Epigenetic control of epithelial-mesenchymal-transition in human cancer (Review)
- Authors:
- Published online on: September 25, 2012 https://doi.org/10.3892/mco.2012.28
- Pages: 3-11
Abstract
Contents
Introduction
The current concept of epithelial-mesenchymal transition
EMT in development and in cancer
Epigenetic regulation of EMT
Therapeutic options targeting epigenetics and EMT
Summary and future directions
Introduction
Epithelial-mesenchymal transition (EMT) as well the reverse process of mesenchymal-epithelial transition (MET) is essential for development and physiological response to injury (such as wound healing) as well in carcinogenesis (1–3).
Under normal conditions, epithelial cells are linked together as well as to the extracellular matrix environment by different types of intercellular junctions (desmosomes, adherens and tight junctions) enabling tissue maintenance and stability. Epithelial cells can gain the potency to acquire a mesenchymal phenotype to allow for physiological circadian tissue changes but also of tissue loss or damage (4). Interestingly, this process is also associated with an intermediate stem cell phenotype, thus reflecting the highly conserved mechanisms during embryogenesis (5–7).
Over the last few years, the interest in understanding EMT and MET has significantly increased since we further understand the essential role of EMT in cancer progression, particularly during the complex initial processes of tissue invasion and extravasation (8). The regulatory mechanisms of EMT have been intensively investigated and can be described by networks of activating/deactivating signalling pathways. Furthermore, EMT is additionally influenced and regulated by epigenetic mechanisms, such as DNA methylation and histone modifications as well as microRNAs (miRNAs, see below). This epigenetic regulation is particularly important as it accounts for the observed reversibility of EMT-associated processes and the plasticity of (cancer) cells to react upon various external and internal stimuli.
Taken together, these data highlight the complex nature of regulations involved in EMT and provide the basis for development of a new types of drugs specifically targeting EMT in human cancers. In this context, we provide a concise review of the current concepts of EMT in human carcinogenesis and an outlook on therapeutic anti-cancer approaches on the epigenetic level.
The current concept of epithelial-mesenchymal transition
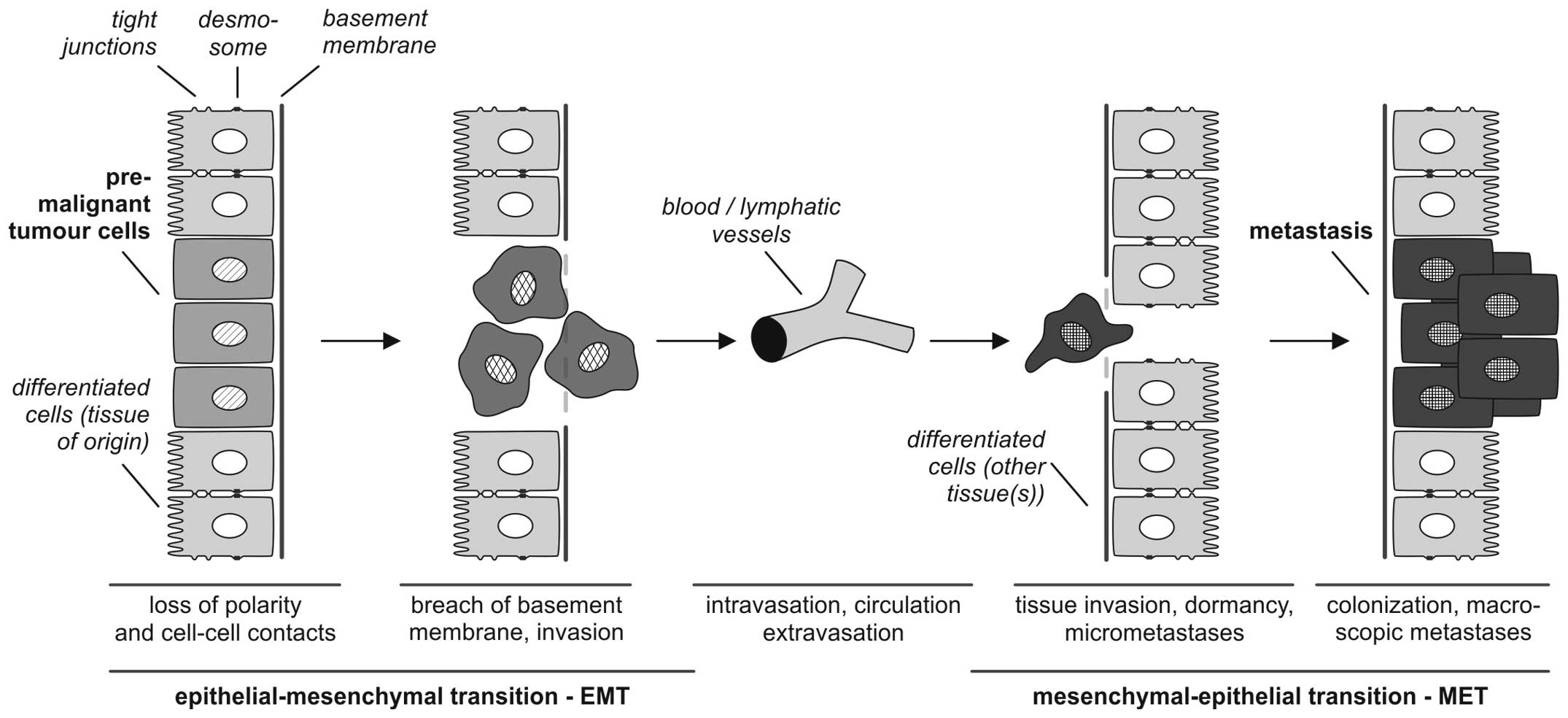
According to Kalluri and Weinberg (9) the biological process of EMT is described as follows: i) epithelial cells are tightly integrated in their cellular environment by tight junctions or desmosomes; ii) under the influence of different EMT mediators (such as growth factors or cytokines, discussed in detail below) epithelial cells gain a mesenchymal status, which iii) is associated with different biological properties, particularly the ability to invade and metastasize. EMT refers to a collective series of transcriptional and post-translational events that cause epithelial cells to take on mesenchymal features, thus allowing the cells to separate from the tissue context, lose baso-apical polarity and gain motility (3,10–12) (Fig. 1).
It is of central importance, that EMT processes are reversible so that mesenchymal cells can undergo MET to differentiate back to epithelial phenotypes. This reverse transition plays a key role in the formation of macroscopic metastases in different organs (13).
For experimental approaches it is important to characterize the EMT or MET status of tumor cells to investigate the influence of agonistic or antagonistic acting drugs. Different markers of extracellular (fibronectin, vitronectin) and cellular localization (vimentin, E-cadherin) are suitable to identify the EMT-MET-related differentiation status (1,12,14,15) (Table I). Cellular markers are either cytoplasmic membrane proteins (such as E-cadherin, claudins, occludin, desmoplakin) or cytoplasmic proteins (cytokeratins, vimentin or mucins). In particular, the epithelial phenotype is typically characterized by cytokeratin expression which stabilizes the cytoskeleton of epithelial cells. Additionally, these cytokeratins hierarchically classify the epithelial differentiation status depending on the tissue/organ context as described by Moll et al(16).
One of the fundamental molecular aspects of EMT in converting differentiated epithelial tumor cells into de-differentiated, migratory mesenchymal cells is the repression of epithelial genes, such as E-cadherin, which results in the loss of epithelial cell-cell contacts. For tumor progression, the loss of E-cadherin is a central feature in the early stages of metastasis (17–21), further supporting the involvement of an EMT-like process in metastatic tissue invasion. During EMT, E-cadherin is replaced by N-cadherin, a process referred to as ‘cadherin switching’ (22,23).
Additionally, intermediary filaments, such as vimentin or smooth muscle cells are used as mesenchymal markers. The intercellular connection status described by the expression pattern of E-cadherin, claudins, occludins or desmoplakin indicate a tissue-integrative epithelial status, whereas the linkage to the extracellular matrix is mediated by glycoproteins, such as fibronectin and vitronectins connecting the extracellular matrix with cellular integrins.
The initiation of the complex process of EMT is triggered by multiple cellular signaling mechanisms including ‘classical’ developmental pathways, such as Hedgehog, Wnt and Notch, as well as signaling by growth factors including transforming growth factor β (TGFβ), fibroblast growth factor (FGF), epidermal growth factor (EGF), and, platelet-derived growth factor (PDGF) (1,11,12) (Table II). Additionally, epigenetic mechanisms (discussed below) as well as miRNA-based regulation have been reported (24,25). Insight into the underlying mechanism of the transcriptional regulation of EMT came from the initial identification of the transcription factor, Snai1 (Snail), as a target of the above-mentioned EMT-promoting signaling pathways, which acts as a direct transcriptional repressor of the E-cadherin gene (26,27). In recent years, additional transcription factors have been identified which repress E-cadherin and mediate the transcriptional initiation of EMT: zinc finger protein Snai2 (Slug) (28), the two-handed zinc finger/homeodomain proteins ZEB1 (δEF1 or ZFHX1A) (29) and ZEB2 (SIP1 or ZFHX1B) (30), the basic helix-loop-helix protein E12/E47 (Tcf3) (31), and Twist (although it is not clear whether the latter directly binds the E-boxes within the E-cadherin promoter).
EMT in development and in cancer
EMT represents the intersection of different aspects of human development which are sequential rather than parallel processes (2). During the early phase of human development, EMT is involved in morphogenesis and stem cell plasticity required for correct implantation, gastrulation and organogenesis (32,33). In the adult organism, subsequent processes relying on regulated EMT or MET are tissue maintenance allowing for reconstruction or maintenance of tissue, as well as cell homeostasis after inflammatory or degenerative insults. In the case of chronic inflammatory and degenerative diseases, such as organ fibrosis, the EMT/MET system is over-regulated which may lead to organ insufficiency or failure (34). Finally, another cancer-related function of EMT was ascribed to cancer stem cells which are centrally involved in tumor progression, metastasis and recurrence after therapy (35,36).
The involved molecular mechanisms of EMT are summarized in the following paragraphs for i) morphogenesis, ii) chronic diseases and finally for iii) cancer.
EMT in development
In order to enable cells to move to new localities, EMT (and the opposite process, MET) is a central aspect in the developing embryo and has been shown to contribute initially to implantation, gastrulation and subsequently to the development of somites, chondrocytes, cardiac valves, and to nephrogenesis (37,38). The associated molecular steps regulating EMT are highly conserved: as mentioned above, the key players of EMT are the transcription factors, Snail, Twist and ZEB and their important repressor target, E-cadherin. The primary goals of all these EMT-related processes are loss of cell-cell adhesion and polarity and changes in the cell shape, as well as enhanced cell motility and ‘invasiveness’ during embryonic development for organ maturation as reviewed in detail by Thiery et al(2). Although the upstream regulatory inputs seem heterogeneous at a first glance, some of the master pathways [such as Hedgehog, Wnt, TGF-β/bone morphogenetic protein (BMP), FGF and EGF] involved in cancer associated-EMT (Table II) also govern EMT during the different phases of embryogenesis. This emphasizes the biological robustness of these pathways (7) and supports the theory that cancer may be viewed as a deregulated program of development (39). Therefore, it is important to further investigate the role of EMT/MET-related processes in development as this knowledge may be transferred to pathophysiological states, such as chronic disease and carcinogenesis which may subsequently aid in the development of new therapeutic approaches.
EMT in chronic diseases
Our knowledge regarding EMT in chronic diseases has increased over the last few years, leading to a new comprehension of chronic diseases. Again, since physiological regeneration and reparation share the same molecular mechanism as EMT/MET in development, it is fascinating to hypothesize that EMT/MET may also play a role in chronic disease caused by over-regulated regeneration and inflammation. This idea is supported by different cellular tracing studies indicating that chronic disease-related interstitial fibrosis is produced by myofibroblasts derived not only from orthotopic fibroblasts, but from epithelial cells via EMT (2). For example, hepatocytes or alveolar epithelial cells differentiate into myofibroblastic cells during carbon tetrachloride (CCl4)-induced liver fibrosis or TGF-β treatment, respectively (40,41). Furthermore, endothelial and mesothelial cells have the potency to trans-differentiate into mesenchymal cells relevant for cardiac (42), renal (43) or peritoneal fibrosis (44). As another example, we have previously demonstrated that vascular smooth muscle cells exhibit a reverse molecular epithelial phenotype in human atherosclerosis associated with progressive atherosclerotic lesions (45). The major driving force behind such a type of chronic disease-related fibrosis is TGF-β signaling and the Snail cascade which may be inhibited by Smad7 gene transfer (46), as well as a systemic vitamin D analogue (47) or BMP-7 application in vivo(48). This observation may be useful for future therapeutic approaches aiming at protection from progressive organ fibrosis and the associated end stage organ failure.
EMT in cancer
As recently reviewed in depth by Brabletz (8), the de-differentiation processes mediated by EMT are now accepted as a hallmark of cancer. EMT plays a key role in the initial steps of tumor cell dissemination and metastasis. In this context, EMT is related to a current concept of cancer stem cell, i.e. ‘migrating cancer stem cells’ [as termed by Brabletz et al(5) and Jung et al(49)]. In these models, EMT enables cancer cells to trans-differentiate to mesenchymal cancer cells accompanied by the induction of stem cell-like properties.
Typically, EMT is found locally at the tumor front with a characteristically increased expression of vimentin paralleled by a loss of E-cadherin (50,51). Since EMT is not always obvious in tumor specimens due to the enhanced stromal cellularity at the tumor margin, the relevance of EMT is still under debate (2). Nevertheless, experimental and clinical data on solid tumors, such as breast, colorectal and ovarian carcinoma have revealed that the overexpression of the classical transcription markers, SNAIL1 and SNAIL2, is associated with a worse outcome in terms of relapse or survival (52–54). Additionally, the inhibition of EMT signaling pathways can enhance the efficiency of ‘classical’ targeted therapy regimes in the experimental setting of hepatic, pancreatic or lung cancer cells (2,55,56). Therefore, detailed topographic analysis of the distribution of EMT markers within the tumor specimen should be carried out for a better prognostic and predictive stratification of cancer patients.
As reviewed by Thiery et al(2) and Brabletz (8), the molecular EMT ‘machinery’ is synergistically and reciprocally regulated together with other control instances, such as the EMT-inhibiting miRNA-200 and miRNA-34 families influencing differentiation, stemness, proliferation and drug sensitivity. Additionally, the expression of these EMT/MET inducers or inhibitors is under the contextual control of the environment as summarized in Table II.
Taken together, the triggering pathways mentioned in Table II induce Snail gene expression, in turn leading to the repression of E-cadherin by the phosphatidylinositol-3 kinase (PI3K)/mitogen-activated protein kinase (MAPK), Smad, RTK, Notch, β-catenin and glioma-associated oncogene (GLI) signaling cascades, thus further illustrating the complexity of autocrine and paracrine growth factor signaling crosstalk during carcinogenesis and EMT (2,6,11).
For the therapeutic exploitation of these results, different approaches are possible: at the first glance, the EMT transcription factors, TWIST, Snail and the ZEB family, may be targeted to inhibit the EMT process during tumor progression. However, the pharmaceutical potency of available low molecular weight drugs has not been sufficient until now (2,57). RNA interference techniques may represent a promising approach to repress these transcription factors on the mRNA level; however, the in vivo stability and transfer efficiency of this drug technology requires further investigation and optimization (2).
For these reasons, another interesting therapeutic approach may be to target the EMT inducers through small molecular weight inhibitors which have already yielded promising results in an in vitro setting (58). The hierarchical regulatory role of EMT inducers depending on the cancer type should be used as the basis for rational drug selection. Based on the intertwined relationship between EMT processes and cancer stem cells, direct targeting of the latter may also manage the disease-related aspects of EMT in cancer. As an example, a promising CSC-targeting drug, salinomycin, was isolated by Gupta et al form a library of 16,000 small molecules (59) and has yielded interesting preclinical results in several tumor entities (60–64).
Additionally, a systematic approach to influence EMT in cancer progression involves modulating the epigenetic regulation of EMT: in the early 80s Jones et al demonstrated that the differentiation status of cultured cells may effectively be influenced by 5-azacytidine, a hypomethylating agent (65,66). Additionally, it has been shown that the histone-associated chromatin structure, as well as the DNA methylation pattern influence the EMT transcriptional regulation of E-cadherin (26,27,67,68) (described in detail in the following chapter).
Epigenetic regulation of EMT
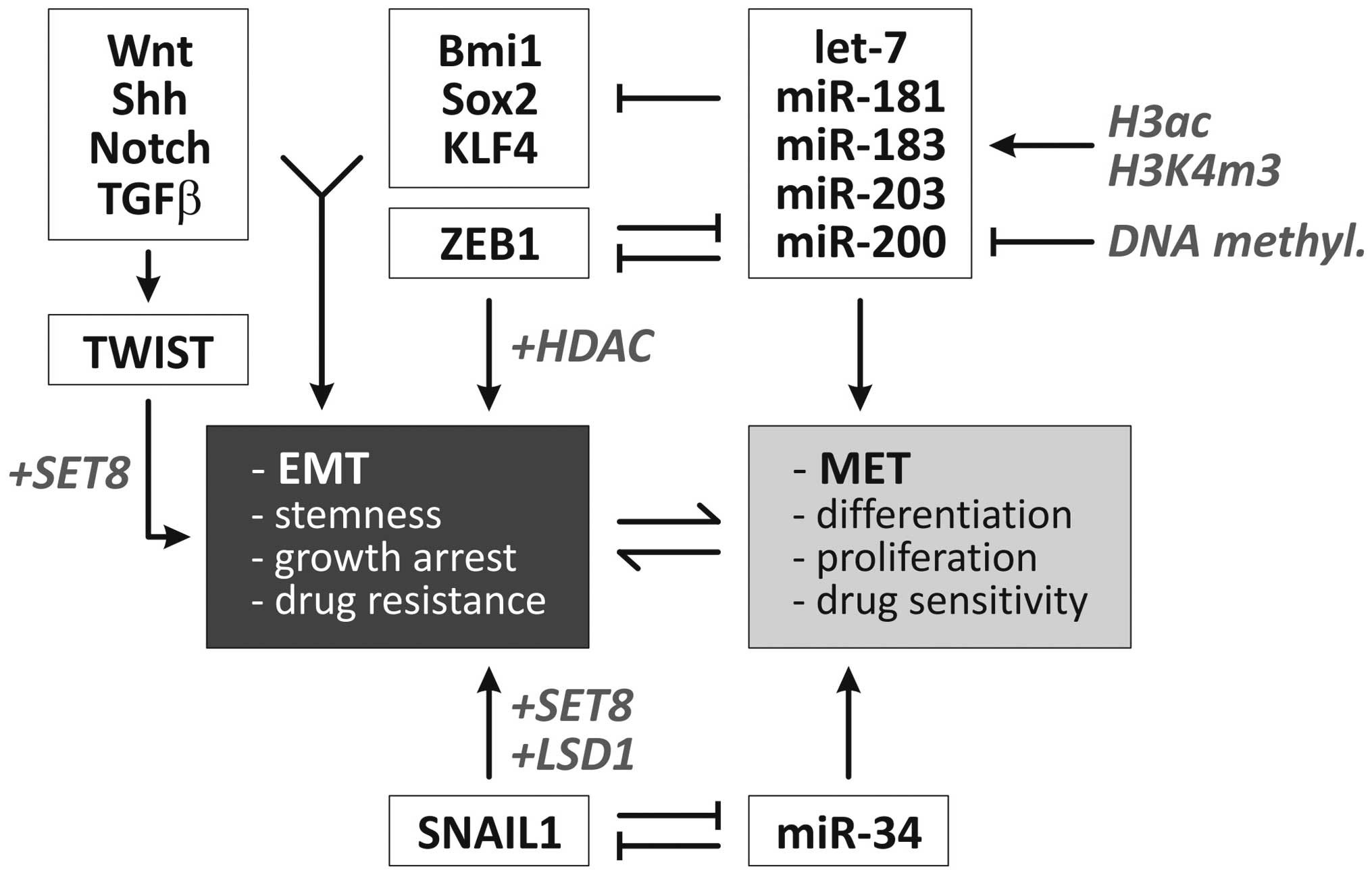
Epigenetic regulatory mechanisms refer to a series of stable but reversible modifications not directly affecting the DNA primary sequence but rather rely on dynamic transcriptional programming effects. Such heritable regulations in the pattern of gene expression are mediated by the DNA methylation of CpG dinucleotides and several post-transcriptional covalent modifications of the NH2 terminal of histone proteins, including acetylation, biotinylation, methylation, phosphorylation and SUMOylation (69). As a general rule, DNA methylation, the di- and trimethylation of H3 lysine 9 (H3K9) and the trimethylation of H3K27 cause chromatin condensation leading to gene silencing mediated by heterochromatin 1 (HP1) and polycomb group (PcG) proteins (70). Several epigenetic events such as global hypomethylation, specific hypermethylation at CpG islands (71,72), as well as aberrations in the histone modification landscape [‘histone onco-modifications’ (73)] have been specifically associated with carcinogenesis. This chapter describes the particular findings on how the EMT and EMT-related markers are regulated via epigenetic events. An overview including some of the epigenetic regulatory mechanisms involved in the control of EMT/MET is presented in Fig. 2.
Multiple epigenetic mechanisms have previously been described that act during the EMT program in the repression of epithelial markers and the conversion of epithelial cells into aggressive, invasive tumor cells. In oral carcinoma cells, hypermethylation at the CDH1 promoter inversely correlates with the expression of E-cadherin and treatment with a demethylating agent (5-azacytidine) causes the re-expression of E-cadherin in cell lines which do not express the SIP1 E-cadherin repressor (74). Similar results have been found in breast tumor cells where CDH1 promoter hypermethylation rather than mutational inactivation caused the reduced expression of E-cadherin. The expression profile of the cell lines complied with fibroblastic (mesenchymal) morphology and CDH1 promoter hypermethylation (75).
A set of transcription factors has been mechanistically linked to the induction of the EMT program, including Twist, Snai1 (Snail), Snai2 (Slug) as mediators of the molecular alterations occurring during EMT (76). In several model systems, epigenetic modifications have been shown to contribute to the repressive function of these transcription factors on epithelial genes. As shown by Lin et al(77), Snai1 recruits the histone demethylase lysine-specific demethylase 1 (LSD1) (KDM1A, AOF2) which removes dimethylation of Lys4 on histone H3 (H2K4m2) and mediates the transcriptional repression of Snai1 target genes, such as CDH1. The short-hairpin RNA-mediated depletion of LSD1 results in partial re-expression of epithelial genes associated with increased levels of H3K4m2 at the CDH1 promoter. These EMT-inducing transcription factors also interact with HDAC1, HDAC2 and the co-repressor mSin3A (74) via their SNAG N-terminal domain as well as polycomb protein repressive complex (PRC2) (77) and cause epigenetic silencing of the CDH1 promoter. Additionally, Yang et al(78) demonstrated that the Twist transcription factor interacts with the monomethyltransferase SET8 which can function both as a repressor (78) or inducer (80) of gene expression. Interestingly, following the interaction of TWIST with SET8, the latter acts as a dual epigenetic modifier on the promoters of E- and N-cadherin to induce the expression of N-cadherin and the repression of E-cadherin via its H4K20 monomethylation activity (78).
As Snail interacts with several repressor complexes including HDAC, PRC2 and Ajuba-PRMT5 (74,77), Snail causes bivalent histone modifications (e.g., coexistence of H3K4m3 and H3K27m3) which render affected genes susceptible to reactivation (81). This is of particular interest as it explains the reversible nature of EMT which, under certain circumstances, can be reversed via MET to generate (e.g., metastasized) cells with epithelial characteristics (1,13).
A large body of evidence demonstrates that the miRNA-200 family and miRNA-205 play an important regulatory role in EMT (82,83). In the context of the epigenetic regulation of EMT, it was found that the CpG island near the miRNA-200c and miRNA-141 transcription start is unmethylated in miRNA-expressing tumor/normal cells and is heavily methylated in miRNA-negative and invasive tumor cells. miRNA expression is further facilitated by the enrichment of chromatin-permissive histone modifications (H3 acetylation and H3K4 trimethylation) (84). Likewise, Davalos et al(85) demonstrated that in epithelial cancer cell lines, the 5′-CpG islands of miRNA-200 family members are unmethylated, whereas the hypermethylation-mediated silencing of these miRNAs was found in transformed mesenchymal cells. The reversibility of this methylation state mediates the shift between EMT and MET (85). Similar results were obtained in bladder cancer (86) and breast cancer cell lines (87). It was further shown that ectopic miRNA-200b and -200c expression inhibits ZEB1 translation and disrupts ZEB1-histone deacetylase repressor complexes. This results in increased histone acetylation and E-cadherin expression. Interestingly, the chemo- and radiosensitivity of these breast cancer cells was increased by enhanced p53-mediated apoptotic pathways (88).
Therapeutic options targeting epigenetics and EMT
The overall aim of an epigenetic therapy is to ‘renew’ the epigenome of the cells by reconstituting the normal expression level of epigenetically misregulated genes (89). Our understanding of the association between modifications of DNA or histones via methylation or acetylation and human diseases has increased over the years, leading to the development of epigenetically functioning drugs, some of which have been approved by the US Food and Drug Administration for the treatment of human cancer (90). As recently reviewed by us, the combination regimen of DNA methyltransferase inhibitors (DNMTi) and histone deacetylase inhibitors (HDACi) yielded promising results in the treatment of myelodysplastic syndrome, a clonal hematological disease (91). Additionally, clinical trials (up to phase IIb) have been performed for other hematological diseases, such as non-Hodgkin’s lymphoma (particularly T-cell lymphoma and diffuse large B-cell lymphoma) and acute myeloid leukemia (90). Nevertheless, the clinical application of epigenetic drugs for solid tumors is still in the pilot phase for e.g., non small-cell lung cancer or only used as experimental therapy in advanced, recurrent or refractory malignancies (90,92).
With respect to the molecular effects of these drugs, the detailed mechanisms of epigenetic therapies were primarily focused on their anti-proliferative and pro-apoptotic effects as well as anti-angiogenic potency as supported by many experimental studies in vitro and in vivo(93,94). Of particular interest, recent investigations revealed that acetylation and de-acetylation are centrally integrated in a cellular network of regulations: the ‘acetylome’ which affects RNA splicing, DNA damage repair, cell cycle control, nuclear transport, actin remodeling, ribosome and chaperone functions (91,95). In our previous studies, we have shown that the cinnamic hydroxamic acid pan-DACi panobinostat (LBH589), a novel potent inhibitor of all HDAC enzymes, influences not only proliferation and apoptosis (96), but also the expression of markers of differentiation and EMT, particularly in an in vivo xenograft model of human hepatoma (97) by upregulation of epithelial markers (cytokeratins) and downregulation of mesenchymal markers (vimentin). Additionally, we demonstrated that the combination of the histone deacetylase inhibitor, SAHA, and the methyltransferase inhibitor, Zebularine, altered the patterns of differentiation in pancreatic cancer models (98). Furthermore, treatment of myelodysplastic syndromes (MDS) and acute myeloid leukemia (AML) with the DNA methylation inhibitor, decitabine (trade name: Vidaza), induced different morphological changes (such as colony forming capacity) and the expression of hematopoietic differentiation markers (99).
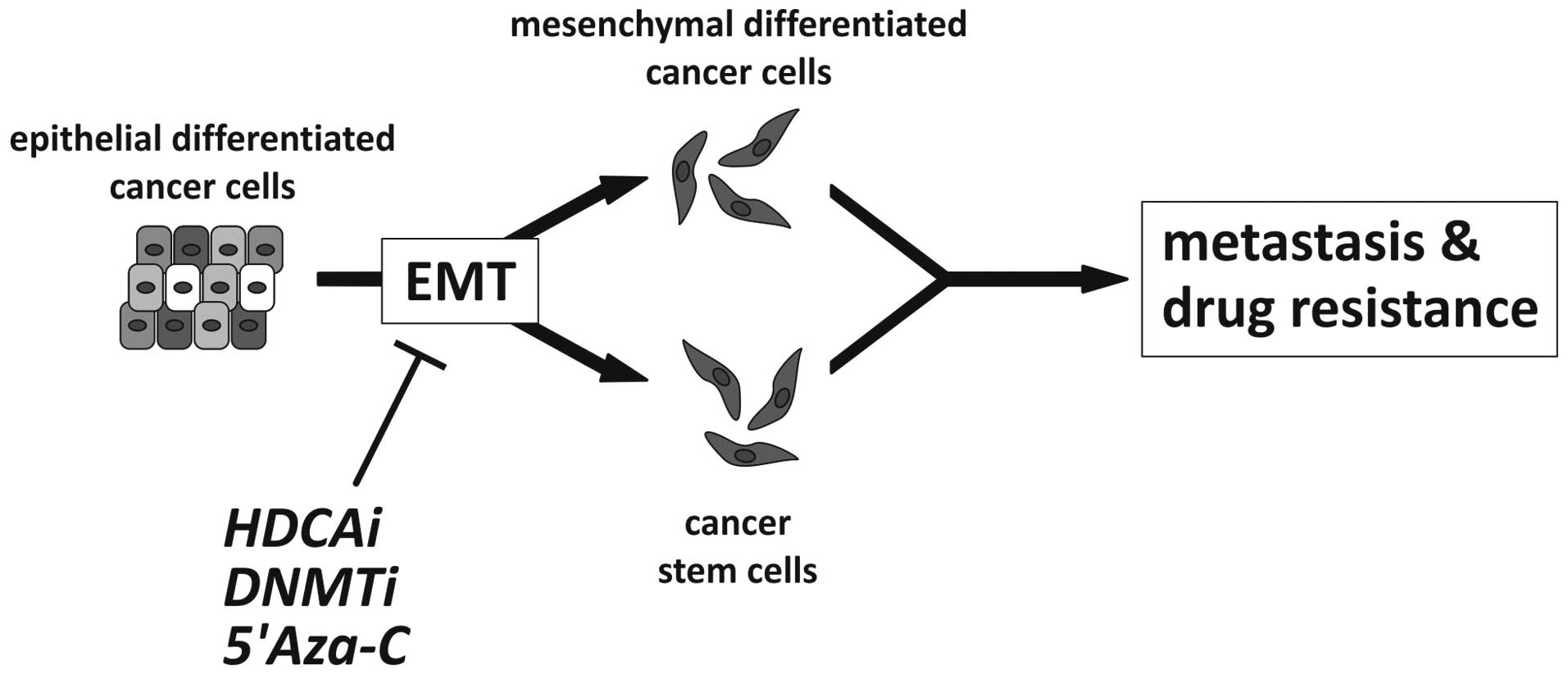
These experimental findings are significant since the differentiation status and the associated EMT/MET status of tumor cells is modulated by classical chemotherapy, selecting transitional, stem cell-like tumor cells which are possibly chemotherapy resistant and are responsible for the clinical recurrence as hypothesized by Todaro et al(100). Therefore, the tumor differentiation status should be characterized in detail prior to, during and after tumor treatment in order to obtain ‘personalized’ predictive, prognostic and therapeutic stratifications. For example, Handra-Luca et al(50) showed that the expression of the ‘basic’ mesenchymal marker, vimentin, in classical pancreatic ductal adenocarcinoma is associated with a worse outcome of patients using immunohistochemistry on a tissue microarray of 387 patients. Additionally, the same group demonstrated that the loss of E-cadherin protein expression was linked to a worse survival of patients with resectable pancreatic adenocarcinomas (51). As clinicopathological investigations of epigenetic treatment and its impact on EMT/MET in cancer specimen are lacking, to date, only experimental data support the theory that epigenetic treatment of cancer cell lines in vitro and in vivo directly influences the Twist-Snail/ZEB-E-cadherin axis and indirectly influences EMT inducers such as Wnt-TGFβ-BMP or other classical pathways (as described in chapter 4). Another interesting recent approach was presented by Ivanova et al(101) who investigated the methylation status of different gastric cancer cell lines, revealing that DNA methylation predicts the responsiveness of these cell lines to treatment with cisplatin, a standard chemotherapy for gastric cancer. Additionally, one of the candidate genes, BMP-4, was epigenetically upregulated in cisplatin-resistant gastric cancer cell lines; therefore, the authors speculated that targeting BMP-4 may improve the sensitivity of such cancer cells to chemotherapy (101). Taken together, several tumorigenic properties initiated/driven by EMT such as invasion, metastasis and drug resistance may be targeted by means of epigenetic therapeutic approaches as illustrated in Fig. 3.
Summary and future directions
Our understanding of the role of EMT/MET-related processes in different phases of human development, homoeostasis, regeneration and reparation, as well as carcinogenesis has dramatically increased. The central molecular pathways associated with the downstream effects on the most important EMT phenotype, i.e., loss of E-cadherin and vimentin expression have been well described. The direct and indirect inducers of EMT/MET are known and we are beginning to decipher their integrated regulatory crosstalk and feedback mechanisms. As reviewed in this article, the role of epigenetics in EMT is being increasingly strengthened by recent experimental data. Nevertheless, further research is required to fully uncover the whole spectrum of the epigenetic regulation of EMT/MET in human cancer. Based on these insights, novel epigenetic therapies that target the EMT-related processes in tumor progression may become feasible.
With respect to basic cell culture experiments showing the influence of epigenetics on therapy responsiveness or resistance (101), recent data demonstrate that the epigenetic pre-treatment of human cancer cells induces differentiation and, therefore, presents us with a chance to improve the efficiency of classical chemotherapies (102). Therefore, we hypothesize that epigenetic therapy may stabilize the epithelial tumor phenotype or induce MET which may subsequently improve tumor sensitivity to conventional chemotherapy (Fig. 3). However, this hypothesis requires further confirmation in appropriate pre-clinical studies and large prospective clinical trials.