chin SF, Dunning MJ, Gale D, Forshew T, Mahler-Araujo B,
et al: Analysis of circulating tumor DNA to monitor metastatic
breast cancer. N Engl J Med. 368:1199–1209. 2013.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Haber DA and Velculescu VE: Blood-based
analyses of cancer: Circulating tumor cells and circulating tumor
DNA. Cancer Discov. 4:650–661. 2014.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Usui K, Yokoyama T, Naka G, Ishida H,
Kishi K, Uemura K, Ohashi Y and Kunitoh H: Plasma ctDNA monitoring
during epidermal growth factor receptor (EGFR)-tyrosine kinase
inhibitor treatment in patients with EGFR-mutant non-small cell
lung cancer (JP-CLEAR trial). Jpn J Clin Oncol. 1:554–558.
2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Zugazagoitia J, Gomez-Rueda A,
Jantus-Lewintre E, Isla D, Camps C, Ramos I, Trigo JM, Bernabé R,
Juan-Vidal O, Sanchez-Torres JM, et al: Clinical utility of
plasma-based digital next-generation sequencing in oncogene-driven
non-small-cell lung cancer patients with tyrosine kinase inhibitor
resistance. Lung Cancer. 134:72–78. 2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Cui S, Ye L, Wang H, Chu T, Zhao Y, Gu A,
Xiong L, Shi C and Jiang L: Use of superARMS EGFR mutation
detection kit to detect EGFR in plasma cell-free DNA of patients
with lung adenocarcinoma. Clin Lung Cancer. 19:e313–e122.
2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Siravegna G, Marsoni S, Siena S and
Bardelli A: Integrating liquid biopsies into the management of
cancer. Nat Rev Clin Oncol. 14:531–548. 2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Diaz LA Jr and Bardelli A: Liquid
biopsies: Genotyping circulating tumor DNA. J Clin Oncol.
32:579–586. 2014.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Nosaki K, Satouchi M, Kurata T, Yoshida T,
Okamoto I, Katakami N, Imamura F, Tanaka K, Yamane Y, Yamamoto N,
et al: Re-biopsy status among non-small cell lung cancer patients
in Japan: A retrospective study. Lung Cancer. 101:1–8.
2016.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Arcila ME, Oxnard GR, Nafa K, Riely GJ,
Solomon SB, Zakowski MF, Kris MG, Pao W, Miller VA and Ladanyi M:
Rebiopsy of lung cancer patients with acquired resistance to EGFR
inhibitors and enhanced detection of the T790M mutation using a
locked nucleic acid-based assay. Clin Cancer Res. 17:1169–1180.
2011.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Li Y, Xu Y, Wu X, He C, Liu Q and Wang F:
Comprehensive analysis of EGFR T790M detection by ddPCR and
ARMS-PCR and the effect of mutant abundance on the efficacy of
osimertinib in NSCLC patients. J Thorac Dis. 11:3004–3014.
2019.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Oxnard GR, Thress KS, Alden RS, Lawrance
R, Paweletz CP, Cantarini M, Yang JCH, Barrett JC and Jänne PA:
Association between plasma genotyping and outcomes of treatment
with osimertinib (AZD9291) in advanced non-small-cell lung cancer.
J Clin Oncol. 34:3375–3382. 2016.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Sakai K, Horiike A, Irwin DL, Kudo K,
Fujita Y, Tanimoto A, Sakatani T, Saito R, Kaburaki K, Yanagitani
N, et al: Detection of epidermal growth factor receptor T790M
mutation in plasma DNA from patients refractory to epidermal growth
factor receptor tyrosine kinase inhibitor. Cancer Sci.
104:1198–1204. 2013.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Wang Z, Chen R, Wang S, Zhong J, Wu M,
Zhao J, Duan J, Zhuo M, An T, Wang Y, et al: Quantification and
dynamic monitoring of EGFR T790M in plasma cell-free DNA by digital
PCR for prognosis of EGFR-TKI treatment in advanced NSCLC. PLoS
One. 9(e110780)2014.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Wu YL, Cheng Y, Zhou X, Lee KH, Nakagawa
K, Niho S, Tsuji F, Linke R, Rosell R, Corral J, et al: Dacomitinib
versus gefitinib as first-line treatment for patients with
EGFR-mutation-positive non-small-cell lung cancer (ARCHER 1050): A
randomised, open-label, phase 3 trial. Lancet Oncol. 18:1454–1466.
2017.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Kuo CS, Huang CH, Liu CY, Pavlidis S, Ko
HW, Chung FT, Lin TY, Wang CL, Guo YK and Yang CT: Prior EGFR-TKI
treatment in EGFR-mutated NSCLC affects the allele frequency
fraction of acquired T790M and the subsequent efficacy of
osimertinib. Target Oncol. 14:433–440. 2019.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Soria JC, Ohe Y, Vansteenkiste J,
Reungwetwattana T, Chewaskulyong B, Lee KH, Dechaphunkul A, Imamura
F, Nogami N, Kurata T, et al: Osimertinib in untreated EGFR-mutated
advanced non-small-cell lung cancer. N Engl J Med. 378:113–125.
2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Wang BX, Ou W, Mao XY, Liu Z, Wu HQ and
Wang SY: Impacts of EGFR mutation and EGFR-TKIs on incidence of
brain metastases in advanced non-squamous NSCLC. Clin Neurol
Neurosurg. 160:96–100. 2017.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Heon S, Yeap BY, Lindeman NI, Joshi VA,
Butaney M, Britt GJ, Costa DB, Rabin MS, Jackman DM and Johnso BW:
The impact of initial gefitinib or erlotinib versus chemotherapy on
central nervous system progression in advanced non-small cell lung
cancer with EGFR mutations. Clin Cancer Res. 18:4406–4414.
2012.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Porta R, Sanchez-Torres JM, Paz-Ares L,
Massuti B, Reguart N, Mayo C, Lianes P, Queralt C, Guillem V,
Salinas P, et al: Brain metastases from lung cancer responding to
erlotinib: The importance of EGFR mutation. Eur Respir J.
37:624–631. 2011.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Niederst MJ, Sequist L, Poirier JT, Mermel
CH, Lockerman EL, Garcia AR, Katayama R, Costa C, Ross KN, Moran T,
et al: RB loss in resistant EGFR mutant lung adenocarcinomas that
transform to small-cell lung cancer. Nat Commun.
11(6377)2015.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Shiroyama T, Nasu S, Tanaka A, Takata S,
Masuhiro K, Takada H, Morita S, Morishita N, Suzuki H, Okamoto N,
et al: Transformation to small cell lung cancer after first-line
afatinib treatment. Respir Med Case Rep. 23:188–190.
2018.PubMed/NCBI View Article : Google Scholar
|