Introduction
It is now widely accepted that the risk of cancer
may increase as a result of chronic inflammation, and that
cytokines and growth factors have very important roles in this
process. Cytokines are secreted proteins necessary for the
co-ordination of immune responses, homeostasis, cell communications
and cancer, which is a cell type context-dependent process.
Understanding the involvement of certain cytokines
in the failed differentiation programs that lead to cancer or a
loss of tumor-suppressive functions is undoubtedly one of the
challenges in modern molecular immuno-oncology (1). The inflammatory cells and regulators
may facilitate angiogenesis and promote the growth, invasion and
metastasis of tumor cells. Previous research regarding
inflammation-associated cancer development has focused on cytokines
and chemokines, as well as their downstream targets in linking
inflammation and cancer.
Chondrosarcoma is cancer of the cartilage. It is the
second most common primary bone malignancy, which affects the
pelvis, long bones and spine, as well as the larynx and the head
and neck, which eventually metastasizes. The signaling events
resulting in mesenchymal cell transformation to sarcoma have yet to
be fully elucidated. Chondrosarcoma does not respond to
chemotherapy or radiation; therefore, the search for novel
therapies is very urgent (2).
This study aimed to characterize the cytokine
expression profile in the human JJ012 chondrosarcoma cell line
compared with C28 chondrocytes. The effect of the mammalian target
of rapamycin complex 1 (mTORC1) inhibitor and anti-proliferative
immunomodulator, proline-rich polypeptide 1 (PRP-1) (3–7), on the
expression of cytokines, tumor suppressors SOCS3, TET1/2 and
oncoproteins of inflammatory oncogenic pathways, including the
Hedgehog and Hippo pathways, was also investigated. PRP-1 is a
powerful upregulator of tumor suppressor microRNAs (miRNAs) and
proteins, and a downregulator of oncoproteins, as reported in
previous publications from our laboratory (8–10). Ten
eleven translocation (TET) enzymes, a family of α-ketoglutarate
(α-KG)-dependent dioxygenases, catalyse the oxidative reactions of
5-methylcytosine (5mC) to promote the DNA demethylation process.
TET (methylcytosine dioxygenase) enzyme activity is inhibited in
IDH1/2-mutated tumors by the oncometabolite, 2-hydroxyglutarate, an
antagonist of α-KG linking 5mC oxidation with cancer development
(11–16). Increased levels of D2
hydroxyglutarate (D-2-HG) have been identified in cartilage tumors
that possess an IDH1 or an IDH2 mutation (17). A recent report has demonstrated that
TET-1 and −2 are present in human chondrocytes, and that TET1
expression was markedly reduced by inflammatory factors (18). Hippo and Hedgehog signaling
contribute to malignancies of mesenchymal origin. These two
pathways are associated with inflammation, and promote
tumorigenesis in numerous diseases, including soft tissue sarcomas,
chondrosarcomas, and so forth (19–23). The
Hedgehog pathway is associated with inflammation and cancerogenesis
(24–26). The network of Hippo signaling
regulates the specific enrichment of genes involved in immune and
inflammatory responses (27).
Materials and methods
Tissue culture
The complete growth medium for the human JJ012
chondrosarcoma cells (obtained from Dr Joel Block's Laboratory,
Rush University, Chicago, IL, USA) and C28 chondrocytes comprised
the following: Modified Eagle's medium (DMEM)/MEM (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with F12, 10%
fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.), 25 µg/ml
ascorbic acid, 100 ng/ml insulin, 100 nM hydrocortisone and 1%
penicillin/streptomycin.
Growth medium for the bone marrow-derived
mesenchymal stem cells (ATCC® PCS-500-012; American Type
Culture Collection, Manassas, VA, USA), Mesenchymal Stem Cell Basal
medium, comprised the following: Fetal bovine serum, 485 ml, 7%
(v/v); recombinant human (rh) insulin-like growth factor-1, 15
ng/ml; rh basic fibroblast growth factor, 125 pg/ml;
L-alanyl-L-glutamine, 2.4 mM; gentamicin, 10 µg/ml; amphotericin B,
0.25 µg/ml; and penicillin/streptomycin, 1% (v/v).
Enzyme-linked immunosorbent assay
(ELISA)
A Human Inflammatory Cytokines Multi-Analyte
ELISArray kit (cat. no. MEH-004A; Qiagen, Valencia, CA, USA) was
used to detect a panel of 12 cytokines following a conventional
ELISA protocol, according to the manufacturer's protocol. The ELISA
panel included: Interleukin (IL)-1α, IL-1β, IL-2, IL-4, IL-6, IL-8,
IL-10, IL-12, IL-17A, interferon-γ (IFN-γ), tumor necrosis factor-α
(TNF-α) and granulocyte-macrophage colony-stimulating factor
(GM-CSF).
Polyacrylamide gel electrophoresis and
western blotting
Upon confluency, the cells were trypsinized and
seeded in 6-well cluster dishes at a concentration of
1×106 cells/ml. The experimental samples were treated
with PRP-1 in corresponding concentrations, whereas control samples
were not treated with the peptide. The cells were incubated for 24
h in a 5% CO2 incubator at 37°C. On the following day,
the cells were washed with ice-cold phosphate-buffered saline. A
protease inhibitor (P8340; Sigma-Aldrich, St. Louis, MO, USA) was
added to the cell lysis buffer (C2978; Sigma-Aldrich) in a 1:100
ratio. The cells were collected with a scraper and centrifuged at
15,000 × g at 4°C. The samples were loaded and run according to the
manufacturer's protocol. The gel running time was ~1 h at 100 V,
after which the gel was transferred for western blotting. PVDF
membranes were briefly treated in methanol, distilled water and
transfer buffer prior to the western blot transfer, which was
performed in a cold room for a further hour.
The membranes were then subjected to incubation with
blocking buffer on the rocker at room temperature for 1 h.
Subsequently, the membranes were incubated with primary antibodies
overnight in the cold room. All primary and secondary antibodies
were diluted in the identical blocking buffer. The following day,
the membranes were washed three times (15 min each wash) in
Tween/PBS wash buffer and subjected to incubation with secondary
antibodies for 2 h at room temperature, followed by three
consecutive washes. Enhanced chemiluminescence (ECL) reagent was
applied to the membrane according the manufacturer's protocol, and
X-ray film was developed in the dark room following exposure for
~2–10 min, depending on the experiment.
Reagents were purchased from Lonza, Inc.
(Walkersville, MD, USA), and all the associated procedures were
performed according to the manufacturer's protocol. The catalog
numbers for the reagents and the suppliers are listed as follows:
Pager gold precast gels (cat. no. 59502; 4% stack, 10%
TRIS-glycine; Lonza, Inc.); ECL reagent (cat. no. RPN2109; GE
Healthcare, Little Chalfont, UK); Western blocker solution (cat.
no. W0138; Sigma-Aldrich); ProSieve™ QuadColor™ Protein Markers
(4.6–300 kDa, cat. no. 00193837; Lonza, Inc.); 20X reducing agent
for ProSieve™ ProTrack™ dual color loading buffer (cat. no.
00193861; Lonza, Inc.); EX running buffer (cat. no. 00200307;
Lonza, Inc.); ProSieve™ EX Western Blot Transfer buffer (cat. no.
00200309; Lonza, Inc.); and Immobilon®-P Polyvinylidene
difluoride membranes (cat. no. P4188; Sigma-Aldrich). The primary
antibodies used were as follows: Rabbit polyclonal anti-suppressor
of cytokine signaling 3 (socs3) antibody [cat. no. ab16030,
molecular weight (M.W.) 30 kDa; Abcam, Cambridge, UK]; rabbit
polyclonal anti-transforming growth factor-β (TGF-β)-1/-2/-3
(H-112; cat. no. sc-7892, M.W. 12–25 kDa); anti-Smad 2 rabbit
polyclonal antibody (cat. no. SAB4300562, M.W. 52 kDa;
SABiosciences, Frederick, MD, USA); anti-Stat3 mouse monoclonal
antibody (mAb; cat. no. sc-293151, M.W. 86–91 kDa, Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA); mouse monoclonal
anti-tubulin (M.W. 52 kDa; cat. no. T5168; Sigma-Aldrich); The
secondary antibodies were as follows: Anti-mouse immunoglobulin G
(IgG; cat. no. A9044; Sigma-Aldrich); goat anti-rabbit IgG
peroxidase conjugate (cat. no. A0545; Sigma-Aldrich); anti-TET1
rabbit polyclonal antibody (cat. no. ab105475, M.W. 235 kDa;
Abcam); and anti-TET2 rabbit polyclonal antibody (cat. no. ab94580,
M.W. 224 kDa, Abcam). All primary antibodies were used at a
dilution of 1:1,000; the secondary antibodies were used at a
dilution of 1:5,000.
Hippo and Hedgehog signaling
antibodies
A Hippo Signaling Antibody Sampler kit (cat. no.
#8579; Cell Signaling Technology, Inc., Danvers, MA, USA) was used
for experiments performed in the present study. The kit comprised
the following: Anti-p-Yap (Ser387) (D1E7Y) rabbit mAb (cat. no.
#13619, M.W 75 kDa); anti-LATS1 (C66B5) rabbit mAb (cat. no. #3477,
M.W. 140 kDa); anti-p-MOB1 (Thr35) (D2F10) rabbit mAb (cat. no.
#8699, M.W. 24 kDa); anti-MOB1 (E1N9D) rabbit mAb (cat. no. #13730,
M.W. 25 kDa); anti-macrophage stimulating 1 (Mst1) rabbit mAb (cat.
no. #3682, M.W. 59 kDa); anti-Mst2 rabbit mAb (cat. no. #3952, M.W.
60 kDa); anti-SAV1 (D6M6X) rabbit mAb (cat. no. #13301, M.W. 45
kDa); anti-p-Yap (Ser127) (D9W21) rabbit mAb (cat. no. #13008, M.W.
65–75 kDa); anti-Yap/Taz (D24E4) rabbit mAb (cat. no. #8418, M.W.
50–70 kDa); and the secondary antibody, horseradish peroxidase
(HRP) -linked goat anti-rabbit IgG (cat. no. #7074).
A Hedgehog Signaling Antibody Sampler kit (cat. no.
#8358; Cell Signaling Technology, Inc.) was also used, comprising
the following antibodies: Anti-Sonic Hedgehog (Shh) (C9C5) rabbit
mAb (cat. no. #2207, M.W. 19–45 kDa); anti-Patch homology 1 (PTCH1)
(C53A3) rabbit mAb (cat. no. #2468, M.W. 180–210 kDa); anti-PTCH2
(G1191) rabbit mAb (cat. no. #2479, M.W 130 kDa); anti-suppressor
of fused homolog (SUFU) (C54G2) rabbit mAb (cat. no. #2520, M.W. 54
kDa); anti-Gli1 (C68H3) rabbit mAb (cat. no. # 3538, M.W 160 kDa);
and the secondary antibody, HRP-linked goat anti-rabbit IgG (cat.
no. #7074).
Nuclear extraction
A nuclear extraction kit (cat. no. #40010; Active
Motif, Carlsbad, CA, USA) was used to prepare the nuclear extracts
from the chondrosarcoma, adult mesenchymal stem and chondrocytic
C28 cells, according to the manufacturer's protocol.
Gel-shift/gel retardation assay
A Gelshift™ Chemiluminescent electrophoretic
mobility shift assay (EMSA) kit (cat. no. #37341; Active Motif) was
used to analyze the protein-DNA interactions. The principle behind
EMSA relies on the fact that DNA-protein complexes migrate slower
than DNA alone in a native polyacrylamide or agarose gel. The
difference in electrophoretic separation of DNA-protein complexes
can be visualized as a ‘shift’ in migration of the labeled DNA
band. Briefly, nuclear extracts were incubated with a biotin 3′- or
5′-end-labeled DNA probe containing the consensus binding site of
interest. Samples were resolved by electrophoresis on a retardation
6% polyacrylamide gel (cat. no. #EC6365BOX; Life Technologies,
Thermo Fisher Scientific, Inc.) and transferred to a nylon membrane
(cat. no. #LC2003; Novex, Thermo Fisher Scientific, Inc.). The
electrophoresis unit was filled with 0.5X Tris/borate EDTA (TBE;
cat. no. #B52, Thermo Fisher Scientific, Inc.) immediately below
the bottom of the wells.
Gels were pre-run at 100 V. During the pre-run,
control DNA and sample DNA binding reactions were performed,
according to the protocol provided in the manual. Once the pre-run
of the gel was finished, 20 µl of sample containing loading buffer
was loaded onto the gel. The gel was run until the Bromophenol blue
dye had migrated three-quarters of the way down the length of the
gel. The free biotin control-DNA duplex migrated immediately behind
the Bromophenol blue. Subsequently, the binding reactions were
transferred to nylon membranes at 380 mA (100 V) for 30 min. This
step was followed by the crosslink transfer of DNA to the membrane
using a transilluminator for 10–15 min. The ECL method was applied
to detect biotin-labeled DNA. The membrane was subsequently placed
into a film cassette and exposed to X-ray film for 2–5 min. The
biotin end-labeled DNA probe was detected using streptavidin
conjugated to HRP and a chemiluminescent substrate. These extracts
were incubated with a biotin 3′-or 5′-end-labeled DNA probe with an
IL-6 consensus sequence.
Duplex IL-6 oligonucleotide sense and
antisense sequences
Oligonucleotide sequences were synthesized by
Integrated DNA Technologies, Inc. (Coralville, IA, USA). The
oligo1-IL-6 sense sequence was prepared with the following
characteristics: M.W. 9.330.1; GC content, 40.0; extinction
coefficient, 321,200 l/(mole.cm); and Mfg ID, M189229415. The
oligo1-IL-6 antisense strand was prepared with these
characteristics: M.W. 8.547.6; GC content, 60.7; extinction
coefficient, 248,800 l/(mole.cm); and Mfg ID, M189229416. The
oligo1-IL-6 sense and antisense strands consisted of the following
sequences, respectively: 5′-CTGAGAAAGGAGACATGTAACAAGAGTAAC-3′ and
3′-CGACTGCTCGACGTCCGTGTCTTGGTCA-5′. The control biotin duplex had
the following characteristics: 100 nmol, M.W. 18,664.5; sequences:
5′-/5Biosg/CTGAGAAAGGAGACATGTAACAAGAGTAAC-3′ (sense) and
5′-/5Biosg/ACTGGTTCTGTGCCTGCAGCTCGTCAG C-3′ (antisense), and those
for the control non-biotin complex were: 100 nmol, M.W. 17,877.7;
sequences: 5′-CTGAGAAAGGAGACATGTAACAAGAGTAAC-3′ (sense) and
5′-ACTGGTTCTGTGCCTGCAGCTCGTCAGC-3′ (antisense). The company Active
Motif also provided unlabeled control DNA target (non-biotin) for
use in competition experiments to verify the specificity of the
DNA-protein complex. The control nuclear extract was also included
in the kit.
Histone H3K9 demethylase activity
assay
The quantification of H3K9-specific histone
demethylase activity was performed using the EpiSeeker Histone H3
(K9) Demethylase Activity Quantification Assay kit (cat. no.
#113458; Abcam), according to the manufacturer's protocol.
Statistical analysis
All experiments were performed in triplicate, and
P<0.05 was considered to indicate a statistically significant
value. Data analysis was performed using a one-way analysis of
variance (ANOVA) unpaired t-test (GraphPad Prism; GraphPad
Software, San Diego, CA, USA).
Results
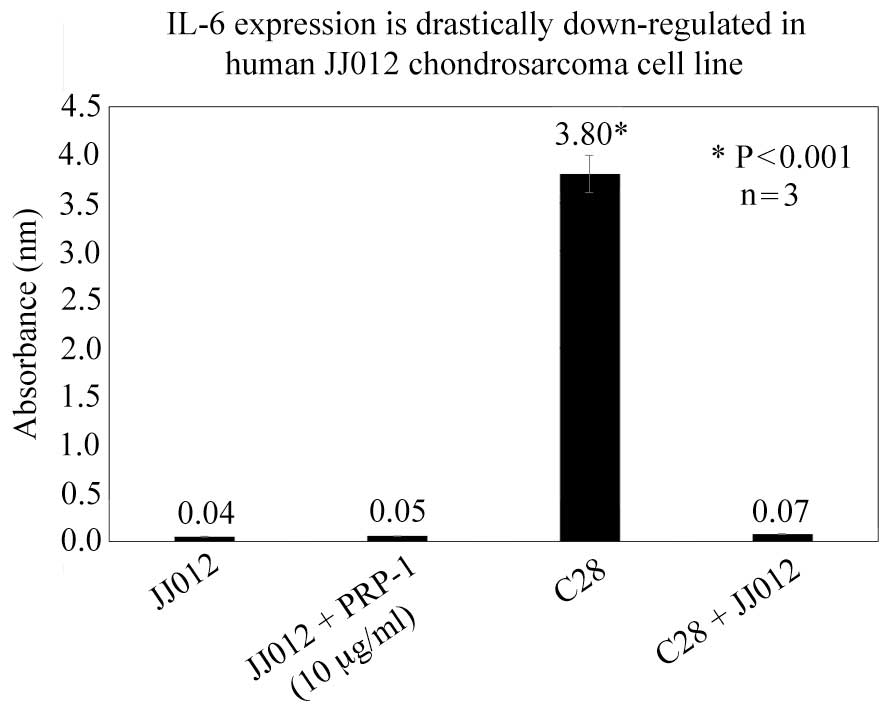
A panel of 12 human inflammatory cytokines from the
Human Inflammatory Cytokines Multi-Analyte ELISArray kit (Qiagen)
were tested. Even though the goal of the present study was to make
a comparison of cytokine expression primarily between the JJ012
human chondrosarcoma cell line and C28 chondrocytes, the results
for the adult mesenchymal cell line were also included in the text,
as well as those of the co-culture experiments. The experimental
data pointed towards an absence of any differences in the
expression of the cytokines among the cell lines, with the
exception of IL-6. The expression of IL-6 in the JJ012 human
chondrosarcoma cells was 86-fold lower compared with the C28
chondrocytes, based on the ELISA results. A 53-fold downregulation
of IL-6 expression in co-culture of chondrosarcoma cells and C28
chondrocytes was demonstrated compared with the C28 chondrocytes
(Fig. 1). The expression of IL-6 was
56-fold lower compared with mesenchymal adult cells, and a 65-fold
downregulation of IL-6 expression in co-culture of mesenchymal stem
cells and JJ012 cells was observed in comparison with its
expression in mesenchymal cells. PRP-1 did not affect the levels of
IL-6 (data not shown). From these experiments, it is possible to
conclude that IL-6 manifested itself as an anti-inflammatory and,
possibly, as an anti-tumorigenic factor.
To explore protein-DNA interactions leading to such
differences, a gel-shift chemiluminescent assay was performed
(Fig. 2). As shown in Fig. 2, the commercially supplied control is
shown in lanes 1–3: In lane 2, the band-shift was observed due to
the presence of biotinylated control and control nuclear extract.
In lane 1, in the absence of the control nuclear extract, no shift
was observed, since the reaction lacked protein to bind to the
oligo-IL-6 DNA, and thereby to cause a shift. In lane 2, the target
protein was present in the control nuclear extract, and bound to
the biotin-labeled control DNA, thereby causing a shift compared
with the reaction in lane 1. In lane 3, no shift was detected: The
excess of unlabeled control DNA competed for binding of the target
protein in the control nuclear extract. This experiment verified
that the signal observed in lane 2 resulted from specific
DNA-protein interactions.
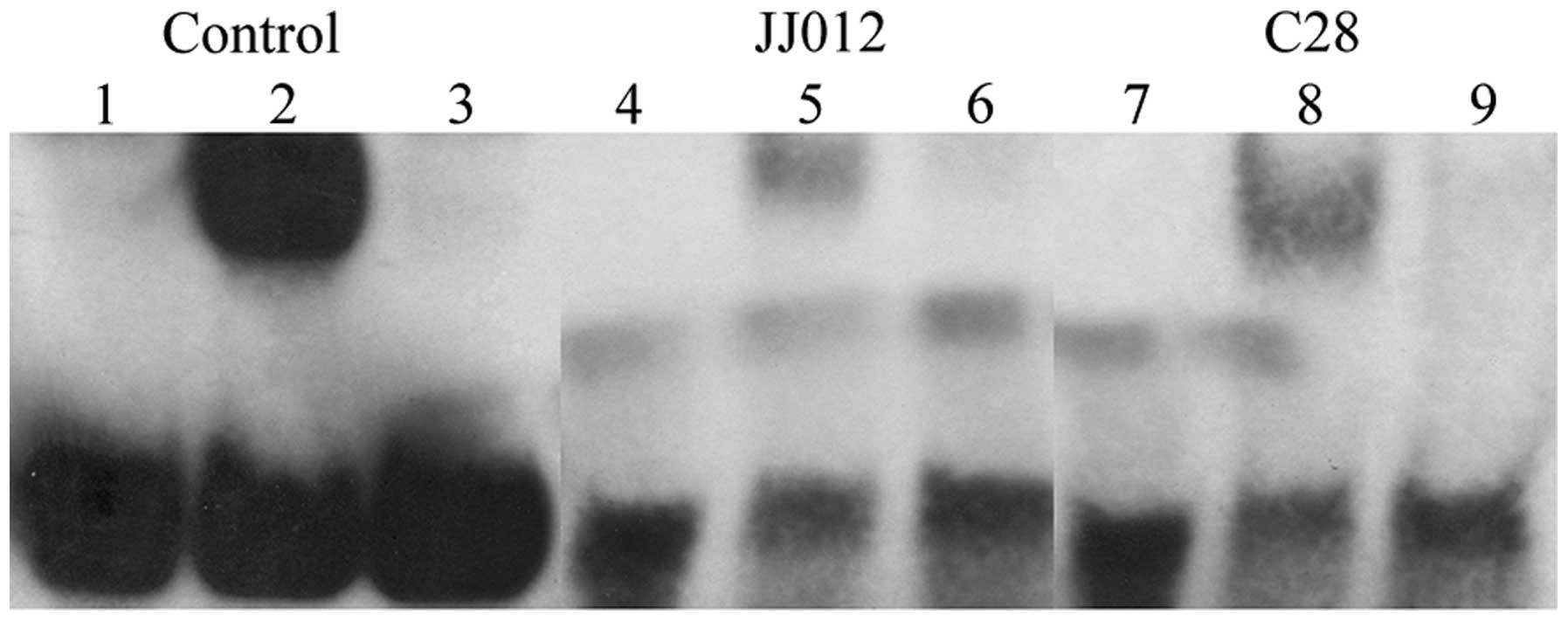 | Figure 2.Electrophoretic mobility shift assay
reveals differences between IL-6 DNA-protein complexes in human
JJ012 chondrosarcoma and C28 chondrocyte nuclear extracts. The
protein IL-6-DNA complex was markedly larger in C28 chondrocytes
compared with the JJ012 cell line. The lanes were loaded as
follows: Lane 1, biotinylated control; lane 2, biotinylated control
+ control nuclear extract; lane 3, biotinylated control + control
nuclear extract + unlabeled control DNA; lane 4, oligo-IL-6 duplex
biotinylated; lane 5, oligo-IL-6 duplex biotinylated + JJ012
extract; lane 6, oligo-IL-6 duplex biotinylated + JJ012 + unlabeled
oligo IL-6 control; lane 7, oligo-IL-6 duplex biotinylated; lane 8,
oligo-IL-6 duplex biotinylated + C28 extract; lane 9, oligo-IL-6
biotinylated DNA + C28 extract + unlabeled oligo-IL-6 control.
IL-6, interleukin-6. |
Gel-shifts were also observed for chondrosarcoma
cells and chondrocytes in the lanes with nuclear cell extract and
oligo-IL-6 DNA. Notably, the DNA-protein complex in C28 cells was
revealed to be markedly larger compared with chondrosarcoma cells:
In the experimental samples, shown in lanes 4–6 for the JJ012
chondrosarcoma cells and lanes 7–9 for the C28 chondrocytes,
band-shifts were observed in lanes 5 and 8 for the JJ012 and the
C28 cells, respectively. Those lanes featuring gel-shifts included
oligo-IL-6-biotinylated duplex with corresponding extracts either
from the JJ012 or C28 cells, and the protein present in those
nuclear extracts was able to bind to the IL-6 DNA. Lanes 4 and 7
featured oligo-IL-6-biotinylated duplex; lanes 6 and 9 featured
oligo-IL-6 biotinylated with corresponding nuclear extract and
unlabeled IL-6 control, respectively, and therefore no shifts were
observed in those lanes. Most importantly, in lane 8 for the C28
chondrocytes, the shifted band pertaining to the DNA-protein
complex was much larger compared with that in lane 5 for the human
JJ012 chondrosarcoma cells.
Western blot analysis demonstrated upregulated
expression levels of SOCS3 in chondrocytes compared with the JJ012
chondrosarcoma cells (Fig. 3).
Addition of PRP-1 restored the expression of SOCS3 in the JJ102
cells in a dose-responsive manner. No marked differences in the
expression levels of signal transducer and activator of
transcription 3 (STAT3) or Smad2 proteins were observed between the
JJ012 cells and the C28 chondrocytes. SOCS3, in certain tumors, has
been revealed to be a tumor suppressor, for example, in breast
cancer cells, and its expression was determined to be independent
of STAT (28,29). In the present study, a marked
upregulation in the level of TGF-β was observed in chondrocytes,
contrasted with a marked downregulation of TGF-β expression in the
chondrosarcoma cells (Fig. 3).
Addition of PRP-1 failed to restore the expression levels of
TGF-β.
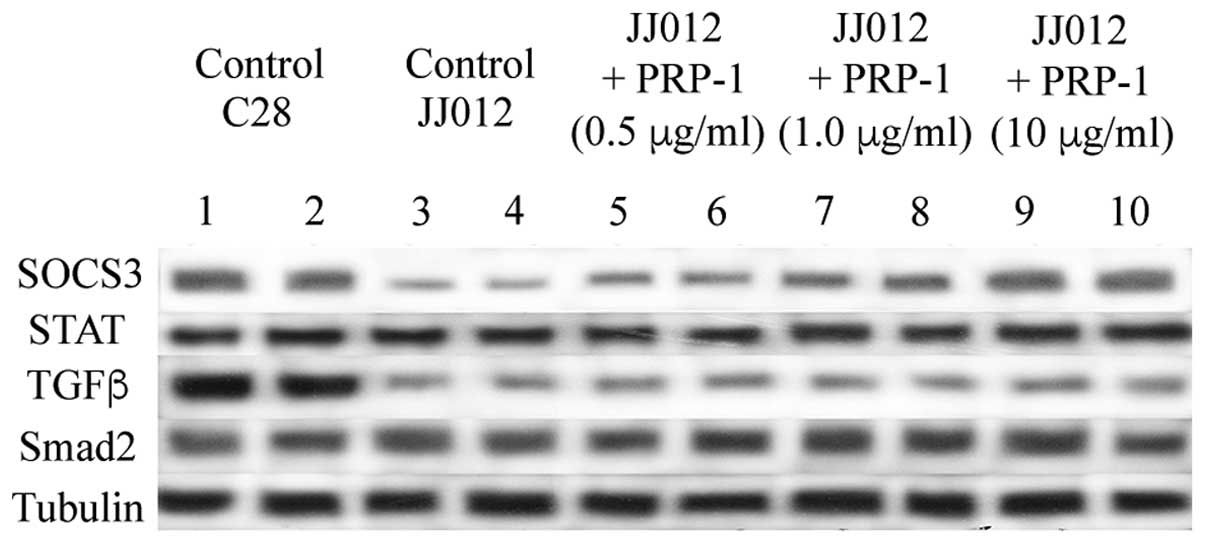 | Figure 3.PRP-1 upregulates SOCS3 expression in
a dose-responsive manner in the human chondrosarcoma JJ012 cell
line. The tumor suppressor, SOC3, was markedly downregulated in the
human JJ012 cell line compared with C28 chondrocytes. Addition of
PRP-1 restored the expression of SOC3 (the band was detected at ~30
kDa). The expression of TGF-β (band detected at 25 kDa) was reduced
in the C28 cells, and although it was markedly higher in C28
chondrocytes, PRP-1 failed to elicit any effects on TGF-β
expression. No differences in STAT or Smad2 protein expression were
identified between the C28 and JJ012 cell lines. All primary
antibodies were used at 1:1,000 dilution, and secondary antibodies
at 1:5,000. α-tubulin was used as a protein loading control, and
each pairing of lanes (1–10) represents the identical experiment
performed in duplicate. SOCS3, suppressor of cytokine signaling 3;
STAT, signal transducer and activator of transcription 3; PRP-1,
proline-rich polypeptide 1; TGF-β, transforming growth
factor-β. |
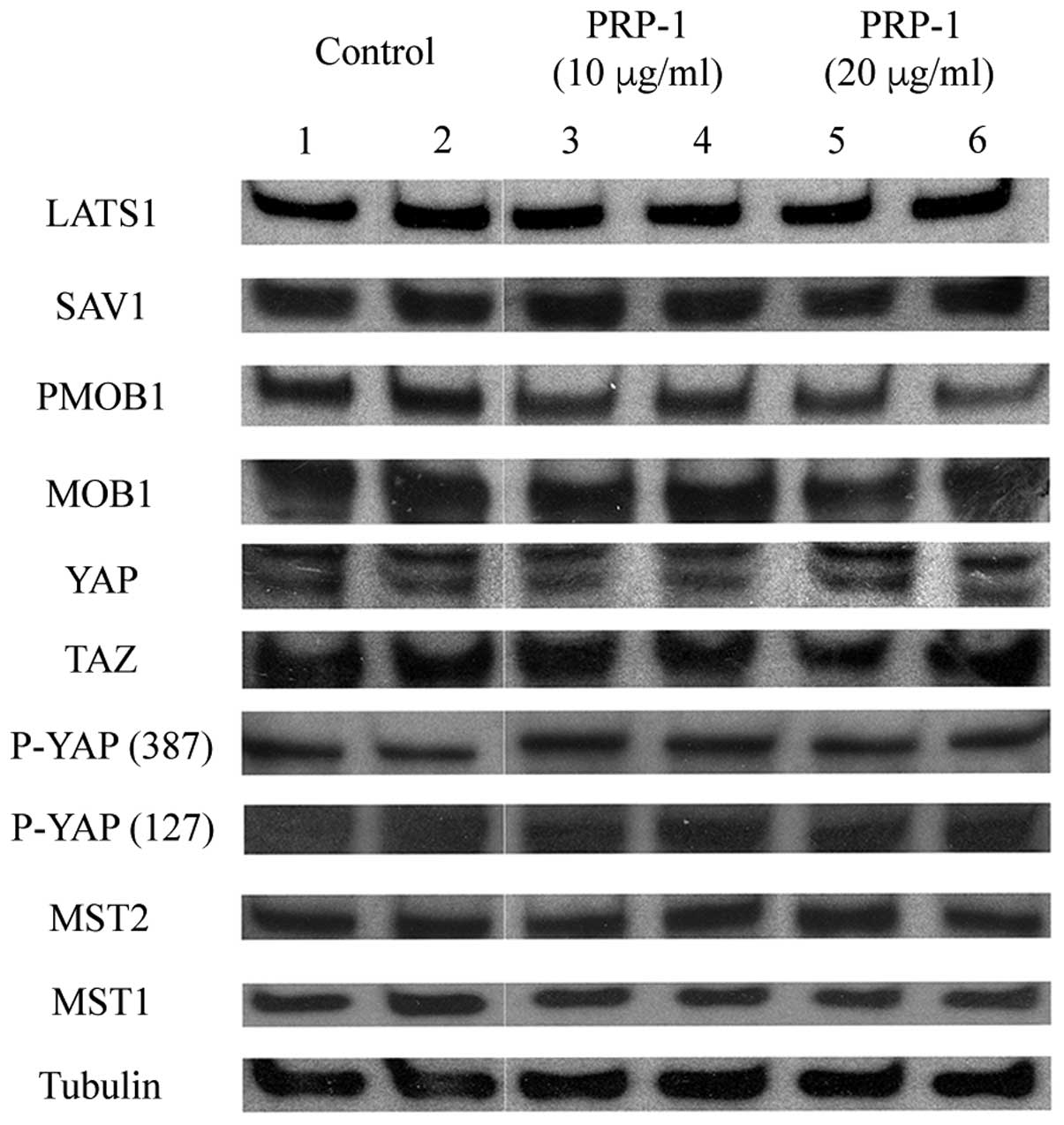
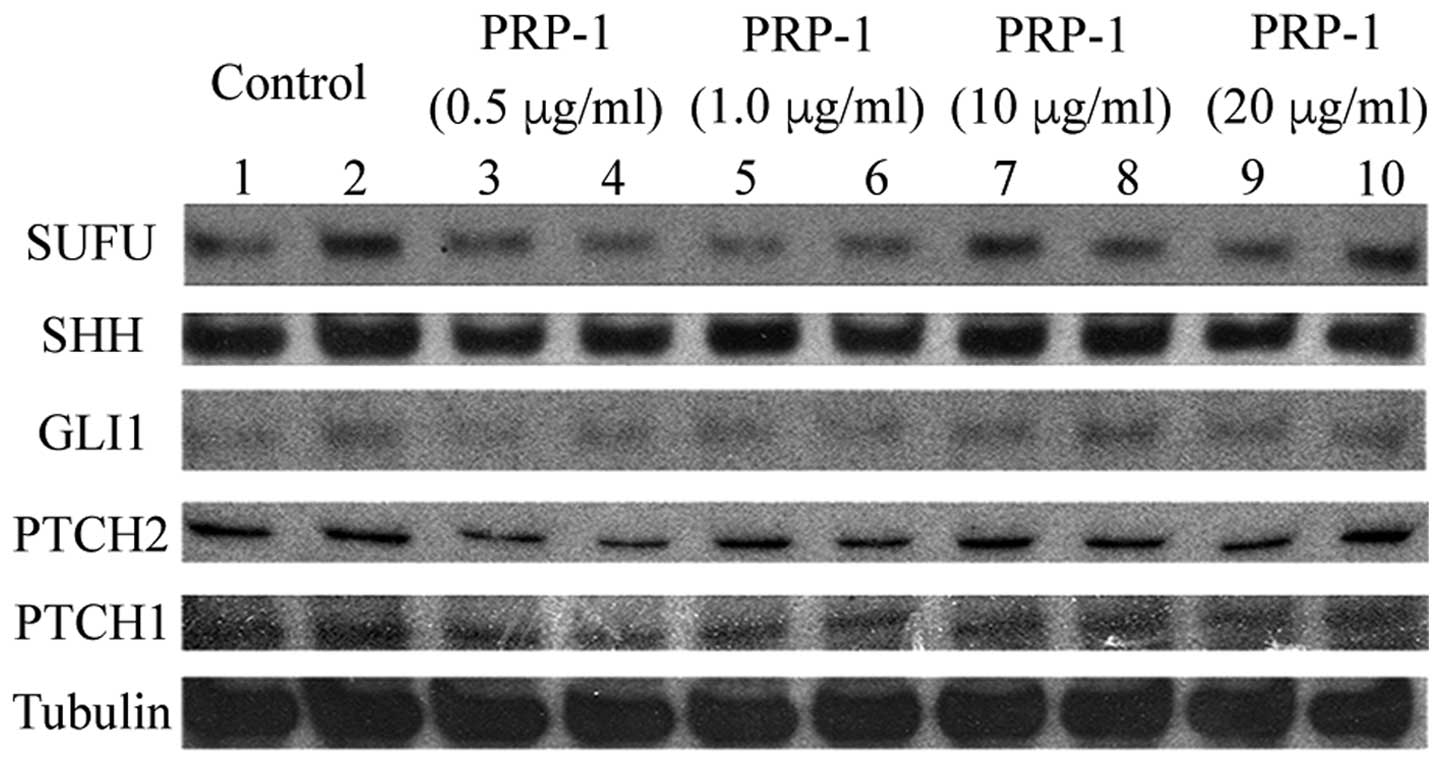
The expression levels of regulatory proteins of the
Hedgehog and Hippo signaling pathways in the human JJ012
chondrosarcoma cell line are shown in Figs. 4 and 5, respectively. PRP-1 did not exert any
effect on the tumor suppressors or oncoproteins of those respective
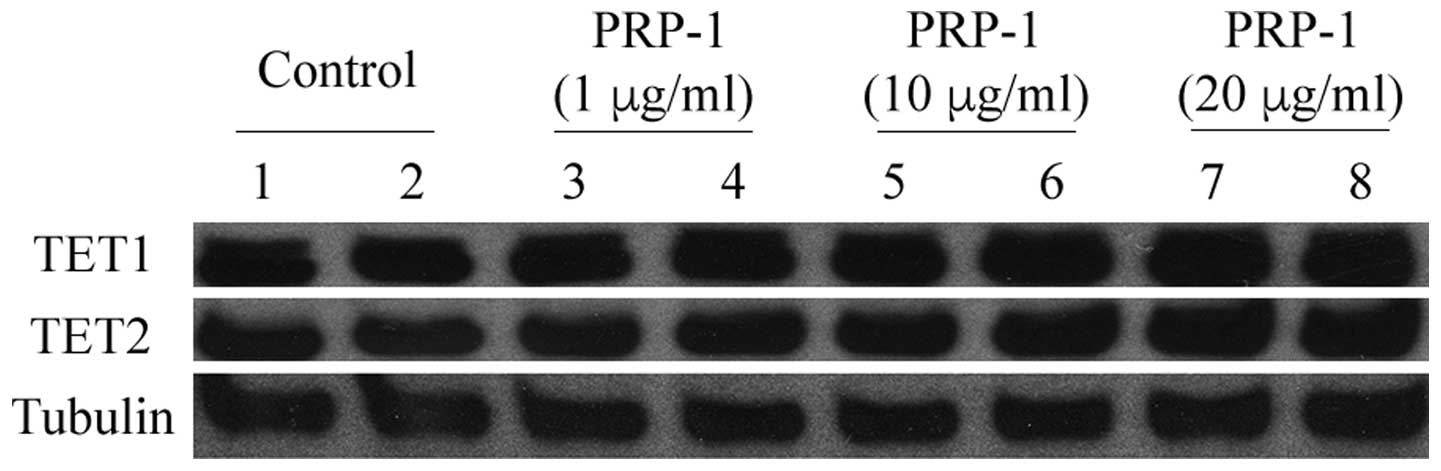
pathways. Other western blot analyses were performed to measure the
expression levels of the proteins TET1 and TET2 in the human JJ012
chondrosarcoma cell line, since their downregulation is an early
event in cell transformation, and these proteins act predominantly
as tumor suppressors. A dose-responsive upregulation in the
expression levels of the TET1 and TET2 proteins was observed
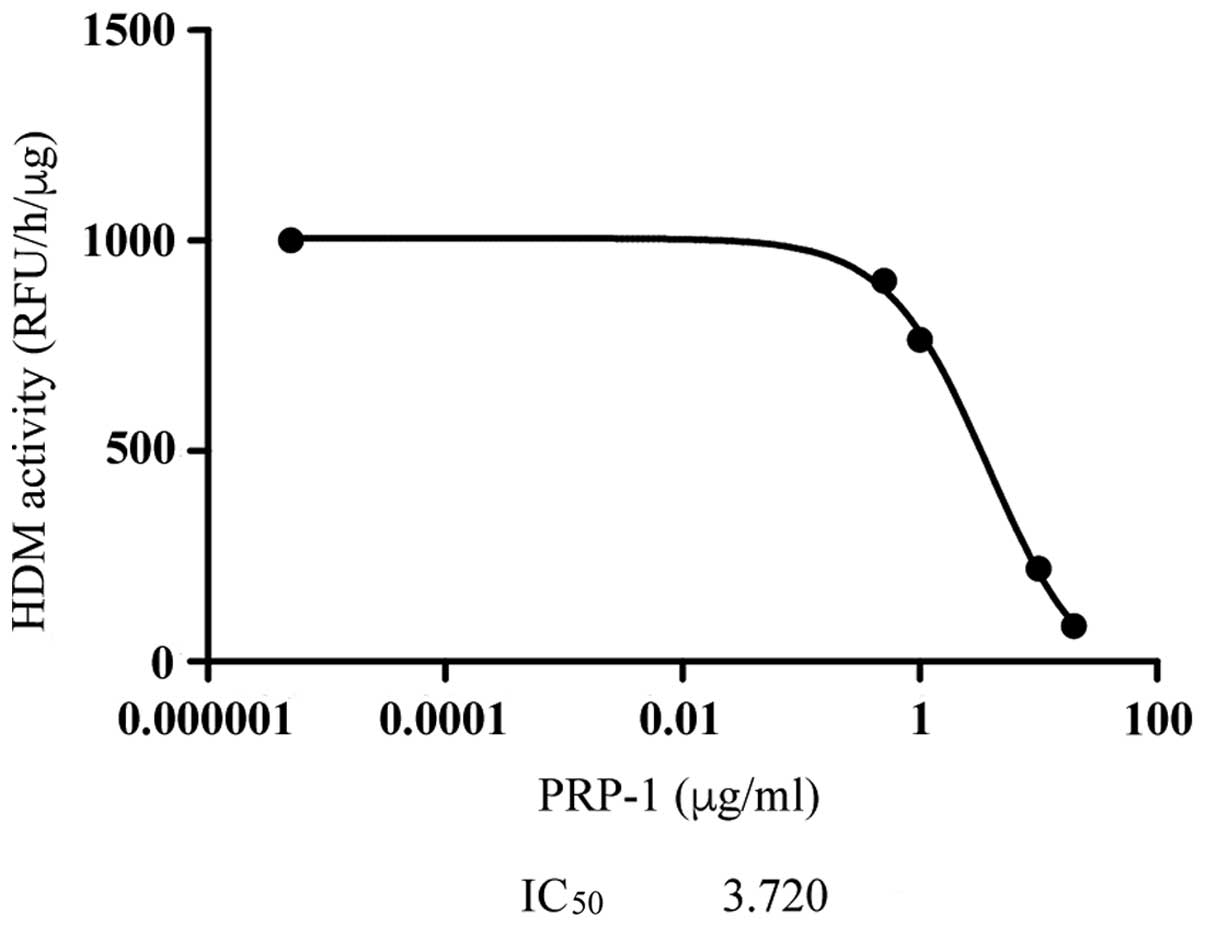
(Fig. 6). From the epigenetic
standpoint, PRP-1 was revealed to inhibit H3K9 demethylase activity
with an IC50 (concentration required to give
half-maximal inhibition) value of 3.72 µg/ml in the human JJ012
chondrosarcoma cell line (Fig.
7).
Discussion
The marked downregulation of IL-6 expression in the
human JJ012 chondrosarcoma cell line compared with C28 chondrocytes
prompted further elucidation of the putative mechanism involved in
this process. Reduced expression of IL-6 as a growth inhibitor and
differentiation factor has been reported in certain tumors, and
this has been linked with the compromised differentiation program
for a number of carcinoma and leukemia cell lines (30–33).
Tumor rejection and long-term survival rates have been reported in
response to IL-6. It is important to mention that IL-6 manifests
itself either as an anti-inflammatory or a pro-inflammatory
cytokine (30–33). IL-6 expression has often been
determined to be within normal levels in well-differentiated forms
of thyroid carcinoma, although it was markedly suppressed in
undifferentiated forms of thyroid carcinomas (34). A sarcomatoid component is present in
these types of tumors. Anaplastic thyroid carcinomas (ATCs) of
sarcomatoid appearance are characterized by spindle cells and giant
cells, which are the most frequent cells observed in ATC. Primary
sarcomas simulating a sarcomatoid ATC have been reported in case
reports for many types of sarcoma, including chondrosarcoma
(35). The gel-shift assay revealed
the presence of a larger protein IL-6-DNA complex in the C28
chondrocytes compared with the chondrosarcoma cells. The
characteristics and origin of the proteins, and the mechanisms
which are responsible for such differences, have yet to be
investigated. Another aspect that deserves further investigation is
the regulation of IL-6 by miRNAs, and their involvement in the
regulation of the differentiation program. Our group has previously
demonstrated that the downregulation of miR181b leads to the
downregulation of its target protein, IL-6 (36).
The upregulation of SOCS3, TET1 and TET2 expression
in a dose-dependent manner by PRP-1 further demonstrates the
ability of this cytokine peptide to upregulate tumor suppressor
genes in general (8–10). Even though PRP-1 did not exert any
effect on tumor suppressors or oncoproteins of the Hippo or
Hedgehog signaling pathways, there was an important conclusion to
consider: The inhibition of tumor suppressors by PRP-1 depends on
the pathway these tumor suppressors represent or are involved in.
TET1 and TET2 act predominantly as tumor suppressors.
Re-introduction and overexpression of these proteins is able to
restore the 5hmC content, and to suppress invasion and growth in
certain tumors (37–41). A previous report revealed that a
deficiency in SOCS3 may promote tumor development, since it has
tumor suppressor functions in numerous cancer cell types (29). It was demonstrated that SOCS3, in
certain circumstances, acts as a regulator of pathways and
processes that are unrelated to the STAT signaling cellular
pathways. SOCS3 attenuates pro-inflammatory signaling (42), and its deficiency promotes
inflammation (43). However, the
role exerted by SOCS3 during bone inflammation is complex, and
effects may work in opposition to each other; therefore, knowledge
of further mechanistic details concerning the SOCS3 pathway are
necessary for a better understanding of the processes of various
bone inflammatory diseases. The epigenetic regulation of
inflammation linked with oncogenesis has a central role. It is
known that H3K9 methylation is eliminated from the promoters of
inflammation-associated genes (44),
and, therefore the mechanisms underpinning the inhibition of H3K9
demethylases merit further attention. The ability of PRP-1 to
inhibit H3K9 demethylase activity was previously reported by our
group (8), and a possible mechanism
of the peptide's function in the upregulation of tumor suppressors
and in the downregulation of oncoproteins was proposed. In the
present study, the PRP-1-mediated inhibition of H3K9 demethylase
was associated with the process of inflammation, and the
IC50 value has been demonstrated.
In summary, cytokines produced in the tumour
microenvironment have an important role in cancer pathogenesis and
in the inhibition of disease progression. IL-6 family cytokines
appear to be double-edged swords, since the development of bone
cancers may be either prevented or enhanced by this family of
cytokines, depending on the cell type, the stage of the tumor and
the bone environment (45). Observed
downregulation of IL-6 in the human JJ012 chondrosarcoma cell line
compared with the chondrocytes supports the hypothesis that IL-6 is
acting as a differentiation/anti-inflammatory factor in this
cellular context, indicating the possibility that a factor(s) or
interacting proteins in the tumor have the ability to inhibit IL-6
expression. Understanding the differences in their expression
patterns between the normal and malignant states, and the signaling
pathways that lead to these differences, require further
investigation as important targets for future therapeutic
interventions. Metastatic chondrosarcoma does not respond to
conventional therapies and the search for new therapeutic
approaches is urgent (2–10,36,46). The
ability of PRP-1 as an antiproliferative agent to restore the
expression of anti-inflammatory cytokines with tumor suppressor
function proves the importance of this neuropeptide for future
clinical consideration.
Acknowledgements
The present study was supported in part by the
Ratcliffe Foundation gift to Miami Center of Orthopedic Research
and Education (Miami CORE).
References
|
1
|
Landskron G, De la Fuente M, Thuwajit P,
Thuwajit C and Hermoso MA: Chronic inflammation and cytokines in
the tumor microenvironment. J Immunol Res. 2014:1491852014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mirra J: Bone Tumors: Clinical,
radiologic, and pathologic correlations. Lea and Febiger;
Philadelphia, PA: 1989
|
|
3
|
Galoian K, Scully S, McNamara G, Flynn P
and Galoyan A: Antitumorigenic effect of brain proline rich
polypeptide-1 in human chondrosarcoma. Neurochem Res. 34:2117–2121.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Galoian K, Scully S and Galoyan A:
Myc-oncogene inactivating effect by proline rich polypeptide
(PRP-1) in chondrosarcoma JJ012 cells. Neurochem Res. 34:379–385.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Galoian K, Temple HT and Galoyan A:
Cytostatic effect of the hypothalamic cytokine PRP-1 is mediated by
mTOR and cMyc inhibition in high grade chondrosarcoma. Neurochem
Res. 36:812–818. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Galoian KA, Temple HT and Galoyan A:
Cytostatic effect of novel mTOR inhibitor, PRP-1 (galarmin) in
MDA231 (ER-) breast carcinoma cell line. PRP-1 inhibits mesenchymal
tumors. Tumour Biol. 32:745–51. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Galoian K, Temple HT and Galoyan A: mTORC1
inhibition and ECM-cell adhesion-independent drug resistance via
Pl3K-AKT and Pl3K-RAS-MAPK feedback loops. Tumour Biol. 33:885–90.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Galoian K, Qureshi A, D'ippolito G,
Schiller PC, Molinari M, Johnstone AL, Brothers SP, Paz AC and
Temple HT: Epigenetic regulation of embryonic stem cell marker
miR302C in human chondrosarcoma as determinant of antiproliferative
activity of proline-rich polypeptide 1. Int J Oncol. 47:465–472.
2015.PubMed/NCBI
|
|
9
|
Galoian K, Qureshi A, Wideroff G and
Temple HT: Restoration of desmosomal junction protein expression
and inhibition of H3K9-specific histone demethylase activity by
cytostatic proline-rich polypeptide-1 leads to suppression of
tumorigenic potential in human chondrosarcoma cells. Mol Clin
Oncol. 3:171–178. 2015.PubMed/NCBI
|
|
10
|
Galoian K, Guettouche T, Issac B, Qureshi
A and Temple HT: Regulation of onco and tumor suppressor miRNAs by
mTORC 1 inhibitor PRP-1 in human chondrosarcoma. Tumour Biol.
35:2335–2341. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang H, Liu Y, Bai F, Zhang JY, Ma SH, Liu
J, Xu ZD, Zhu HG, Ling ZQ, Ye D, et al: Tumor development is
associated with decrease of TET gene expression and
5-methylcytosine hydroxylation. Oncogene. 32:663–669. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q,
Ding J, Jia Y, Chen Z, Li L, et al: Tet-mediated formation of
5-carboxylcytosine and its excision by TDG in mammalian DNA.
Science. 333:1303–1307. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ito S, Shen L, Dai Q, Wu SC, Collins LB,
Swenberg JA, He C and Zhang Y: Tet proteins can convert
5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine.
Science. 333:1300–1303. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chmielecki J and Meyerson M: DNA
sequencing of cancer: What have we learned? Annu Rev Med. 65:63–79.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim
SH, Ito S, Yang C, Wang P, Xiao MT, et al: Oncometabolite
2-hydroxyglutarate is a competitive inhibitor of
α-ketoglutarate-dependent dioxygenases. Cancer Cell. 19:17–30.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tahiliani M, Koh KP, Shen Y, Pastor WA,
Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L and
Rao A: Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in
mammalian DNA by MLL partner TET1. Science. 324:930–935. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Suijker J, Oosting J, Koornneef A, Struys
EA, Salomons GS, Schaap FG, Waaijer CJ, Wijers-Koster PM,
Briaire-de Bruijn IH, Haazen L, et al: Inhibition of mutant IDH1
decreases D-2-HG levels without affecting tumorigenic properties of
chondrosarcoma cell lines. Oncotarget. 6:12505–12519. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Haseeb A, Makki MS and Haqqi TM:
Modulation of ten-eleven translocation 1 (TET1), Isocitrate
Dehydrogenase (IDH) expression, α-Ketoglutarate (α-KG), and DNA
hydroxymethylation levels by interleukin-1β in primary human
chondrocytes. J Biol Chem. 289:6877–6885. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Deel MD, Li JJ, Crose LE and Linardic CM:
A review: Molecular aberrations within hippo signaling in bone and
soft-tissue sarcomas. Front Oncol. 5:1902015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tiet TD, Hopyan S, Nadesan P, Gokgoz N,
Poon R, Lin AC, Yan T, Andrulis IL, Alman BA and Wunder JS:
Constitutive hedgehog signaling in chondrosarcoma up-regulates
tumor cell proliferation. Am J Pathol. 168:321–30. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lu L, Li Y, Kim SM, Bossuyt W, Liu P, Qiu
Q, Wang Y, Halder G, Finegold MJ, Lee JS and Johnson RL: Hippo
signaling is a potent in vivo growth and tumor suppressor pathway
in the mammalian liver. Proc Natl Acad Sci USA. 107:1437–1442.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Eisinger-Mathason TS, Mucaj V, Biju KM,
Nakazawa MS, Gohil M, Cash TP, Yoon SS, Skuli N, Park KM, Gerecht S
and Simon MC: Deregulation of the Hippo pathway in soft-tissue
sarcoma promotes FOXM1 expression and tumorigenesis. Proc Natl Acad
Sci USA. 112:E3402–E3411. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mohamed AD, Tremblay AM, Murray GI and
Wackerhage H: The Hippo signal transduction pathway in soft tissue
sarcomas. Biochim Biophys Acta. 1856:121–129. 2015.PubMed/NCBI
|
|
24
|
Li R, Cai L, Ding J, Hu CM, Wu TN and Hu
XY: Inhibition of hedgehog signal pathway by cyclopamine attenuates
inflammation and articular cartilage damage in rats with
adjuvant-induced arthritis. J Pharm Pharmacol. 67:963–971. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nakashima H, Nakamura M, Yamaguchi H,
Yamanaka N, Akiyoshi T, Koga K, Yamaguchi K, Tsuneyoshi M, Tanaka M
and Katano M: Nuclear factor-kappaB contributes to hedgehog
signaling pathway activation through sonic hedgehog induction in
pancreatic cancer. Cancer Res. 66:7041–7049. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zacharias WJ, Li X, Madison BB, Kretovich
K, Kao JY, Merchant JL and Gumucio DL: Hedgehog is an
anti-inflammatory epithelial signal for the intestinal lamina
propria. Gastroenterology. 138:2368–2377, 2377.e1-e4. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Taniguchi K, Wu LW, Grivennikov SI, de
Jong PR, Lian I, Yu FX, Wang K, Ho SB, Boland BS, Chang JT, et al:
A gp130-Src-YAP module links inflammation to epithelial
regeneration. Nature. 519:57–62. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Duh QY and Grossman RF: Thyroid growth
factors, signal transduction pathways, and oncogenes. Surg Clin
North Am. 75:421–437. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Barclay JL, Anderson ST, Waters MJ and
Curlewis JD: SOCS3 as a tumor suppressor in breast cancer cells,
and its regulation by PRL. Int J Cancer. 124:1756–1766. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Revel M: Growth regulatory functions of
IL6 and antitumor effects. Res Immunol. 143:769–773. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Givon T, Slavin S, Haran-Ghera N,
Michalevicz R and Revel M: Antitumor effects of human recombinant
interleukin-6 on acute myeloid leukemia in mice and in cell
cultures. Blood. 79:2392–2398. 1992.PubMed/NCBI
|
|
32
|
Basolo F, Fiore L, Pollina L, Fontanini G,
Conaldi PG and Toniolo A: Reduced expression of interleukin 6 in
undifferentiated thyroid carcinoma: in vitro and in vivo studies.
Clin Cancer Res. 4:381–387. 1998.PubMed/NCBI
|
|
33
|
Mulé JJ, McIntosh JK, Jablons DM and
Rosenberg SA: Antitumor activity of recombinant interleukin 6 in
mice. J Exp Med. 171:629–636. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ragazzi M, Ciarrocchi A, Sancisi V,
Gandolfi G, Bisagni A and Piana S: Update on anaplastic thyroid
carcinoma: Morphological, molecular, and genetic features of the
most aggressive thyroid cancer. Int J Endocrinol. 2014:7908342014.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tseleni-Balafouta S, Arvanitis D,
Kakaviatos N and Paraskevakou H: Primary myxoid chondrosarcoma of
the thyroid gland. Arch Pathol Lab Med. 112:94–96. 1988.PubMed/NCBI
|
|
36
|
Galoian K, Guettouche T, Issac B, Navarro
L and Temple HT: Lost miRNA surveillance of Notch, IGFR
pathway-road to sarcomagenesis. Tumour Biol. 35:483–492. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sarkar S, Horn G, Moulton K, Oza A, Byler
S, Kokolus S and Longacre M: Cancer development, progression, and
therapy: An epigenetic overview. Int J Mol Sci. 14:21087–21113.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lian CG, Xu Y, Ceol C, Wu F, Larson A,
Dresser K, Xu W, Tan L, Hu Y, Zhan Q, et al: Loss of
5-hydroxymethylcytosine is an epigenetic hallmark of melanoma.
Cell. 150:1135–1146. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Fan M, He X and Xu X: Restored expression
levels of TET1 decrease the proliferation and migration of renal
carcinoma cells. Mol Med Rep. 12:4837–4842. 2015.PubMed/NCBI
|
|
40
|
Cimmino L, Dawlaty MM, Ndiaye-Lobry D, Yap
YS, Bakogianni S, Yu Y, Bhattacharyya S, Shaknovich R, Geng H,
Lobry C, et al: TET1 is a tumor suppressor of hematopoietic
malignancy. Nat Immunol. 16:8892015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang Q, Zhao K, Shen Q, Han Y, Gu Y, Li
X, Zhao D, Liu Y, Wang C, Zhang X, et al: Tet2 is required to
resolve inflammation by recruiting Hdac2 to specifically repress
IL-6. Nature. 525:389–393. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jo D, Liu D, Yao S, Collins RD and Hawiger
J: Intracellular protein therapy with SOCS3 inhibits inflammation
and apoptosis. Nat Med. 11:892–898. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Qin H, Holdbrooks AT, Liu Y, Reynolds SL,
Yanagisawa LL and Benveniste EN: SOCS3 deficiency promotes M1
macrophage polarization and inflammation. J Immunol. 189:3439–3448.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Villeneuve LM, Reddy MA, Lanting LL, Wang
M, Meng L and Natarajan R: Epigenetic histone H3 lysine 9
methylation in metabolic memory and inflammatory phenotype of
vascular smooth muscle cells in diabetes. Proc Natl Acad Sci USA.
105:9047–9052. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Blanchard F, Duplomb L, Baud'huin M and
Brounais B: The dual role of IL-6-type cytokines on bone remodeling
and bone tumors. Cytokine Growth Factor Rev. 20:19–28. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Clark JC, Akiyama T, Dass CR and Choong
PF: New clinically relevant, orthotopic mouse models of human
chondrosarcoma with spontaneous metastasis. Cancer Cell Int.
10:202010. View Article : Google Scholar : PubMed/NCBI
|