Introduction
Lung cancer is currently the most common malignancy
and is a leading cause of mortality worldwide (1–2). The
mechanism of this type of carcinogenesis remains to be fully
elucidated. Tobacco smoking has been suspected to be the most
significant cause of lung cancer. However, only 1 in 10 smokers
develop lung cancer, demonstrating that it is likely that genetic
susceptibility also plays a significant role (3). Therefore, the detection of genetic
polymorphisms should be taken into consideration to explain
individual differences in lung cancer susceptibility. Non-small
cell lung cancer (NSCLC) represents ~80% of primary lung cancer
cases and approximately two-thirds of these patients are diagnosed
at an advanced stage (4). Several
effective chemotherapeutic agents are available for treatment and
platinum-based chemotherapy, including cisplatin and carboplatin,
is the standard initial treatment regimen for NSCLC (5). Evidence from NSCLC trials involving
unselected patients has shown that the efficacy of the regimen used
is reported to be only 30–40% (6).
Although the disease stage at diagnosis is the major prognostic
predictor, there are variations in survival rates among patients
who begin treatment at a similar disease status and undergo similar
treatment regimens. Findings of a previous study have indicated
that genetic factors may also affect the effectiveness of treatment
(7).
Pharmacogenetics plays a significant role in current
cancer chemotherapy and it has been reported that the prognosis can
be partly influenced by genes (8).
These antineoplastic agents contribute to the inhibition of DNA
replication and transcription due to the formation of adducts and
covalent cross-links between DNA-double strands that lead to DNA
damage. These adduct and cross-links can be repaired by complex
molecules in the nucleotide excision repair (NER) pathway,
including ERCC1, XPD, XPF and XPG
gene-encoded proteins. Therefore, NER gene polymorphisms may be
able to predict the outcome and prognosis in individual patients
with NSCLC undergoing platinum chemotherapy.
Mutations are early events in carcinogenesis and for
various types of cancer the defect of DNA repair is a risk factor
(9). The maintenance of genome
integrity is extremely significant for the survival of all
organisms, but DNA is regularly damaged by various types of
endogenous and exogenous mutagens. The DNA repair gene system is
crucial in protecting against gene mutation caused by
carcinogenesis. The deficiency in the capacity of DNA repair may
result in birth defects, cancer and a reduced lifespan (10). DNA repair consists of at least four
types, including damaged base excision repair, DNA-NER, mismatch
repair and double-strand break repair (11). Among these, NER is a highly
adaptable and advanced DNA damage removal pathway that impedes the
deleterious effects of a multitude of DNA lesions, including major
types of environmental-induced damage. In eukaryotic cells, the
process requires >30 proteins to perform at various steps
(12).
In recent years, an increasing body of
epidemiological studies (1,3,5) have
demonstrated that the potential role of polymorphisms of the genes
in the NER pathway may be associated with the clinical prognosis of
patients with NSCLC receiving platinum-based chemotherapies in
China. However, the results were shown to be inconclusive. In order
to investigate the effect of these genetic factors, including
ERCC1, XPD/ERCC2, XPA and XPG, on the
prognosis of platinum-based chemotherapy, a meta-analysis was
performed, to the best of our knowledge, for the first time with
regards to the key genes of DNA repair and metabolism in the
Chinese population.
Materials and methods
Literature search strategy and selection
criteria
A comprehensive literature search was performed
using the PubMed, Embase, Chinese National Knowledge Infrastructure
(CNKI) (http://www.cnki.net/) and Wanfang
databases (http://www.wanfangdata.com.cn/) to identify the
studies investigating the associations between the NER gene
variants and NSCLC risk in the Chinese population that were
published prior to October 1, 2013. The following terms were used
in the search: Lung cancer, non-small cell lung cancer or NSCLC; in
combination with polymorphisms, variants or mutation; ERCC1,
XPD/ERCC2, XPA or XPG; platinum or cisplatin
and carboplatin; and in combination with China or Chinese. The
searches were limited to human studies and the Chinese population.
The gender and average ages of patients in each original study were
not taken into consideration. All the references of the review and
original articles on this topic were also checked. When multiple
publications reported the same or overlapping data, only the most
updated study with the largest sample size was selected.
The studies included in the meta-analysis had to
meet all the following inclusion criteria: i) Cancer should be
confirmed as NSCLC; ii) treatment regimens were platinum-based
chemotherapies; and iii) the original data were presented with the
calculation of risk ratios (RRs) with corresponding 95% confidence
intervals (CIs) or other available data for estimating RR (95%
CI).
Exclusion criteria were: Studies without genotype or
allele data, case reports, studies pertaining to small cell lung
cancer, studies containing overlapping data, non-human studies,
interim analyses, comparisons of laboratory methods, editorials and
review articles (including meta-analyses). Any missing information
was obtained by contacting the corresponding authors in all cases
and the studies were not considered if critical missing information
could not be obtained following repeated requests. The method for
how the polymorphism was detected was not limited and the
evaluation criteria for the tumor response were accepted for all
subjects [the World Health Organization (WHO) criteria or the
Response Evaluation Criteria in Solid Tumors (RECIST)]. In terms of
the definition of the tumor response, two different standards were
allowed and they were based on the aforementioned evaluation. If
certain studies did not obtain the crucial information of drug
response and/or state of survival rate, the corresponding authors
were contacted to request the relevant data.
Data extraction
Data were extracted and entered into a database. Two
investigators (DH and YZ) searched the initially relevant
literature with keywords in the titles or abstracts and eligible
studies were determined. When extracting data from each eligible
study independently to ensure the accuracy of data, any discordance
with regard to results was resolved when agreement was reached by
the two investigators. All the studies were evaluated by titles and
abstracts initially, prior to further evaluation for particular
studies.
Data were collected with regards to the genotypes of
ERCC1 C118T/C8092A, ERCC2/XPD A751C, XPA G23A
and XPG C46T, and the following information was extracted
from each of the eligible studies: First author, year of
publication, sample size of genotyped cases, gender (male/female),
median (or mean) age and (range) year, smoking/no-smoking,
genotyping methods, chemotherapy regimens, evaluation criteria,
histology, clinical stage and genotype studied.
Statistical analysis
The RR for the tumor response [complete response
(CR) + partial response (PR) vs. stable disease (SD) + progressive
disease (PD) or CR + PR + SD vs. PD] by adopting the WHO or the
RECIST criteria (13) was estimated
subsequent to accepting the aforementioned chemotherapy
treatment.
Two models of meta-analysis, the random-effects
[DerSimonian and Laird (14)] and
the fixed-effects models (Mantel-Haenszel), were performed to
calculate the pooled RRs in the present study. For each comparison,
statistical heterogeneity among the studies was evaluated by
calculating the χ2-based Q statistical test (Cochran’s Q
statistics), when P<0.1 heterogeneity existed (15). I2 statistics were
calculated to assess the degree of between-study inconsistency due
to heterogeneity rather than by chance when I2>50%
indicated the statistical significance (16). When heterogeneity existed, the
random-effects model was chosen to evaluate the overall or pooled
estimate of risk (RRs). The fixed-effects model was chosen when
heterogeneity detected between studies had no significance.
An evaluation of potential publication bias was
performed by visual inspection with the funnel plots and
statistical evaluation with Begg and Egger’s unweighted regression
tests (17–18). Possible publication bias was
indicated by an asymmetric plot. Stata version 9.0 (StataCorp,
College Station, TX, USA) was implemented for statistical analyses.
All the P-values were two-tailed and P<0.05 was considered to
indicate a statistically significant difference.
Results
Identification and characteristics of
included studies
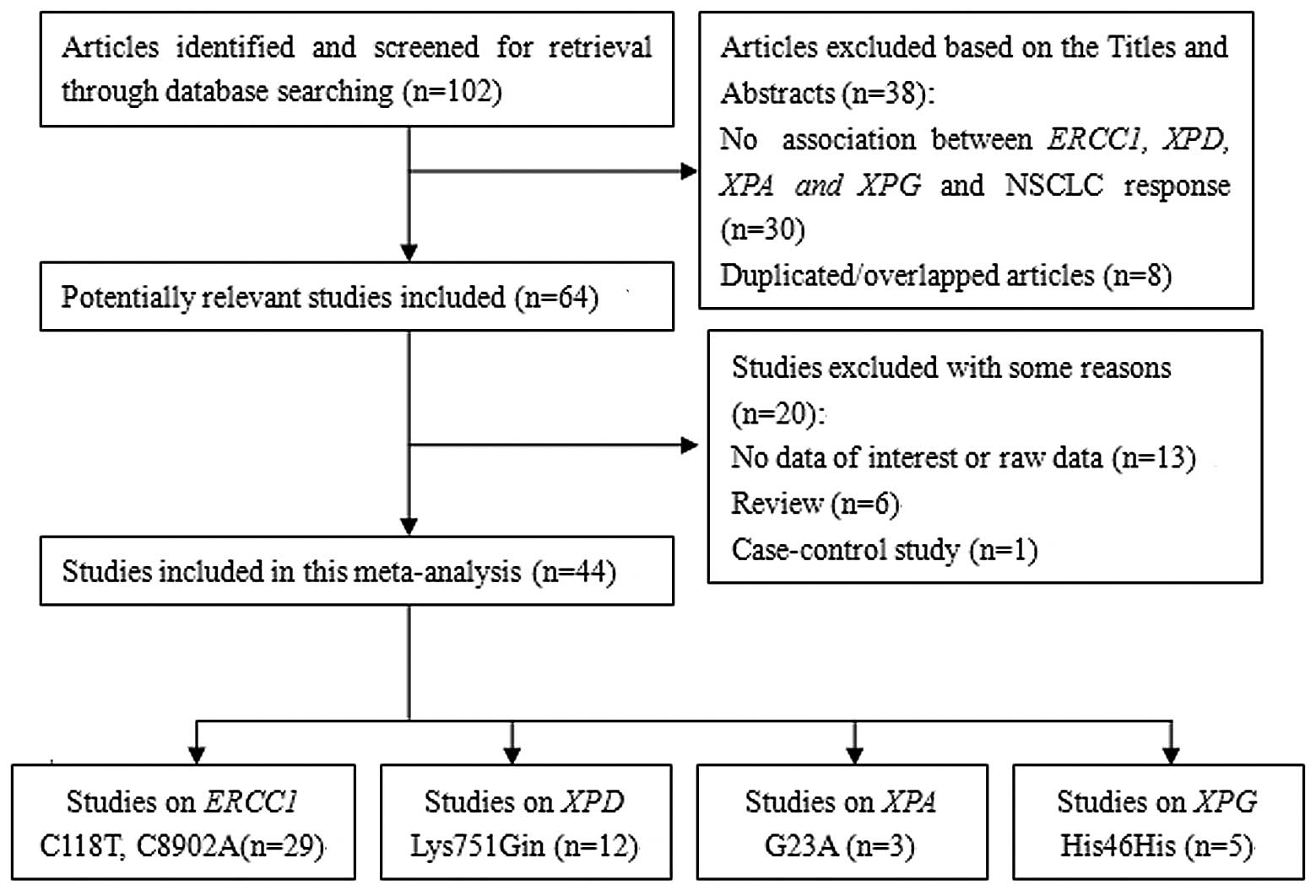
There were 102 relevant publications identified
through the literature search and selection was based on the
inclusion criteria following the initial screening. Among these, 64
potentially relevant studies were identified subsequent to
carefully reading titles and abstracts. A total of 20 studies were
excluded for the following reasons: One was a case-control study,
six were reviews, 13 had no data of interest or raw data and eight
studies were excluded due to duplicated/overlapping studies. No
additional studies were identified from the references cited in the
published studies that were searched for manually. Twenty-nine
studies concerning the ERCC1 C118T/C8092A polymorphism
(19–47), eight with the C8092A
polymorphism (19,22,24,31,39,40,48–49),
12 with the XPD A751C polymorphism (26,27,39,47,49–56),
three with XPA G23A (57–59)
and five studies with XPG His46His (58–62).
The specific process of the inclusion and exclusion of eligible
studies is shown in Fig. 1. The
main characteristics of the studies identified are shown in
Table I.
 | Table ICharacteristics of the studies on
platinum-based chemotherapy for ERCC1, XPD,
XPA and XPG polymorphisms about NSCLC identified in
the meta-analysis. |
Table I
Characteristics of the studies on
platinum-based chemotherapy for ERCC1, XPD,
XPA and XPG polymorphisms about NSCLC identified in
the meta-analysis.
| First author (year)
(ref.) | Sample size | Male/female | Median (or mean)
age and (range) year |
Smoking/no-smoking | Genotyping
methods | Chemotherapy | Evaluation
criteriaa | Histology | Clinical stage | Genotype
studied |
|---|
| Ren et al
(2010) (19) | 117 | 88/29 | 61 (21–81) | NA | PCR-RFLP | DDP + NVB, DDP +
TXT, DDP + GEM | WHO | SCC, 28; ACA, 80;
other, 9 | III: 84, IV:
33 |
ERCClC8092A/C118T |
| Han et al
(2011) (20) | 91 | 55/36 | 56 (NA) | NA | Direct
sequencing | DDP +TAX, DDP +
GEM | RECIST | SCC, 43; ACA,
48 | NA | ERCCl C118T |
| Chen and Wang
(2011) (21) | 54 | 38/16 | 56 (30–73) | NA | PCR-RFLP | DDP +TAX, DDP +
DOC | RECIST | SCC, 34; ACA, 16;
other, 4 | III: 20, IV:
34 | ERCCl C118T |
| Zhang and Li (2009)
(22) | 68 | NA | NA | NA | Direct
sequencing | GP or TP | RECIST | NA | NA |
ERCClC8092A/C118T |
| Cheng et al
(2012) (23) | 142 | 89/53 | 62 (43–81) | NA | Direct
sequencing | DDP + NVB, DDP +
TAX | RECIST | SCC, 82; ACA,
60 | III: 96, IV:
46 | ERCCl C118T |
| Wang et al
(2012) (24) | 130 | 90/40 | 62 (28–83) | NA | PCR-RFLP | DDP + NVB, DDP +
TAX, DDP + GEM | RECIST | SCC, 46; ACA, 76;
other, 8 | III: 58, IV:
72 |
ERCClC8092A/C118T |
| Lu et al
(2013) (25) | 100 | 54/46 | 61 (41–82) | NA | PCR-RFLP | DDP + NVB, DDP +
TAX | RECIST | SCC, 60; ACA,
40 | III: 44, IV:
56 | ERCCl C118T |
| Zhang et al
(2013) (26) | 78 | 52/26 | 58 (29–81) | 48/30 | PCR-RFLP | DDP + NVB, DDP +
TAX, DDP + GEM | RECIST | SCC, 33; ACA, 42;
other, 3 | III: 49, IV:
29 | ERCCl C118T,
XPDLys751Gin |
| Li et al
(2012) (27) | 89 | 64/25 | 59 (21–84) | NA | PCR-RFLP | DDP + NVB, DDP +
TAX, DDP + GEM, DDP + DOC | RECIST | SCC, 30; ACA, 43;
other, 16 | III: 18, IV:
59 | ERCCl C118T,
XPDLys751Gin |
| Wang et al
(2011) (28) | 142 | 89/53 | 62 (43–81) | 79/63 | PCR-RFLP | DDP + NVB, DDP +
TAX | RECIST | SCC, 82; ACA,
60 | III: 96, IV:
46 | ERCCl C118T |
| Yang and Hang
(2011) (29) | 62 | 44/16 | 59 (35–77) | NA | PCR-LDR | DDP + GEM | RECIST | SCC, 23; ACA,
37 | III: 26, IV:
34 | ERCCl C118T |
| Xu et al
(2012) (30) | 149 | 99/50 | 62 (28–83) | NA | PCR-LDR | DDP + NVB, DDP +
TAX, DDP + GEM, DDP + DOC | RECIST | SCC, 52; ACA, 83;
other, 14 | III: 57, IV:
92 | ERCCl C118T |
| Wang et al
(2010) (31) | 90 | 63/27 | 55 (33–73) | NA | Direct
sequencing | DDP + NVB, DDP +
TAX, DDP + GEM, DDP + DOC | RECIST | SCC, 20; ACA, 69;
other, 1 | III: 30, IV:
60 |
ERCClC8092A/C118T |
| Chen and Xu (2009)
(32) | 95 | 76/19 | 58 (35–77) | 67/28 | PCR-LDR | DDP + NVB, DDP +
TAX, DDP + GEM | RECIST | SCC, 51; ACA, 39;
other, 5 | III: 40, IV:
55 | ERCCl C118T |
| Ren et al
(2009) (33) | 130 | 74/56 | 61 (30–78) | NA | Direct
sequencing | DDP + NVB, DDP +
TAX, DDP + GEM | RECIST | SCC, 49; ACA,
81 | III: 40, IV:
90 | ERCCl C118T |
| Hua et al
(2011) (34) | 64 | 42/22 | 58 (31–78) | NA | Direct
sequencing | NA | RECIST | SCC, 22; ACA, 40;
other, 2 | III: 18, IV:
46 | ERCCl C118T |
| Zhou et al
(2010) (35) | 130 | 74/56 | 61 (30–78) | 73/57 | TaqMan | DDP/CBP + NVB,
DDP/CBP + TAX, DDP/CBP + GEM | RECIST | SCC, 49; ACA,
81 | III: 40, IV:
90 | ERCCl C118T |
| Zhong et al
(2008) (36) | 210 | 158/51 | 57 (25–81) | NA | MALDI-TOF | DDP + NVB, DDP +
TAX, DDP + GEM, DDP + DOC, DDP + GEM | RECIST | SCC, 38; ACA, 121;
other, 40 | III: 80, IV:
129 | ERCCl C118T |
| Li et al
(2010) (37) | 115 | 78/37 | 60 (NA) | 68/47 | PCR-RFLP | DDP + NVB, DDP +
TAX, DDP + GEM, DDP + DOC | WHO | SCC, 30; ACA, 82;
other, 3 | IIIb–IV: 115 | ERCCl C118T,
XPDLys751Gin |
| Liu et al
(2008) (38) | 94 | 71/23 | 60 (32–79) | NA | PCR-RFLP | DDP + TAX | WHO | SCC, 33; ACA, 42;
other, 19 | I–II: 12, III–IV:
82 | ERCCl C118T |
| Liao et al
(2012) (39) | 62 | 35/27 | 57 (36–78) | NA | TaqMan | DDP + GEM | RECIST | SCC, 4; ACA, 52;
other, 6 | III: 10, IV:
52 | ERCCl C118T/C8092A,
XPDLys751Gin |
| Hong et al
(2013) (40) | 135 | 90/45 | 56 (25–72) | 77/58 | TaqMan | DDP + GEM | RECIST | SCC, 39; ACA, 80;
other, 16 | III: 12, IV:
123 |
ERCClC8092A/C118T |
| Gao et al
(2010) (41) | 57 | 44/13 | 59 (38–77) | NA | PCR-RFLP | DDP + GEM | WHO | SCC, 12; ACA, 40;
other, 5 | II: 1, III: 22, IV:
34 | ERCCl C118T |
| Jin et al
(2010) (42) | 73 | 59/14 | 59 (24–82) | NA | PCR-RFLP | DDP + NVB, DDP +
TAX, DDP + GEM, DDP + DOC | WHO | SCC, 42; ACA, 28;
other, 3 | III: 30, IV:
43 | ERCCl C118T |
| Su and Cao (2012)
(43) | 161 | 108/53 | NA | 88/73 | TaqMan | DDP/CBP + NVB,
DDP/CBP + DOC/TAX, DDP + GEM, DDP + PEM | WHO | SCC, 52; ACA, 82;
other, 27 | III: 55, IV:
106 | ERCCl C118T |
| Wang et al
(2012) (44) | 101 | 60/41 | 53 (36–76) | 54/47 | PCR-RFLP | DDP + NVB, DDP +
TAX, DDP + GEM | RECIST | SCC, 31; ACA, 68;
other, 2 | III: 17, IV:
84 | ERCCl C118T |
| Li et al
(2010) (45) | 60 | 41/19 | NA | 37/23 | TDI-FP | DDP + NVB, DDP +
TAX, DDP + GEM | RECIST | SCC, 19; ACA, 37;
other, 4 | III: 35, IV:
25 | ERCCl C118T |
| Zhou et al
(2013) (46) | 204 | 120/84 | 61 (45–75) | NA | MALDI- TOF-MS | DDP | RECIST | SCC, 89; ACA,
115 | NA | ERCCl C118T |
| Sun et al
(2009) (47) | 113 | 76/37 | 60 (34–84) | NA | PCR-gene chip | DDP/CBP + GEM,
DDP/CBP + NVB, DDP/CBP + TAX/TXT/DOC | WHO | SCC, 30; ACA, 80;
other, 3 | NA | ERCC1 C118T,
XPDLys751Gin |
| KimCurran et
al (2011) (48) | 300 | 201/99 | 60 (33–78) | 157/143 | PCR-RFLP | DDP + NVB, DDP +
TAX, DDP + GEM | RECIST | SCC, 88; ACA, 168;
other, 44 | NA | ERCC1 C8092A |
| Yuan et al
(2006) (49) | 200 | 130/70 | 56 (30–74) | NA | PCR-RFLP | DDP + NVB, CBP +
NVB, CBP + TAX, DDP + TAX/TXT | WHO | SCC, 37; ACA, 143;
other, 20 | III: 49, IV:
151 | XPDLys751Gin,
ERCC1C8092A |
| Chen et al
(2012) (50) | 355 | 248/107 | 60 (32–78) | 202/153 | TaqMan | DDP/CBP + NVB,
DDP/CBP + GEM, DDP/CBP + TAX/TXT | RECIST | SCC, 99; ACA, 201;
other, 55 | III: 118, IV:
237 | XPDLys751Gin |
| Yao et al
(2009) (51) | 108 | 71/37 | 61 (39–79) | NA | PCR-RFLP | DDP + NVB, DDP +
TAX, DDP + GEM, DDP + TXT | WHO | SCC, 28; ACA, 77;
other, 3 | III: 37, IV:
71 | XPDLys751Gin |
| Wu et al
(2012) (52) | 353 | 246/107 | 57 (32–80) | 199/154 | Direct
sequencing | DDP + NVB, DDP +
TAX, DDP + GEM, DDP + TXT | NA | SCC, 75; ACA, 213;
other, 67 | III: 141, IV:
212 | XPDLys751Gin |
| Zhang et al
(2013) (53) | 62 | 38/24 | 58 (37–72) | NA | Direct
sequencing | DDP + NVB, DDP +
TAX, DDP + GEM, DDP + MTA | RECIST | SCC, 13; ACA, 39;
other, 10 | NA | XPDLys751Gin |
| Fan et al
(2008) (54) | 81 | 63/18 | 63 (55–80) | NA | PCR-RFLP | DDP + NVB, DDP +
TAX/TXT, DDP + GEM | WHO | SCC, 21; ACA, 44;
other, 16 | III: 34, IV:
47 | XPDLys751Gin |
| Chen et al
(2011) (55) | 87 | 54/33 | 60 (43–81) | NA | PCR-RFLP | DDP + NVB, DDP +
TAX, DDP + GEM, DDP + MTA | WHO | SCC, 52; ACA,
35 | III: 58, IV:
29 | XPDLys751Gin |
| Ren et al
(2012) (56) | 340 | 232/108 | 60 (30–78) | 184/156 | TaqMan | DDP + NVB, DDP +
GEM, DDP + TAX/TXT | RECIST | SCC, 145; No-SCC,
195 | III: 111, IV:
229 | XPDLys751Gin |
| Sun et al
(2007) (57) | 96 | 62/34 | 58 (34–77) | NA | PCR-cDNA chip | NA | WHO | SCC, 39; ACA,
57 | NA | XPAG23A |
| Jia et al
(2011) (58) | 89 | 45/44 | NA | NA | DNA sequencing | DDP/CBP + DOC,
DDP/CBP + GEM | RECIST | SCC, 39; ACA, 44;
other, 6 | III: 22, IV:
67 | XPAG23A,
XPGHis46His |
| Feng et al
(2009) (59) | 115 | 78/37 | 60 (34–84) | NA | DNA microarray
genotyping | DDP/CBP + TAX,
DDP/CBP + GEM, DDP/CBP + NVB | WHO | SCC, 30; ACA, 82;
other, 3 | NA | XPAG23A,
XPGHis46His |
| Zhang et al
(2013) (60) | 475 | 306/145 | 64 (32–76) | NA | TaqMan | DDP + DOC, DDP/CBP
+ GEM, DDP/CBP + NVB | EORTC | SCC, 169; ACA, 256;
other, 27 | III: 159, IV:
292 | XPGHis46His |
| Lv et al
(2012) (61) | 85 | 49/36 | 56 (36–71) | NA | DNA sequencing | DDP + DOC, DDP +
GEM, DDP + NVB, DDP + MTA | RECIST | SCC, 34; ACA, 43;
other, 8 | NA | XPGHis46His |
| Sun et al
(2009) (62) | 82 | 53/29 | 59 (34–79) | NA | DNA microarray
genotyping | DDP/CBP +
TAX/TAT/DOC, DDP/CBP + GEM, DDP/CBP + NVB | WHO | SCC, 16; ACA, 61;
other, 5 | IV: 82 | XPGHis46His |
Overall analysis of data
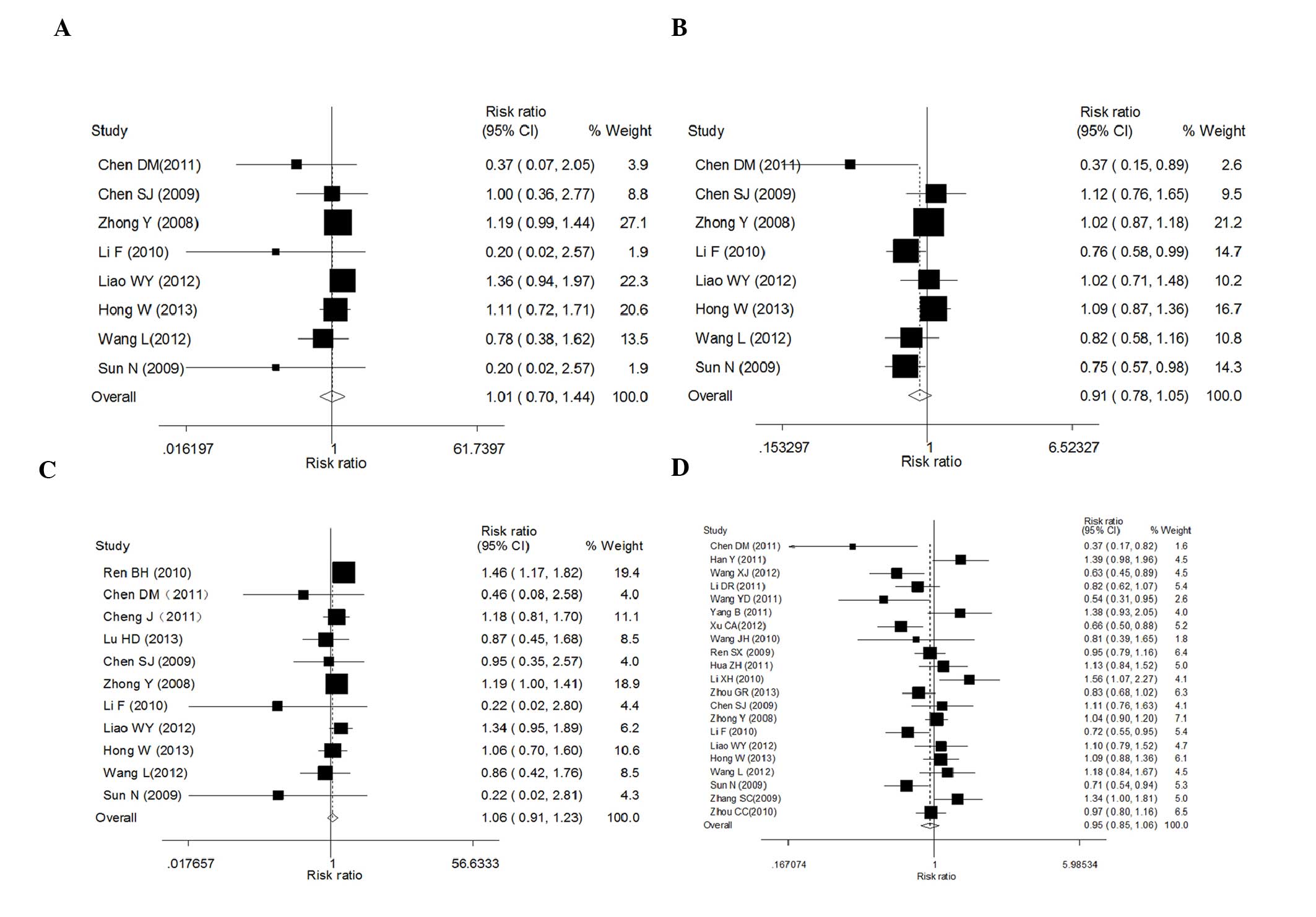
Tumor response of ERCC1 C118T/C8092A
polymorphisms (non-response: SD or PD)
A total of 24 studies were eventually included with
a total sample size of 2,585 patients. Data showed that there was
no significant association between ERCC1 C118T polymorphisms
and tumor response in the present study analyses under the four
comparison models [TT vs. CC: RR (95% CI), 1.01 (0.7–1.45),
P=0.976; CT vs. CC: RR (95% CI), 0.91 (0.78–1.05), P=0.199; CT/TT
vs. CC: RR (95% CI), 0.95 (0.85–1.06), P=0.386; and TT vs. CC/CT:
RR (95% CI), 1.06 (0.91–1.23), P=0.463; Table II and Fig. 2]. There was significant
heterogeneity observed between studies in the initial fixed model
in three genetic contrasts for which a random model was required
(TT vs. CC: I2=61%; CT vs. CC: I2=51.1%; and
CT/TT vs. CC: I2=66.9%; Table II), except for the recessive model
(TT vs. CC/CT: I2=39.8%; Table II). In addition, publication bias
was detected in Egger’s test (TT vs. CC: P=0.028; and TT vs. CC/CT:
P=0.029), but not in the other models (CT vs. CC: P=0.251; and
CT/TT vs. CC: P=0.973) (data not shown).
 | Table IISummary of the risk ratios (RRs) and
95% confidence intervals (CIs) of the non-small cell lung cancer
risk for contrasts (non-response, stable disease or progressive
disease). |
Table II
Summary of the risk ratios (RRs) and
95% confidence intervals (CIs) of the non-small cell lung cancer
risk for contrasts (non-response, stable disease or progressive
disease).
| Genetic model | No. of studies | Pooled RR (95%
CI) | P-value | Heterogeneity | Begg’s test
(P-value) | Egger’s test
(P-value) |
|---|
|
|---|
| P-value | I2
(%) |
|---|
| ERCC1 C118T |
| Homozygote
comparison (TT vs. CC) | 8 | 1.01
(0.70–1.45) | 0.976 | 0.0112 | 61.00 | 0.174 | 0.028 |
| Heterozygote
comparison (CT vs. CC) | 8 | 0.91
(0.78–1.05) | 0.199 | 0.046 | 51.10 | 0.711 | 0.251 |
| Dominant (CT/TT
vs. CC) | 21 | 0.95
(0.85–1.06) | 0.386 | 0.0 | 66.90 | 0.786 | 0.973 |
| Recessive (TT vs.
CC/CT) | 11 | 1.06
(0.91–1.23) | 0.463 | 0.083 | 39.80 | 0.213 | 0.029 |
| ERCC1 C8092A |
| Homozygote
comparison (AA vs. CC) | 3 | 1.09
(0.88–1.35) | 0.412 | 0.899 | 0.0 | 1.000 | 0.53 |
| Heterozygote
comparison (CA vs. CC) | 3 | 0.94
(0.81–1.1) | 0.431 | 0.447 | 0.0 | 0.296 | 0.106 |
| Dominant (AA/CA
vs. CC) | 8 | 1.11
(0.91–1.35) | 0.328 | 0.754 | 0.0 | 0.296 | 0.044 |
| Recessive (AA vs.
CC/CA) | 3 | 0.96
(0.88–1.05) | 0.377 | 0.959 | 0.0 | 0.266 | 0.397 |
| XPD Lys751Gln |
| Homozygote
comparison (GlnGln vs. LysLys) | 3 | 1.21
(0.92–1.59) | 0.183 | 0.9 | 0.00 | 1.000 | 0.065 |
| Heterozygote
comparison (LysGln vs. LysLys) | 7 | 1.08
(0.96–1.22) | 0.182 | 0.175 | 33.10 | 0.548 | 0.828 |
| Dominant
(GlnGln/LysGln vs. LysLys) | 12 | 1.02
(0.92–1.13) | 0.714 | 0.052 | 43.70 | 0.837 | 0.665 |
| Recessive (GlnGln
vs. LysLys/LysGln) | 3 | 1.16
(0.89–1.52) | 0.272 | 0.925 | 0.0 | 1.000 | 0.022 |
Eight studies were included with a total sample size
of 1,102 patients. Data showed that the ERCC1 C8092A
polymorphism was not associated with tumor response in this
analysis [AA vs. CC: RR (95% CI), 1.09 (0.88–1.35), P=0.412; CA vs.
CC: RR (95% CI), 0.94 (0.81–1.1), P=0.431; AA/CA vs. CC: RR (95%
CI), 1.11 (0.91–1.35), P=0.328; and AA vs. CC/CA: RR (95% CI), 0.96
(0.88–1.05), P=0.377; Table II].
No significant between-study heterogeneity was observed in the
initial fixed model and therefore a random model was not performed,
resulting in P-values of 0. In addition, no publication bias was
detected in Egger’s test (AA vs. CC: P=0.53; CA vs. CC: P=0.106;
and AA vs. CC/CA: P=0.397), but not in the dominant model (AA/CA
vs. CC: P=0.044) (data not shown).
Tumor response of ERCC1 C118T
polymorphisms (non-response: PD)
Seven studies were included with a total sample size
of 729 patients. Data showed that ERCC1 C118T polymorphisms
were not associated with the tumor response in this analysis under
the four comparison models (TT vs. CC: RR (95% CI), 0.65
(0.3–1.41), P=0.276; CT vs. CC: RR (95% CI), 1 (0.5–2), P=0.995;
CT/TT vs. CC: RR (95% CI), 0.83 (0.64–1.07), P=0.149; and TT vs.
CC/CT: RR (95% CI), 0.67 (0.31–1.44), P=0.304; Table III). Significant between-study
heterogeneity was observed in the initial fixed model and then a
random model was performed (I2=75%) under the
heterozygote comparison model. In addition, no publication bias was
detected with a P>0.05 in Egger’s test, and no significant
outcome of influence analysis was observed (data not shown).
 | Table IIISummary of the risk ratios (RRs) and
95% confidence intervals (CIs) of the non-small cell lung cancer
risk for contrasts (non-response, progressive disease). |
Table III
Summary of the risk ratios (RRs) and
95% confidence intervals (CIs) of the non-small cell lung cancer
risk for contrasts (non-response, progressive disease).
| Genetic model | No. of studies | Pooled RR (95%
CI) | P-value | Heterogeneity | Begg’s test
(P-value) | Egger’s test
(P-value) |
|---|
|
|---|
| P-value | I2
(%) |
|---|
| ERCC1 C118T |
| Homozygote
comparison (TT vs. CC) | 3 | 0.65
(0.30–1.41) | 0.276 | 0.234 | 31.20 | 1.000 | 0.687 |
| Heterozygote
comparison (CT vs. CC) | 3 | 1.00
(0.50–2.00) | 0.995 | 0.018 | 75.00 | 0.296 | 0.245 |
| Dominant (CT/TT
vs. CC) | 7 | 0.83
(0.64–1.07) | 0.149 | 0.142 | 37.60 | 0.368 | 0.283 |
| Recessive (TT vs.
CC/CT) | 3 | 0.67
(0.31–1.44) | 0.304 | 0.545 | 0.00 | 1.000 | 0.803 |
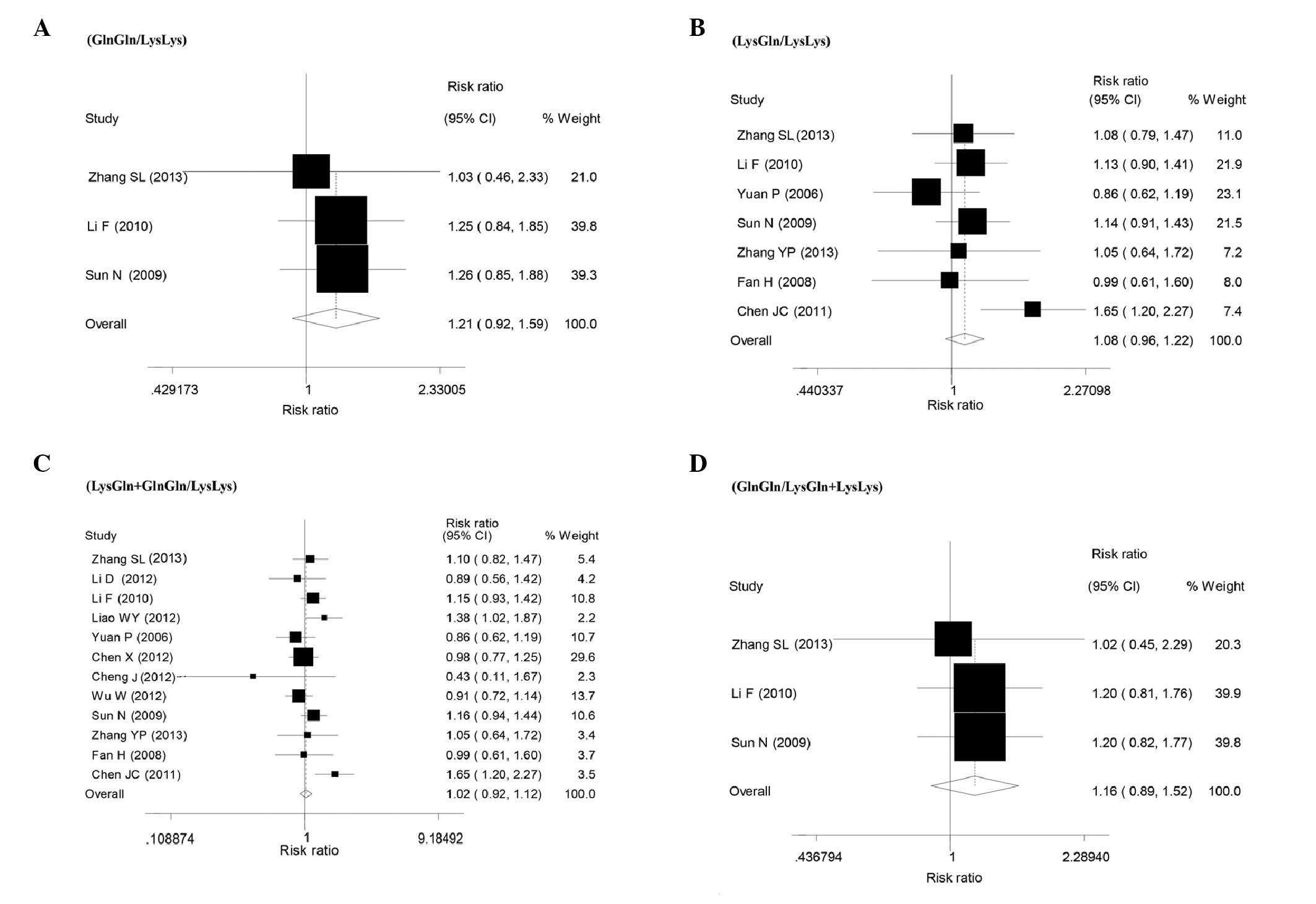
Tumor response of XPD A751C
polymorphisms (non-response: SD or PD)
Twelve studies were included with a total sample
size of 2,043 patients. Overall, the meta-analysis showed that
there was no statistically significant association between the XPD
A751C polymorphism and tumor response for all the genetic models
(GlnGln vs. LysLys: RR=1.21, 95% CI: 0.92–1.59, P=0.183,
I2=0.0% for heterogeneity test; LysGln vs. LysLys:
RR=1.08, 95% CI: 0.96–1.22 P=0.182, I2=33.1% for
heterogeneity test; the recessive model, GlnGln vs. LysLys/LysGln:
RR=1.16, 95% CI: 0.89–1.52, P=0.272, I2=0% for
heterogeneity test; and the dominant model, GlnGln/LysGln vs.
LysLy: RR=1.02, 95% CI: 0.92–1.13, P=0.714, I2=43.7% for
heterogeneity test; Table II and
Fig. 3). In addition, no
publication bias was detected in Egger’s test with P>0.05
(Table II). The shape of the
funnel plot did not reveal any evidence of clear asymmetry (data
not shown).
Tumor response of XPA A23G and XPG
C46T polymorphisms (non-response: SD or PD)
Three studies were included with a total sample size
of 300 patients. Overall, this meta-analysis showed that there was
no statistically significant association between the XPA
A23G polymorphism and tumor response for the recessive model
(GG/AG vs. AA: RR=0.99, 95% CI: 0.65–1.52, P=0.969,
I2=80.8% for heterogeneity test; Table IV). The other three models were not
performed as there was no data of interest or raw data.
 | Table IVSummary of the risk ratios (RRs) and
95% confidence intervals (CIs) of the non-small cell lung cancer
risk for contrasts (non-response, stable disease or progressive
disease). |
Table IV
Summary of the risk ratios (RRs) and
95% confidence intervals (CIs) of the non-small cell lung cancer
risk for contrasts (non-response, stable disease or progressive
disease).
| Genetic model | No. of studies | Pooled RR (95%
CI) | P-value | Heterogeneity | Begg’s test
(P-value) | Egger’s test
(P-value) |
|---|
|
|---|
| P-value | I2
(%) |
|---|
| XPA A23G |
| Recessive (GG/AG
vs. AA) | 3 | 0.99
(0.65–1.52) | 0.969 | 0.001 | 80.80 | 0.734 | 0.694 |
| XPG C46T |
| Homozygote
comparison (TT vs. CC) | 3 | 1.31
(1.14–1.5) | 0.000 | 0.915 | 0.00 | 0.296 | 0.136 |
| Heterozygote
comparison (CT vs. CC) | 3 | 1.10
(0.97–1.25) | 0.136 | 0.651 | 0.00 | 1.000 | 0.791 |
| Dominant (TT/CT
vs. CC) | 5 | 1.23
(1.11–1.36) | 0.000 | 0.480 | 0.00 | 0.806 | 0.143 |
| Recessive (TT vs.
CC/CT) | 3 | 1.22
(1.11–1.36) | 0.000 | 0.564 | 0.00 | 1.000 | 0.907 |
For the XPG C46T polymorphism, five studies
were included with a total sample size of 846 patients. Overall,
the meta-analysis showed that there was an increase in the
statistically significant association between the XPG C46T
polymorphism and tumor response for the three genetic models (TT
vs. CC: RR=1.31, 95% CI: 1.14–1.5, P=0.00, I2=0% for
heterogeneity test; the recessive model, TT vs. CC/CT: RR=1.22, 95%
CI: 1.11–1.36, P=0.00, I2=0% for heterogeneity test; and
the dominant model, TT/CT vs. CC: RR=1.23, 95% CI: 1.11–1.36,
P=0.00, I2=0% for heterogeneity test, Table IV). The XPG C46T
polymorphism had no association with tumor response in the
heterozygote comparison (CT vs. CC: RR=1.1, 95% CI: 0.97–1.25,
P=0.136, I2=0% for heterogeneity test). In addition, no
publication bias was detected in Egger’s test with all P>0.05
(Table IV). The shape of the
funnel plot did not reveal any evidence of clear asymmetry (data
not shown).
Discussion
A total of 44 studies with 5,944 NSCLC patients were
included that examined the ERCC1 C118T/C8092A, ERCC2/XPD
A751C, XPA G23A and XPG C46T polymorphisms. To
the best of our knowledge, this is the first meta-analysis
elaborating the role of NER gene polymorphisms on the prognosis of
platinum-based chemotherapy in Chinese NSCLC patients. However, the
overall combined RRs did not support any appreciable association
between the ERCC1 C118T/C8092A, XPD A751C and XPA
G23A polymorphisms on the prognosis of platinum-based
chemotherapy under the four genetic contrast models when SD or PD
was defined as non-response, which was consistent with previous
meta-analyses (63,64). Additionally, no significant
association was obtained for the ERCC1 C118T polymorphisms
when only PD was considered as non-response. However, elevated
associations were observed for the homozygote, dominant and
recessive comparisons in the XPG C46T polymorphism,
indicating that such gene carriers were more susceptible to
platinum-based chemotherapy in Chinese patients with NSCLC.
NER is a highly versatile pathway that is primarily
responsible for repairing DNA damage by removing the majority of
DNA damage through incisions on both sides of the lesion. The NER
pathway is a significant defense mechanism in humans for protection
from two major carcinogens; sunlight and cigarette smoke (65). NER has two systems: The repair of
strand distortions of the genome by global genome repair and the
removal of distorted lesions that block elongating RNA polymerases
by transcription-coupled repair (66). There are >30 proteins involved in
the NER pathway (67). The removal
of these platinum adducts, which results in resistance to
chemotherapy, is mainly carried out through NER and due to the
deficiency of NER, cells are hypersensitive to platinum (68). For the platinum-chemotherapy
resistance of NSCLC, there is still no definite predictive
biomarker. Regarding the ERCC1 C118T/C8092A and XPD
A751C polymorphisms on the prognosis of platinum-based
chemotherapy in patients with NSCLC, several meta-analyses are
available although with inconsistent conclusions from each other
(1,63–64,69–70),
and certain evidence was found to support the use of the ERCC1
C118T/C8092A and XPD A751C polymorphisms as prognostic
predictors of platinum-based chemotherapies in NSCLC. However, its
role in Chinese NSCLC patients has not been well-established.
Therefore, a meta-analysis of published studies was performed with
the aim of clarifying the correlation between five common
polymorphisms of four NER genes and prognosis of platinum-based
chemotherapy among Chinese NSCLC patients. However, the result
showed that the tumor response rate was not significant in patients
for ERCC1 C118T/C8092A, XPD A751C and XPA G23A
polymorphisms. More interactions of genetic variations and the
gene-environment may also be attributed to the prognosis regarding
ERCC1, XPD and XPA. Due to these similar
mechanisms, the linkage disequilibrium with other genes should be
taken into account. The combination with other gene polymorphisms
may result in no significant difference. Additionally, unknown
regions may contribute to the potential mechanism of ERCC1,
XPD and XPA polymorphisms. Regarding the XPG
polymorphism, the total sample size was limited and the positive
results required further confirmation.
Potential limitations of the current study should be
acknowledged. First, heterogeneity is a noteworthy issue in a
meta-analysis and one of the most significant goals of
meta-analysis is identifying the sources of heterogeneity. The
study on the XPD A751C and XPA A23G polymorphism
observed evidence of significant heterogeneity. A large
heterogeneity between studies was always identified in certain
comparisons, which could interfere with the interpretation of the
findings of a meta-analysis. There may be additional potential
sources of heterogeneity besides the aforementioned reason,
however, owing to a lack of access to original source data, these
sources were not investigated further in subgroup analyses
according to ethnicity. Second, although evaluation of specific
potential confounding factors was attempted, including age
distribution, gender, nutrition, alcohol abuse, family history,
lifestyle, dietary habits, body mass index, smoking status and
stress, the similar environmental conditions and the definition of
each stratum varied among studies and were reported in only a
limited number of studies. A more precise analysis should be
conducted based on the adjusted estimates. Third, owing to only
published studies being included in the meta-analysis, it is
extremely possible that other unpublished studies and published
studies in languages other than English and Chinese may have been
omitted. Therefore, the possibility of a larger sample size and
increased statistical power may have been missed. Fourth, the
meta-analysis is based on unadjusted estimates, and the
availability of individual date potentially allows for more precise
analysis. Owing to a lack of interest/complementary data, the
opportunity allowing for an adjustment estimate was lost (at least
for age and smoking). Fifth, little observation on the gene-gene
and gene-environment interactions may be responsible for the
unstable results. Notably, the majority of these studies were
retrospective studies. Additionally, as a significant impact
factor, the tumor description, including classification and stages,
may also be accountable for the inconsistent results. The analysis
also failed to demonstrate the influence of the ERCC1,
XPD, XPG and XPA polymorphisms on survival and
progression-free survival rates with a lack of
interest/complementary data. Taking these potential limitations
into consideration, the results may not have enough statistical
power to explore the association of these polymorphisms with NSCLC
susceptibility.
In conclusion, no evidence was found to support the
use of the ERCC1 C118T/C8092A, XPD Lys751Gln and
XPA A23G polymorphisms as prognostic predictors of
platinum-based chemotherapies in NSCLC treatment based on the
current published data in the meta-analysis. For the XPG
C46T polymorphism, there was a significant association with an
objective response detected. However, occasional studies could not
be ruled out due to the limited number of subjects examined and
observation of between-study heterogeneity. Additional genetic
epidemiological investigations that are well designed and have
large samples for these findings are required for the association
between biomarkers and tumor prognosis.
Acknowledgements
The authors would like to thank all the participants
of the study. The present study was not supported by any
grants.
References
|
1
|
Xu TP, Shen H, Liu LX and Shu YQ:
Association of ERCC1-C118T and -C8092A polymorphisms with lung
cancer risk and survival of advanced-stage non-small cell lung
cancer patients receiving platinum-based chemotherapy: a pooled
analysis based on 39 reports. Gene. 526:265–274. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Siegel R, Ward E, et al: Cancer
statistics, 2007. CA Cancer J Clin. 57:43–66. 2007. View Article : Google Scholar
|
|
3
|
Xing D, Tan W, Wei Q and Lin D:
Polymorphisms of the DNA repair gene XPD and risk of lung cancer in
a Chinese population. Lung Cancer. 38:123–129. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jemal A, Siegel R, Ward E, et al: Cancer
statistics, 2009. CA Cancer J Clin. 59:225–249. 2009. View Article : Google Scholar
|
|
5
|
Pujol JL, Barlesi F and Daurès JP: Should
chemotherapy combinations for advanced non-small cell lung cancer
be platinum-based? A meta-analysis of phase III randomized trials.
Lung Cancer. 51:335–345. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ohe Y, Ohashi Y, Kubota K, et al:
Randomized phase III study of cisplatin plus irinotecan versus
carboplatin plus paclitaxel, cisplatin plus gemcitabine, and
cisplatin plus vinorelbine for advanced non-small-cell lung cancer:
Four-Arm Cooperative Study in Japan. Ann Oncol. 18:317–323. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rosell R, Cecere F, Santarpia M, et al:
Predicting the outcome of chemotherapy for lung cancer. Curr Opin
Pharmacol. 6:323–331. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jun L, Haiping Z and Beibei Y: Genetic
polymorphisms of GSTP1 related to response to
5-FU-oxaliplatin-based chemotherapy and clinical outcome in
advanced colorectal cancer patients. Swiss Med Wkly. 139:724–728.
2009.PubMed/NCBI
|
|
9
|
Ronen A and Glickman BW: Human DNA repair
genes. Environ Mol Mutagen. 37:241–283. 2001. View Article : Google Scholar
|
|
10
|
Wei Q and Spitz MR: The role of DNA repair
capacity in susceptibility to lung cancer: a review. Cancer
Metastasis Rev. 16:295–307. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Goode EL, Ulrich CM and Potter JD:
Polymorphisms in DNA repair genes and associations with cancer
risk. Cancer Epidemiol Biomarkers Prev. 11:1513–1530.
2002.PubMed/NCBI
|
|
12
|
van Hoffen A, Balajee AS, van Zeeland AA
and Mullenders LH: Nucleotide excision repair and its interplay
with transcription. Toxicology. 193:79–90. 2003.PubMed/NCBI
|
|
13
|
Choi JH, Ahn MJ, Rhim HC, et al:
Comparison of WHO and RECIST criteria for response in metastatic
colorectal carcinoma. Cancer Res Treat. 37:290–293. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
DerSimonian R and Laird N: Meta-analysis
in clinical trials. Control Clin Trials. 7:177–188. 1986.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cochran WG: The comparison of percentages
in matched samples. Biometrika. 37:256–266. 1950. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Higgins JP, Thompson SG, Deeks JJ and
Altman DG: Measuring inconsistency in meta-analyses. BMJ.
327:557–560. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Begg CB and Mazumdar M: Operating
characteristics of a rank correlation test for publication bias.
Biometrics. 50:1088–1101. 1994. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Egger M, Davey Smith G, Schneider M and
Minder C: Bias in meta-analysis detected by a simple, graphical
test. BMJ. 315:629–634. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ren BH, Yang X, Li M, et al: Single
nucleotide polymorphisms in DNA repair gene ERCCl predict clinical
response to platinum-based chemotherapy in non-small cell lung
cancer. Chin J Exp Surg. 27:1200–1202. 2010.
|
|
20
|
Han Y, Liang J, Lv HY, et al: ERCC1 and
XRCC1 gene polymorphisms and clinical response to platinum-based
chemotherapy in advanced non-small cell lung cancer. Chin J Prac
Intern Med. 31:638–639. 2011.
|
|
21
|
Chen DM and Wang NJ: Association of the
protein expression and gene polymorphisms in DNA repair genes
XRCC1, ERCC1, RRM1 and the effect of platinum-based regimens
chemotherapy in advanced non-small cell lung cancer patients. PhD
dissertation. Michigan State University. MM dissertation, Ningxia
Med Univ, ProQuest; 2011
|
|
22
|
Zhang SC and Li Q: Clinical significance
of expression of ERCC1, class III β-tubulin in resected patients
with non-small cell lung cancer and association between
polymorphisms of ERCC1 and response and survival in advanced
non-small cell lung cancer patients treated with cisplatin-based
chemotherapy. PhD dissertation. The Tuberculosis Thoracic Tumor
Research Institute of Beijing, ProQuest; 2009
|
|
23
|
Cheng J, Ha M, Wang Y, et al: A C118T
polymorphism of ERCC1 and response to cisplatin chemotherapy in
patients with late-stage non-small cell lung cancer. J Cancer Res
Clin Oncol. 138:231–238. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang XJ, Xu CA, Zhang Y, et al:
Relationship between ERCC1 and XPD genetic polymorphism and
sensitivity of platinum-based chemotherapy in advanced non-small
cell lung cancer. Chin Gen Pract. 15:1010–1014. 2012.
|
|
25
|
Lu HD, Cui EH and Hua F: The association
between the susceptibility to platinum drugs and the genetic
polymorphisms of ERCC l and BAG-1 in patients with advanced
non-small cell lung cancer. China Modern Doctor. 51:65–67.
2013.
|
|
26
|
Zhang SL, Zhang ZL and Shi MH: Association
between polymorphisms of ERCC1 and XPD and sensitivity to
platinum-based chemotherapy in advanced non-small cell lung cancer.
Jiangsu Med J. 39:303–305. 2013.
|
|
27
|
Li D, Zhou Q, Liu Y, et al: DNA repair
gene polymorphism associated with sensitivity of lung cancer to
therapy. Med Oncol. 29:1622–1628. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang YD, Cheng J, Chen JC, et al: The
association of genetic polymorphisms of BAG-1 and ERCCl with the
clinical response to platinum drugs in patients with advanced
non-small cell lung cancer. Tumor. 31:824–829. 2011.
|
|
29
|
Yang B and Hang FC: Polymorphisms in
ERCC1, RRM1 genes and sensitivity to gemcitabine/cisplatin
chemotherapy in advanced non-small cell lung cancer. MM
dissertation. Shanxi Med Univ, ProQuest; 2011
|
|
30
|
Xu CA, Li Q, Wang XJ, et al: Sensitivity
relationship of ERCC1 and XRCC1 genetic polymorphism on response of
platinum-based chemotherapy in advanced NSCLC. J Pract Oncol.
27:245–250. 2012.
|
|
31
|
Wang J, Zhang Q, Zhang H, et al:
Association between polymorphisms of ERCC1 and response in patients
with advanced non-small cell lung cancer receiving cisplatin-based
chemotherapy. Zhongguo Fei Ai Za Zhi. 13:337–341. 2010.(In
Chinese).
|
|
32
|
Chen SJ and Xu XJ: The relationship
between MDR1 and ERCC1 genetic polymorphisms and haplotype response
rate of platinum-based chemotherapy and side effect on non-small
cell lung cancer therapy. MM dissertation. Shantou University,
ProQuest; 2009
|
|
33
|
Ren SX, Zhou CC, Zhou SW, et al:
Predictive role of ERCC1 and XRCC3 gene polymorphism on response of
platinum-based chemotherapy in advanced NSCLC. J Oncol. 15:706–710.
2009.
|
|
34
|
Hua ZH, Fang WZ, Zhao ZQ, et al:
Association study of ERCC1 and ERCC4 genetic polymorphism with
response and survival in non-small cell lung cancer patients
treated with cisplatin-based chemotherapy. Acta Metallurgica
Sinica. 16:772–778. 2011.
|
|
35
|
Zhou C, Ren S, Zhou S, et al: Predictive
effects of ERCC1 and XRCC3 SNP on efficacy of platinum-based
chemotherapy in advanced NSCLC patients. Jpn J Clin Oncol.
40:954–960. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhong Y, Lu DR, Han BH, et al: The
association study of ERCCl polymorphism and chemotherapy
response/toxicity of lung cancer. MM dissertation. Fudan
University, ProQuest; 2008
|
|
37
|
Li F, Sun X, Sun N, et al: Association
between polymorphisms of ERCC1 and XPD and clinical response to
platinum-based chemotherapy in advanced non-small cell lung cancer.
Am J Clin Oncol. 33:489–494. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu YF, Guan XX, Chen LB, et al: Study on
ERCC1, XPD and XPA polymorphisms for prediction of platinum-based
chemotherapy sensitivity in non-small cell lung cancer. Chin J
Cancer Prev Treat. 15:1285–1288. 2008.
|
|
39
|
Liao WY, Shih JY, Chang GC, et al: Genetic
polymorphism of XRCC1 Arg399Gln is associated with survival in
non-small-cell lung cancer patients treated with
gemcitabine/platinum. J Thorac Oncol. 7:973–981. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hong W, Wang K, Zhang YP, et al:
Methylenetetrahydrofolate reductase C677T polymorphism predicts
response and time to progression to gemcitabine-based chemotherapy
for advanced non-small cell lung cancer in a Chinese Han
population. J Zhejiang Univ Sci B. 14:207–215. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gao CM, Shi MQ, Wu JZ, et al:
Polymorphisms in ERCC1, XRCC1 genes and sensitivity to
gemcitabine/cisplatin chemotherapy in non-small cell lung cancer.
Chin J Cancer Prev Treat. 16:27–30. 2010.
|
|
42
|
Jin YF, Li TC, Wang Y, et al: Single
nucleotide polymorphisms in ERCCl and XPD genes and sensitivity to
platinum-based chemotherapy in non-small-cell Iung cancer.
Carcinogenesis, Teratogenesis & Mutagenesis. 22:374–378.
2010.
|
|
43
|
Su T and Cao GW: Associations of DNA
repair gene, MDRl, and PTPRD polymorphisms with chemotherapy and
survival in lung cancer patients. MM dissertation. Second Military
Medical University, ProQuest; 2012
|
|
44
|
Wang L, Dai XF, Wu G, et al: Predictive
value of excision repair cross-complementation group 1 (ERCCl)
polymorphism on chemotherapy response and survival of advanced
non-small cell lung cancer patients treated with cisplatin. Herald
Med. 31:720–724. 2012.
|
|
45
|
Li XH, Jiang WF, Zhang HL, et al:
Relationship between SNPs of ERCC1 in peripheral blood of NSCLC and
effects of cisplatin-based chemotherapy. Chin J Cancer Prev Treat.
17:169–172. 2010.
|
|
46
|
Zhou GR, Feng JF, Lu JW, et al: On
sensitivity of XRCC1 and ERCC1 single nucleotide polymorphism and
non-small cell lung cancer to platinum-based chemotherapy. J Hainan
Med Univ. 20:20–24. 2013.
|
|
47
|
Sun N, Chen BA and Sun XC: Association
between single nucleotide polymorphisms of some genes and clinical
response to platinum-based chemotherapy in advanced non-small cell
lung cancer (PhD thesis). Southeast University. 2009.
|
|
48
|
KimCurran V, Zhou C, Schmid-Bindert G, et
al: Lack of correlation between ERCC1 (C8092A) single nucleotide
polymorphism and efficacy/toxicity of platinum based chemotherapy
in Chinese patients with advanced non-small cell lung cancer. Adv
Med Sci. 56:30–38. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yuan P, Miao XP, Zhang XM, et al: XRCCl
and XPD genetic polymorphisms predict clinical responses to
platinum-based chemotherapy in advanced non-small cell lung cancer.
Zhonghua Zhong Liu Za Zhi. 28:196–199. 2006.(In Chinese).
|
|
50
|
Chen X, Sun H, Ren S, et al: Association
of XRCC3 and XPD751 SNP with efficacy of platinum-based
chemotherapy in advanced NSCLC patients. Clin Transl Oncol.
14:207–213. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yao CY, Huang XE, Li C, et al: Lack of
influence of XRCC1 and XPD gene polymorphisms on outcome of
platinum-based chemotherapy for advanced non small cell lung
cancers. Asian Pac J Cancer Prev. 10:859–864. 2009.PubMed/NCBI
|
|
52
|
Wu W, Li H, Wang H, et al: Effect of
polymorphisms in XPD on clinical outcomes of platinum-based
chemotherapy for Chinese non-small cell lung cancer patients. PLoS
One. 7:e332002012. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhang YP, Xue HB, Ling Y, et al:
Correlation between the polymorphisms in XPD751 gene and
sensitivity and toxicity of chemotherapy in advanced non-small cell
lung cancer. Hainan Med J. 24:648–650. 2013.
|
|
54
|
Fan H, Huang XE, Zhang Q, et al:
Relationship of XRCC1 and XPD gene polymorphisms with
chemosensitivity to platinum-based chemotherapy in advanced
non-small cell lung cancer. Pract Geriatr. 22:306–314. 2008.(In
Chinese).
|
|
55
|
Chen JC, Liu ZL, Cheng J, et al:
Relationship between XPD single nudeotide polymorphisms and
platinum sensitivity in advanced non-small cell lung Cancer. Chin J
Gerontol. 31:1114–1117. 2011.(In Chinese).
|
|
56
|
Ren S, Zhou S, Wu F, et al: Association
between polymorphisms of DNA repair genes and survival of advanced
NSCLC patients treated with platinum-based chemotherapy. Lung
Cancer. 75:102–109. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Sun XC, Su N, Cheng HY, et al: XPA A23G
polymorphism and clinical response to platin-based chemotherapy in
advanced non-small cell lung cancer. J Med Postgrad. 20:1271–1273.
2007.
|
|
58
|
Jia XF, Liang J, Lv HY, et al:
Relationship between XPA and XPG polymorphisms and platinum-based
chemotherapy outcomes in advanced non-small cell lung cancer. Prog
Mod Biomed. 11:1718–1722. 2011.
|
|
59
|
Feng J, Sun X, Sun N, et al: XPA A23G
polymorphism is associated with the elevated response to
platinum-based chemotherapy in advanced non-small cell lung cancer.
Acta Biochim Biophys Sin (Shanghai). 41:429–435. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zhang T, Sun J, Lv M, et al: XPG is
predictive gene of clinical outcome in advanced non-small-cell lung
cancer with platinum drug therapy. Asian Pac J Cancer Prev.
14:701–705. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Lv HY, Li QC, Wei HJ, et al: Relationship
between GSTP1 and XPG genetic polymorphisms and survival of
platinum-based chemotherapy in advanced non-small cell lung cancer
patients. China Oncol. 22:609–617. 2012.
|
|
62
|
Sun X, Li F, Sun N, et al: Polymorphisms
in XRCC1 and XPG and response to platinum-based chemotherapy in
advanced non-small cell lung cancer patients. Lung Cancer.
65:230–236. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Yin M, Yan J, Voutsina A, et al: No
evidence of an association of ERCC1 and ERCC2 polymorphisms with
clinical outcomes of platinum-based chemotherapies in non-small
cell lung cancer: a meta-analysis. Lung Cancer. 72:370–377. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Yu D, Shi J, Sun T, et al: Pharmacogenetic
role of ERCC1 genetic variants in treatment response of
platinum-based chemotherapy among advanced non-small cell lung
cancer patients. Tumour Biol. 33:877–884. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Machius M, Henry L, Palnitkar M and
Deisenhofer J: Crystal structure of the DNA nucleotide excision
repair enzyme UvrB from Thermus thermophilus. Proc Natl Acad
Sci USA. 96:11717–11722. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Hoeijmakers JH: DNA damage, aging, and
cancer. N Engl J Med. 361:1475–1485. 2009. View Article : Google Scholar
|
|
67
|
Friedberg EC: DNA damage and repair.
Nature. 421:436–440. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Furuta T, Ueda T, Aune G, et al:
Transcription-coupled nucleotide excision repair as a determinant
of cisplatin sensitivity of human cells. Cancer Res. 62:4899–4902.
2002.PubMed/NCBI
|
|
69
|
Wei HB, Hu J, Shang LH, et al: A
meta-analytic review of ERCC1/MDR1 polymorphism and
chemosensitivity to platinum in patients with advanced non-small
cell lung cancer. Chin Med J (Engl). 125:2902–2907. 2012.PubMed/NCBI
|
|
70
|
Wei SZ, Zhan P, Shi MQ, et al: Predictive
value of ERCC1 and XPD polymorphism in patients with advanced
non-small cell lung cancer receiving platinum-based chemotherapy: a
systematic review and meta-analysis. Med Oncol. 28:315–321. 2011.
View Article : Google Scholar : PubMed/NCBI
|