Introduction
Huntington’s disease (HD) is a progressive,
autosomal dominant, neurodegenerative disorder caused by the
expansion of a CAG trinucleotide repeat (>35 repeats) in the
huntingtin (Htt) gene, which is translated into an expanded
polyglutamine tract in the N-terminus of the Htt protein (1). The most obvious characteristic of HD
is uncontrolled movement, dementia, emotional disturbance and early
death, which often occur in middle age (2,3). These
symptoms are associated with neuronal loss that preferentially
occurs in medium-sized spiny neurons of the striatum and also
extends to other brain regions during the late stages of the
disease.
Wild-type Htt is considered to possess a number of
cellular functions, such as protein transport, vesicle trafficking
and cytoskeletal anchoring. When Htt is mutated with an expanded
polyglutamine tract (>36 glutamines), Htt becomes misfolded to
induce gain of toxic function and may also lose its normal
function. Mutant Htt appears to affect a variety of cellular
functions, including intracellular signaling pathways (4), the protein degradation pathway
(5) and the nuclear transcription
and axon transport (6,7), resulting in degeneration of neuronal
cells in the striatum and other brain regions.
Clearing mutant Htt may be of value in preventing
the pathogenesis and development of HD. In eukaryotic cells, there
exist two systems of protein degradation, the ubiquitin-proteasome
system and autophagy. Autophagy is a non-specific bulk degradation
pathway for long-lived cytoplasmic proteins, protein complexes, or
damaged organelles. There are three types of autophagy, namely
macroautophagy, microautophagy and chaperone-mediated autophagy
(CMA). Macroautophagy is considered to promote the degradation of
Htt and clearance of aggregates. This is supported by several lines
of evidence that rapamycin (an activator of autophagy) (8) and its several small molecular
enhancers (9–11) may enhance the degradation of mutant
Htt and reduce its toxicity. It is also important to investigate
whether mutant Htt can damage autophagic function. It was reported
that mutant Htt did not alter the expression of
microtubule-associated protein 1 light chain 3, suggesting the
activity of macroautophagy is not changed by Htt (12).
CMA is another type of autophagy, which is distinct
from macroautophagy and microautophagy, as a subset of long-lived
cytosolic soluble proteins are directly delivered to the lysosomal
lumen via specific receptors (13).
The basic machinery consists of at least three types of proteins,
including i) chaperone proteins, which are responsible for
recognizing substrates based on their specific motifs and
delivering them to the lysosomes; ii) receptor proteins, which bind
and transport/pull substrates into the lysosomal lumen; and iii)
substrate proteins, which are a subset of soluble cytosolic
proteins containing specific motifs associated with KFERQ. When CMA
is activated, the expression levels of two important proteins,
lysosomal-associated membrane protein 2a (Lamp2a) and heat-shock
cognate protein 70 (Hsc70) are significantly increased (14–16).
It is known that heat-shock protein β-8 may participate in protein
quality control through a non-chaperone-like mechanism, which
requires the phosphorylation of eIF2α (17). It was recently demonstrated that CMA
contributes to the degradation of mutant Htt in cultured cells
(18). However, it remains unclear
whether mutant Htt alters CMA activity. Elucidating this issue may
help determine whether modulating CMA activity is an effective
therapeutic approach for the management of HD.
In this study, we aimed to investigate the effect of
mutant Htt on the basal activity of CMA in cultured cells and
determine whether enhancing CMA activity is an effective way to
reduce the toxicity of mutant Htt.
Materials and methods
Plasmids, antibodies and reagents
Enhanced green fluorescent protein
(EGFP)-Htt-exon1-20Q and EGFP-Htt-exon1–120Q plasmids were gifts
from Professor Xiaojiang Li from the Department of Human Genetics
at Emory University School of Medicine (Atlanta, GA, USA). XL1-Blue
Competent cells were obtained from Beijing TransGen Biotech Co.,
Ltd., Beijing, China; the Plasmid Mini kit was purchased from Omega
Bio-Tek, Inc., Norcross, GA, USA; the Endotoxin-Free Plasmid Maxi
kit was purchased from Tiangen Biotech Co., Ltd., Beijing, China;
and the BamHI and EcoRI restriction nucleotide
enzymes used in this study were purchased from Fermentas, Inc.,
Glen Burnie, MD, USA.
HEK293T cells were purchased from ATTC (Teddington,
UK) and cultured with high-glucose Dulbecco’s modified Eagle’s
medium (DMEM) (HyClone, Logan, USA) supplemented with
penicillin-streptomycin, fetal bovine serum (FBS) and 0.25%
trypsin-EDTA (Gibco-BRL, Carlsbad, CA, USA). SuperFectin™ In Vitro
DNA Transfection reagent (Shanghai Pufei Biotech Co., Ltd,
Shanghai, China) was used for DNA transfection.
The following antibodies were used in the study:
mouse monoclonal anti-green fluorescent protein (GFP) antibody
(1:20,000, Proteintech Group, Inc., Chicago, IL, USA); rabbit
polyclonal anti-Lamp2a antibody (1:600; Abcam, Cambridge, MA, USA);
rabbit monoclonal anti-Hsc70 antibody (1:500; Abcam); mouse
anti-β-actin antibody (1:2,000; Kangwei Century Biology Ltd., Co.,
Beijing, China). Enhanced chemiluminescence (ECL) developer
(Tiangen Biotech Co.) was used for western blotting.
Cell culture and Htt transfection
Two plasmids (EGFP-Htt-exon1-20Q and
EGFP-Htt-exon1–120Q) were used for transfection of HEK293T cells.
The HEK293T cells were maintained in DMEM supplemented with 10%
FBS, 100 U/ml penicillin and 100 mg/ml streptomycin sulfate at 37°C
in an atmosphere of 5% CO2 and 95% humidity. The cells
were seeded onto 6-well or 12-well plates and, after reaching a
confluency of 80%, they were transiently transfected with 0.75 or
1.0 μg wild-type (20Q) or mutant (120Q) Htt plasmids using
SuperFectin™ In Vitro DNA Transfection reagent. After 24, 36 and 48
h of culture, the cells were used for western blotting and
fluorescence microscopy analysis.
Western blot analysis and
immunofluorescent microcopy
After 36 or 48 h of transfection, the cultured cells
were rinsed with phosphate-buffered saline (PBS) three times at
room temperature and 1× SDS loading buffer (250 μl) was added
immediately to the cell pellet. The cell lysates were sonicated for
5–8 sec and heated at 100°C for 5 min in 1× gel-loading buffer
containing 2% SDS for western blot analysis. The samples (20 μg of
protein) were separated on a 10% (for Htt and other proteins)
Tris-glycine SDS-polyacrylamide gel. The proteins were transferred
to a nitrocellulose membrane (Amersham Pharmacia Biotech,
Piscataway, NJ, USA). The nitrocellulose membranes were blocked
with 5% non-fat dry milk/PBS with 0.1% Tris-buffered saline with
Tween-20 (TBST) for 30 min and the blots were rinsed with 1× TBST
and incubated with the primary antibodies overnight at 4°C.
Secondary horseradish peroxidase-conjugated anti-rabbit or
anti-mouse IgG were incubated with the blot in 5% non-fat dry
milk/PBS for 1 h at room temperature. The ECL developer was then
used to reveal immunoreactive bands on the blots. To verify the
protein loading amounts, the blots were also re-probed with mouse
anti-β-actin antibody.
After 24 h of transfection, immunofluorescent
staining of cultured cells with antibodies against Htt and Lamp2a
or Hsc70 was performed. Non-transfected and Htt-transfected HEK293T
cells were fixed for 7 min with 4% paraformaldehyde/PBS and then
examined under a immunofluorescent microscope. The cells were also
stained with Hoechst dye (blue) for the nuclei and the ratios of
Lamp2a (red) or Hsc70 (red) to blue color were quantified. These
ratios in Htt-transfected (green) or non-transfected cells (not
green) were compared to determine whether mutant Htt was able to
reduce Lamp2a or Hsc70. For the control, HEK293T cells were also
transfected with normal Htt (20Q).
Statistical analysis
All the data are expressed as means ± standard error
of the mean. The statistical results were analyzed by GraphPad
Prism software, version 5 (GraphPad Software, Inc., San Diego, CA,
USA) and the statistical significance (P<0.05) was assessed
using the Student’s t-test or one-way analysis of variance,
followed when appropriate by a post hoc analysis using the
Dunnett’s test.
Results
High-efficiency expression of Htt in
HEK293T cells
The expression of wild-type or mutant Htt was
achieved by transient transfection of wild-type Htt
(EGFP-Htt-exon1-20Q) and mutant Htt (EGFP-Htt-exon1–120Q) in
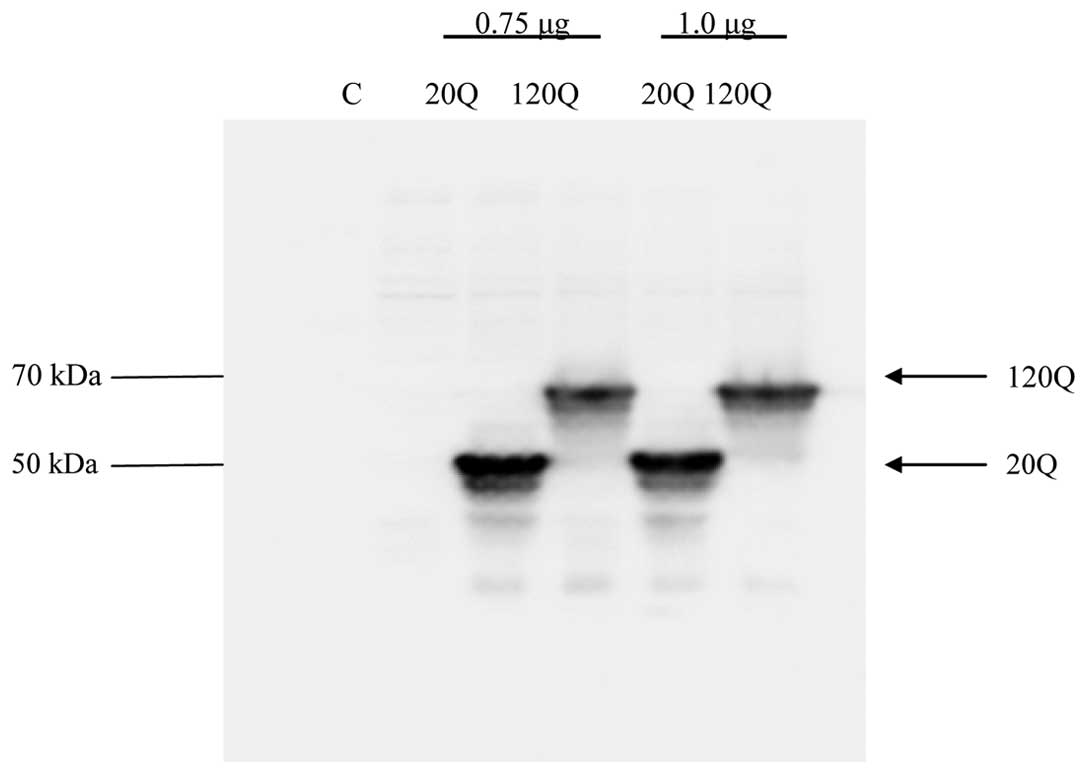
HEK293T cells. We performed western blotting using GFP antibody to
detect wild-type and mutant Htt expression. As expected, wild-type
Htt migrated at 50 kDa, while mutant Htt migrated at 70 kDa, as it
carries a larger polyglutamine repeat (Fig. 1).
Mutant Htt did not reduce the activity of
CMA
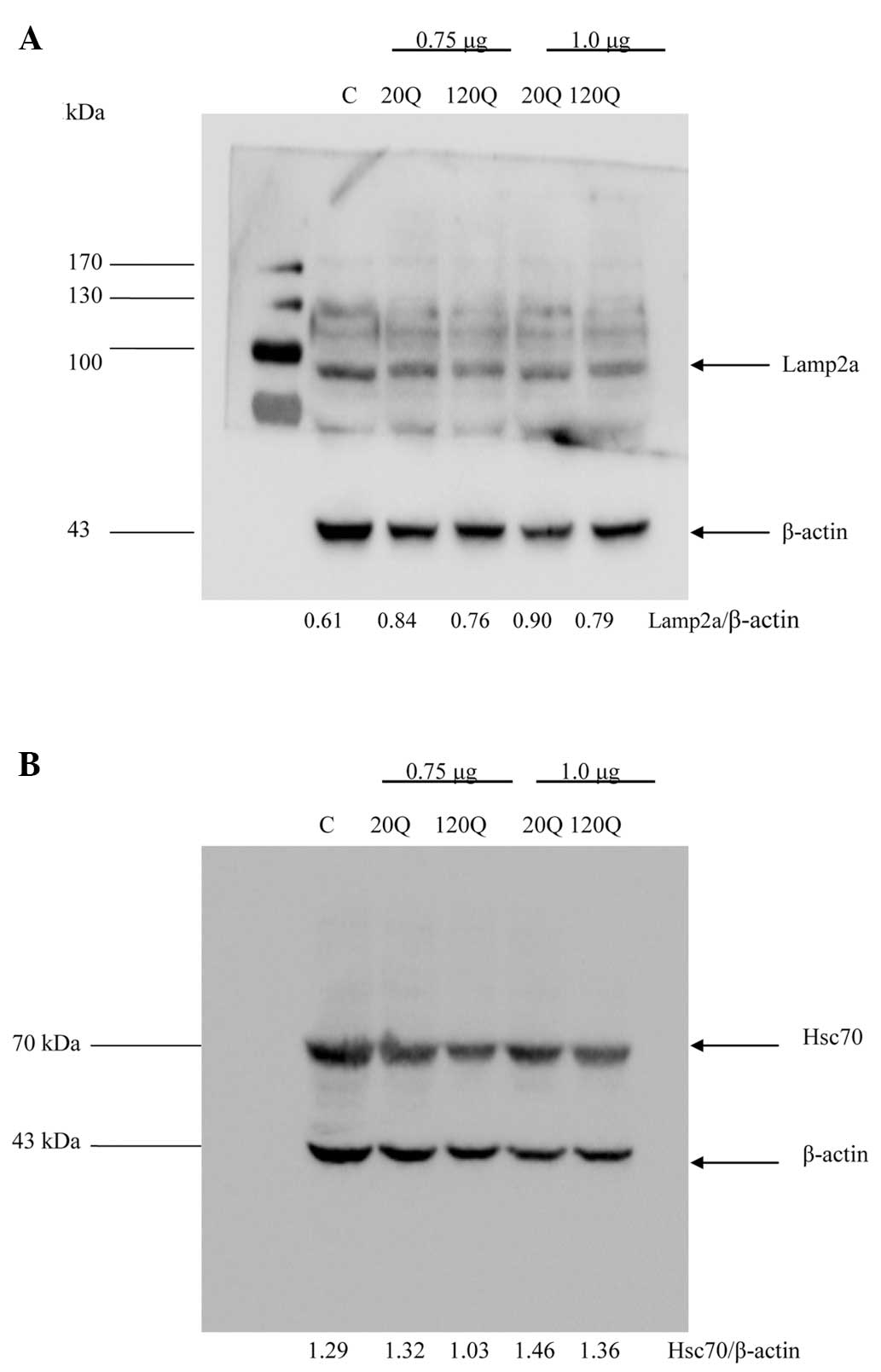
The expression levels of two important proteins,
Lamp2a and Hsc70, represents CMA activation. When CMA is activated,
the expression levels of Lamp2a and Hsc70 are significantly
increased (14–16). We used the ratios of these proteins
to β-actin on the western blots to assess their levels and observed
no significant difference in the Lamp2a/β-actin ratio between the
cells transfected with Htt-20Q and those transfected with Htt-120Q
at a dose of 0.75 and 1.0 μg (Fig.
2A). In addition, there was no significant difference in the
Hsc70/β-actin ratio between the cells transfected with Htt-20Q and
those transfected with Htt-120Q (Fig.
2B). We also performed immunofluorescent double staining of the
transfected cells and did not identify significant differences in
the staining signals for Hsc70 and Lamp2a between Htt-20Q- and
Htt-120Q-transfected cells (data not shown). These results suggest
that the expression of mutant Htt did not reduce the CMA activity
in HEK293T cells.
Discussion
HD is one of nine polyQ expansion diseases that
shares the age-dependent and selective neurodegeneration
characteristic with Alzheimer’s disease, Parkinson’s disease and
amyotrophic lateral sclerosis (19). All these diseases are characterized
by late-onset accumulation of toxic proteins that are misfolded and
form aggregates. Thus, clearance of misfolded proteins is key to
effectively treating these neurodegenerative diseases.
Autophagy is an important intracellular metabolic
pathway that includes macroautophagy, microautophagy and CMA. All
the substrates are degraded by hydrolytic enzymes. It was reported
that the activity of CMA displayed constitutive compensatory
upregulation in different cellular and transgenic mouse models of
HD (20). Further investigations
indicated that the Lamp2a and Hsc70 proteins played an important
role in clearing Htt through the CMA pathway (21,22).
Although the involvement of CMA in Htt metabolism has been
suggested by other investigators, the essential effect of mutant
Htt on CMA remains to be elucidated. Thus, our study aimed to
provide evidence regarding the effect of Htt on CMA.
Using HEK293T cells transfected with wild-type and
mutant Htt, we were able to define whether mutant Htt specifically
affects CMA activity. Our results suggested that mutant Htt did not
significantly alter the activity of CMA. Consistent with a recent
finding that mutant Htt does not affect the basal levels of
autophagy in the hypothalamus of HD transgenic mice (23), it appears that the expression of
mutant Htt may not affect the basal activity of CMA in cells. Thus,
enhancing CMA activity may effectively remove the toxic and
misfolded Htt and alleviate its toxicity.
Acknowledgements
This study was supported by a grant from the Natural
Science Foundation of Jiangxi Province (no. 20122BAB215034). We
would like to thank Professor Xiaojiang Li for providing the Htt
plasmids and Xiaoyu Wang, De Kang and Qiang Qiu for their technical
assistance.
References
|
1
|
No authors listed. A novel gene containing
a trinucleotide repeat that is expanded and unstable on
Huntington’s disease chromosomes. The Huntington’s Disease
Collaborative Research Group. Cell. 72:971–983. 1993.
|
|
2
|
Gusella JF, MacDonald ME, Ambrose CM and
Duyao MP: Molecular genetics of Huntington’s disease. Arch Neurol.
50:1157–1163. 1993.
|
|
3
|
Vonsattel JP and DiFiglia M: Huntington
disease. J Neuropathol Exp Neurol. 57:369–384. 1998. View Article : Google Scholar
|
|
4
|
Kim YJ, Yi Y, Sapp E, et al: Caspase
3-cleaved N-terminal fragments of wild-type and mutant huntingtin
are present in normal and Huntington’s disease brains, associate
with membranes, and undergo calpain-dependent proteolysis. Proc
Natl Acad Sci USA. 98:12784–12789. 2001.PubMed/NCBI
|
|
5
|
Ho LW, Carmichael J, Swartz J, Wyttenbach
A, Rankin J and Rubinsztein DC: The molecular biology of
Huntington’s disease. Psychol Med. 31:3–14. 2001.
|
|
6
|
Gauthier LR, Charrin BC, Borrell-Pages M,
et al: Huntingtin controls neurotrophic support and survival of
neurons by enhancing BDNF vesicular transport along microtubules.
Cell. 118:127–138. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Morton AJ and Edwardson JM: Progressive
depletion of complexin II in a transgenic mouse model of
Huntington’s disease. J Neurochem. 76:166–172. 2001.PubMed/NCBI
|
|
8
|
Berger Z, Ravikumar B, Menzies FM, et al:
Rapamycin alleviates toxicity of different aggregate-prone
proteins. Hum Mol Genet. 15:433–442. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Floto RA, Sarkar S, Perlstein EO, Kampmann
B, Schreiber SL and Rubinsztein DC: Small molecule enhancers of
rapamycin-induced TOR inhibition promote autophagy, reduce toxicity
in Huntington’s disease models and enhance killing of mycobacteria
by macrophages. Autophagy. 3:620–622. 2007.PubMed/NCBI
|
|
10
|
Sarkar S, Davies JE, Huang Z, Tunnacliffe
A and Rubinsztein DC: Trehalose, a novel mTOR-independent autophagy
enhancer, accelerates the clearance of mutant huntingtin and
alpha-synuclein. J Biol Chem. 282:5641–5652. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sarkar S, Perlstein EO, Imarisio S, et al:
Small molecules enhance autophagy and reduce toxicity in
Huntington’s disease models. Nat Chem Biol. 3:331–338.
2007.PubMed/NCBI
|
|
12
|
Li X, Wang CE, Huang S, Xu X, Li XJ, Li H
and Li S: Inhibiting the ubiquitin-proteasome system leads to
preferential accumulation of toxic N-terminal mutant huntingtin
fragments. Hum Mol Genet. 19:2445–2455. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li W, Yang Q and Mao Z: Chaperone-mediated
autophagy: machinery, regulation and biological consequences. Cell
Mol Life Sci. 68:749–763. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cuervo AM, Knecht E, Terlecky SR and Dice
JF: Activation of a selective pathway of lysosomal proteolysis in
rat liver by prolonged starvation. Am J Physiol. 269:C1200–C1208.
1995.PubMed/NCBI
|
|
15
|
Agarraberes FA, Terlecky SR and Dice JF:
An intralysosomal hsp70 is required for a selective pathway of
lysosomal protein degradation. J Cell Biol. 137:825–834. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cuervo AM and Dice JF: Regulation of
lamp2a levels in the lysosomal membrane. Traffic. 1:570–583. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Carra S, Brunsting JF, Lambert H, Landry J
and Kampinga HH: HspB8 participates in protein quality control by a
non-chaperone-like mechanism that requires eIF2{alpha}
phosphorylation. J Biol Chem. 284:5523–5532. 2009.PubMed/NCBI
|
|
18
|
Bauer PO, Goswami A, Wong HK, et al:
Harnessing chaperone-mediated autophagy for the selective
degradation of mutant huntingtin protein. Nat Biotechnol.
28:256–263. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li X, Li H and Li XJ: Intracellular
degradation of misfolded proteins in polyglutamine
neurodegenerative diseases. Brain Res Rev. 59:245–252. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Koga H, Martinez-Vicente M, Arias E,
Kaushik S, Sulzer D and Cuervo AM: Constitutive upregulation of
chaperone-mediated autophagy in Huntington’s disease. J Neurosci.
31:18492–18505. 2011.PubMed/NCBI
|
|
21
|
Qi L, Zhang XD, Wu JC, Lin F, Wang J,
DiFiglia M and Qin ZH: The role of chaperone-mediated autophagy in
huntingtin degradation. PLoS One. 7:e468342012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Qi L and Zhang XD: Role of
chaperone-mediated autophagy in degrading Huntington’s
disease-associated huntingtin protein. Acta Biochim Biophys Sin
(Shanghai). 46:83–91. 2014.
|
|
23
|
Baldo B, Soylu R and Petersen A:
Maintenance of basal levels of autophagy in Huntington’s disease
mouse models displaying metabolic dysfunction. PLoS One.
8:e830502013.PubMed/NCBI
|