Introduction
Lumbar intervertebral disc herniation (LDH) is one
of the most common orthopedic conditions that can cause lower back
pain and sciatica. It is estimated that ~70% of the population
experiences lower back pain during their lifespan, and at any given
time, 55% suffer from lower back pain associated with radicular
syndromes (1–3). Sciatica symptoms are extremely
persistent and up to one-third of all patients with sciatica
undergo lumbar surgery (4). A
number of studies have discussed the mechanical and biochemical
factors involved in the induction of radicular pain due to LDH
(5) and numerous trials of
potential molecular and biological therapies have been conducted in
this context. Previous studies have evaluated three main
biochemical changes: Decreased matrix synthesis, increased
catabolism, and changed levels of growth factors and cytokines
associated with altered disc cell phenotypes during disc aging and
degeneration (6–11). However, sciatica of a radicular
origin represents an undetermined clinical problem. The study by
Bogduk (12) stated that discogenic
pain cannot be diagnosed clinically and relies on supposition. Thus
far, the pathogenesis of LDH is poorly understood and, aside from
surgical intervention, no effective therapy is available.
Proteomic techniques are powerful tools that can
provide detailed information regarding changes to the expression of
protein profiles. Studies on proteomics have been carried out in
certain diseases, including malignant tumors and neuropathy
(13–17). A specific marker for a particular
disease can be identified by comparing the protein spectrum of a
patient with the disease with that of a healthy subject or with the
protein spectrum from the gene bank (18). Proteomics may become one of the best
measures for tracking disease markers and drug targets. Although
there are certain studies describing the use of proteomics for
marker detection and evaluation of pathogenesis in rheumatoid
arthritis, few studies have been published regarding proteomics
research in the osteoarthritis system (19,20),
particularly in LDH. The pathogenesis and development of LDH
undergoes a multi-stage process, which is based on gene expression
changes. Proteins are the key bridges that connect genes with the
biological function outcomes. Therefore, more proteomic studies are
required to help improve the understanding of LDH pathogenesis.
Proteomic analysis studies of the cerebrospinal fluid (CSF) of
patients with LDH have documented quantitative differences in the
expression of CSF proteins from patients with LDH compared to
controls. Thus, these changes can be considered to be biomarkers of
a condition, such as LDH (21).
However, it is difficult to obtain the CSF and therefore it is
difficult to identify such biomarkers. Therefore, the aim of the
present study was to use proteomics to identify differentially
expressed proteins in the sera of LDH patients compared to control
subjects. The validity of the differential protein expression in
the patients with LDH was further examined and confirmed with an
enzyme-linked immunosorbent assay (ELISA). We suggest that these
differentially expressed proteins may be useful as biomarkers for
LDH, and in addition, they may lead to the development of targeted
therapies for this disease.
Materials and methods
Diagnostic criteria for LDH
The diagnosis of LDH was based on the symptoms and
objective signs of sciatica. Lower back pain, severe lower leg pain
and the Lasègue sign were observed in all the patients. In
addition, magnetic resonance imaging confirmed LDH. Subjects with
liver or kidney diseases were excluded from the study. Three
orthopedic surgeons conducted independent clinical diagnoses on the
basis of the physical and radiological examinations, which were
confirmed by surgery.
Subjects
A total of 30 patients with LDH (15 females and 15
males, 28.1±2.3 years old, who had never received any prior
surgical treatment and had not possessed any disease-related or
predisposing risk for LDH) and 30 healthy subjects who lacked any
symptoms of disc herniation (15 females and 15 males, 25.2±3.1
years old) were included in the proteomics study. Another 10
patients with LDH (5 females and 5 males, 26.3±2.3 years old) and
10 healthy subjects (5 females and 5 males, 30.1±3.3 years old)
were enrolled for the ELISA test.
Serum samples
Venous blood samples were collected from all
subjects and phenylmethylsulfonyl fluoride was added to the
samples. The serum was separated from the samples and stored at
−80°C until use according to the instructions of the ProteoPrep
Blue Albumin and IgG Depletion kit (Sigma) and 2-D clean up kit
(Amersham Biosciences). To minimize the variation among
individuals, the gender and age of the patients were matched in the
control and LDH groups in the proteomic study. The study was
approved by the local ethics committee of the Third Affiliated
Hospital of Sun Yat-sen University and it was carried out in
compliance with the principles stipulated by the Declaration of
Helsinki. All patients and control subjects gave their informed
consent prior to enrollment in the study.
A 30-μl serum sample was loaded onto a 1-ml column.
According to the manufacturer’s instructions, a
ProteoPrep® Blue Albumin and immunoglobulin G (IgG)
Depletion kit (Sigma-Aldrich, St. Louis, MO, USA) was used to
remove serum albumin and IgG. A 2-D clean-up kit (Amersham
Biosciences Corp., Piscataway, NJ, USA) was utilized to discard the
salt and sulfatide in the serum. Protein concentrations in the
serum were determined prior to conducting two-dimensional
electrophoresis (2-DE), using Bradford’s method (22) with bovine serum albumin as the
standard.
Two-dimensional polyacrylamide gel
electrophoresis (2D-PAGE)
2D-PAGE was performed according to the method of
Görg et al (23) and the
manufacturer’s instructions of the Bio-Rad (Hercules, CA, USA)
electrophoresis device. Non-linear immobiline pH 4–7 gradient (IPG)
strips (Bio-Rad) were used for isoelectric focusing (IEF). A 0.3-mg
sample of the total protein was loaded onto the IPG strips. The
strips were rehydrated at 50 V for 12 h, followed by IEF at 150 V
for 1 h, 500 V for 3 h, 1,000 V for 1 h, 5,000 V for 1 h, 7,000 V
for 2 h and 10,000 V until the total voltage x hours of exposure
reached 50,000 Vh. Following this, the strips were equilibrated for
15 min in equilibration buffer I [6 M urea, 2% sodium dodecyl
sulfate, 50 M Tris-HCl (pH 8.8), 30% glycerol, 2% dithiothreitol
(DTT) and bromophenol blue]. The strips were subsequently
equilibrated in the same buffer containing 3% iodoacetamide instead
of DTT for 15 min, and were then transferred to a 12% glycerol
gradient gel for separation. The second dimensional separation was
performed at 12 mA per strip for 30 min, subsequently switching to
20 mA per strip until the bromophenol blue indicator reached the
bottom of the gel. Following 2-DE, the proteins in the gel were
visualized by silver staining (20).
Image analysis
The stained gel was scanned by an Image Scanner type
II (Amersham Biosciences Corp.) and the 2-D electrophoregram was
analyzed with the application of ImageMaster 2D Elite 5.0 software
(Amersham Biosciences Corp.). The data were statistically processed
on the Windows program SPSS v15.0 (SPSS, Inc., Chicago, IL, USA).
P<0.05 was considered to indicate a statistically significant
difference.
Identification of proteins by
matrix-assisted laser desorption/ionization (MALDI) time-of-flight
(TOF) mass spectrometry (MS)
Protein spots exhibiting significant expressional
differences (up- or downregulated by >3-fold) were excised from
the gel and sliced into sections of ~1 mm3. The spots
were destained and dehydrated. Trypsin digestion solution, which
was treated with N-tosyl-L-phenylalanine chloromethyl ketone, was
added to the tube containing the spots for digestion overnight at
37°C. Following digestion, the peptides were obtained from the
extraction buffer, which contained 50% acetonitrile and 0.1%
trifluoroacetic acid (TFA), and were dried under N2. The
extracted peptides were mixed with α-cyano-4-hydroxycinnamic acid
solution in 0.1% TFA and 50% acetonitrile. The solution obtained
was placed on the metal plate of the MALDI-TOF target and dried at
room temperature. The samples were analyzed using MALDI-TOF-MS
(Applied Biosystems, Foster City, CA, USA). The spectrum was
recorded in the reflector positive ion mode. The standard
conditions consisted of a 355-nm wavelength laser and an
accelerating voltage of 20,000 V. The matrix and peptides were
selected in the mass range of 700–3,500 Da. Subsequent to MS
analysis, the peptides that were different from the matrix peptide
mass fingerprint were selected and the mass spectrum/mass spectrum
was achieved.
Database retrieval
The mass spectrum obtained from the MALDI-TOF was
assessed by searching the Mascot software from NCBInr (www.matrixscience.com) with the following restrictions
to the initial retrieval: Species, Homo Sapiens; mass range,
800–4,000 Da; isoelectric point (pI) range, 4–7; maximum tolerance
error of peptide fragment, ±0.5 Da; ion selection,
(M+H)+ and monoisotopic; a minimum of 4 matched
peptides; fixed modifications, carbamidomethyl; and variable
modifications, oxidation.
ELISA
The levels of the aforementioned identified proteins
(associated with LDH pathology) were measured in the blood serum
samples using an ELISA kit (BMA Biomedicals AG, Augst, Switzerland)
according to the manufacturer’s instructions. The blood serum was
obtained from another 10 patients with LDH and 10 healthy
subjects.
Results
Analysis of the protein profiles
The protein profiles from the sera of 30 patients
with LDH and 30 healthy subjects were analyzed by 2-DE with
silver-staining of proteins in the gel. Several protein spots were
separated on 2-DE gels from the experimental and control groups,
totaling 950±50 and 920±50 spots, respectively. The majority of
protein spots were clustered between 10 and 120 kDa and between pI
4–7. The overall pattern of the protein expression in all 2-DE gels
was extremely similar. The proteins that were up- or downregulated
by >3-fold and appeared in all patients with LDH pathology were
selected for identification by peptide mass fingerprinting, using
MALDI-TOF-MS and protein database searching. The results from this
identification were of high confidence if the protein had a
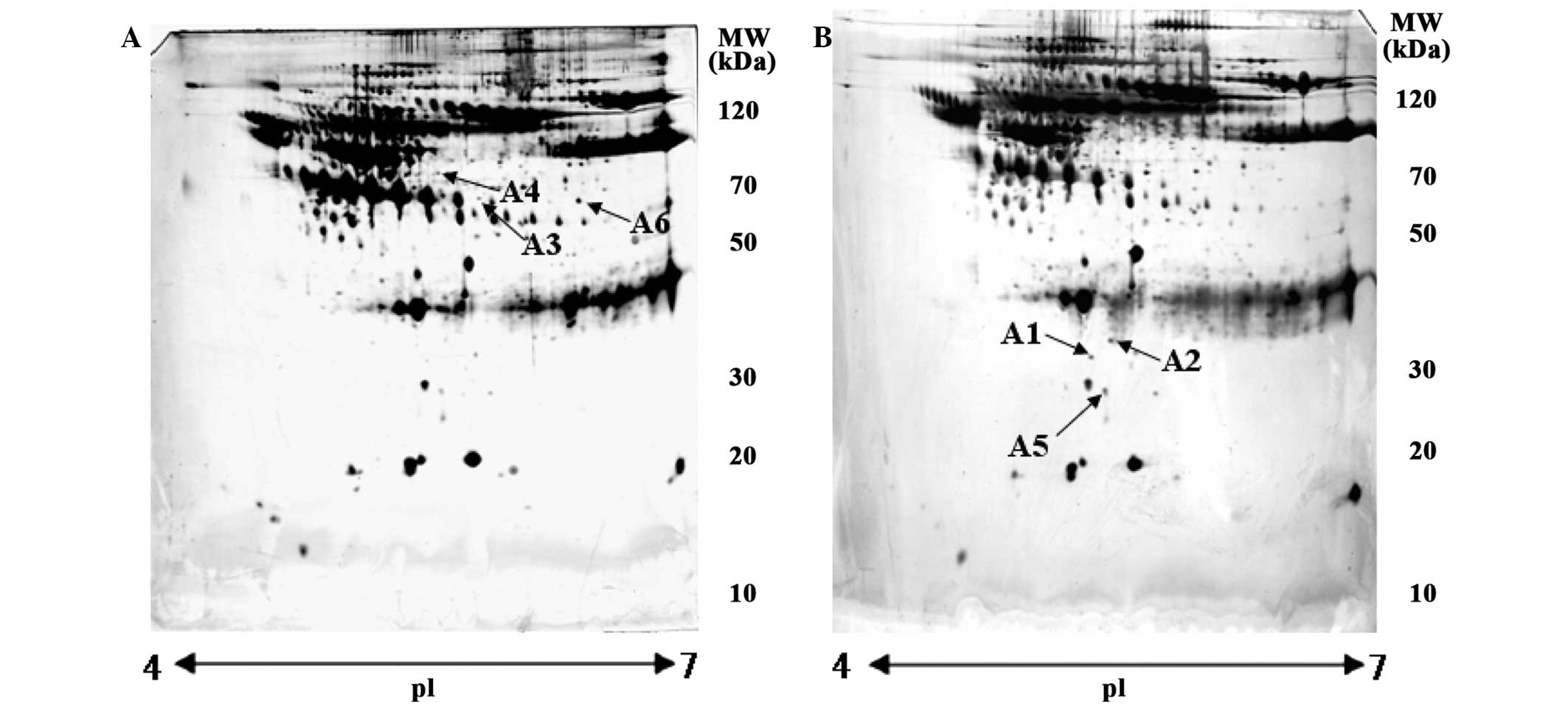
significant score and high-sequence coverage. A total of 6 protein
spots were identified as being associated with LDH pathology
(Table I). The identities of these
6 spots were found to be upregulated apolipoprotein-L1 (APO-L1) and
two serum albumin precursors, and downregulated apolipoprotein M
(APO-M), tetranectin (TN) and immunoglobulin light chain (IGL). The
6 proteins were found to be significantly and consistently
different in the sera of patients with LDH compared to healthy
subjects, according to 2-DE gel separation and proteomic analysis
(Fig. 1).
 | Table IMatrix-assisted laser
desorption/ionization time-of-flight mass spectrometry analysis of
differentially expressed proteins in patients with lumbar
intervertebral disc herniation (LDH) compared to healthy
controls. |
Table I
Matrix-assisted laser
desorption/ionization time-of-flight mass spectrometry analysis of
differentially expressed proteins in patients with lumbar
intervertebral disc herniation (LDH) compared to healthy
controls.
| Spot no. | Accession no. | Protein name | MW, Da | pI | Protein score
intensity in LDH | Optical density |
|---|
| A1 | 645213 | Apolipoprotein M | 13042.4 | 7.66 | 210 | Decreased |
| A2 | 796167 | Immunoglobulin light
chain | 14669.1 | 6.25 | 207 | Decreased |
| A3 | 384697 | Serum albumin
precursor | 47329.7 | 5.97 | 371 | Increased |
| A4 | 745872 | Serum albumin
precursor | 69321.5 | 5.92 | 107 | Increased |
| A5 | 9028 | Tetranectin | 22552.3 | 5.52 | 307 | Decreased |
| A6 | 514475 | Apolipoprotein
L1 | 43900.0 | 5.84 | 234 | Increased |
The protein expression of APO-L1 was not detected in
the sera of healthy subjects but was only found in patients with
LDH pathology. In addition, the protein expression levels of APO-M,
TN and IGL in the sera from patients with LDH were all
downregulated by 22±3 (P<0.01), 37±5 (P<0.01) and 27±3%
(P<0.01), respectively, compared to the sera from healthy
subjects.
The ELISA results support the proteomic analysis
results. The ELISA demonstrated that the mean serum concentrations
of APO-M, TN and IGL were significantly lower in LDH patients
compared to the healthy control group (P<0.05). By contrast, the
plasma levels of APO-L1 in LDH patients were significantly higher
than the values recorded for the healthy control group
(P<0.01).
Discussion
Proteomics is a powerful tool in the analysis of
protein expression and composition of various tissues, as it
provides useful information regarding biochemical pathways and
biological processes and connects the relevant genes with protein
expression. Proteomics primarily involves the application of 2-DE,
MS and database retrieval systems (24). Due to its reproducibility, accuracy
and high-throughput, proteomics has been extensively used in
studies pertaining to tumors, inflammation and immunity, as well as
other complex areas of study. Although proteomics is becoming
increasingly available to investigators, the application of
proteomics to LDH is only in the initial stages (21).
In the present study, proteomics was applied to
identify specific biomarkers associated with the pathogenesis of
LDH. A total of 6 proteins were found that were differentially
expressed in the sera of 30 patients with LDH compared to 30
healthy subjects. Furthermore, 4 of these 6 proteins were
statistically different in terms of their serum level between the
experimental and control groups as determined by the ELISA
measurements on the serum samples. These 4 proteins were APO-L1,
TN, APO-M and IGL.
Extremely little is known with regard to APO-L, a
protein first characterized in 1997 as a new human high-density
lipoprotein (25). APO-L was found
only in the pancreas and is described as a 383-amino acid residue
protein with no significant homology to any other known protein
sequence. In healthy subjects, free APO-L is not detected in the
plasma; instead, it binds to other proteins, mainly large
APO-A1-containing lipoproteins. The cloning of CG12_1 in 1999
suggested that other APO-L proteins may exist. APO-L is
specifically expressed in endothelial cells lining healthy
atherosclerotic iliac arteries and the aorta, and it is responsive
to changes in the tumor necrosis factor (TNF)-α level (26). The study by Horrevoets et al
(26) also identified and
classified novel members of the APO-L protein family. The
similarities of these proteins suggest that they have arisen
through local gene duplication. Although they do not possess
classical signal peptide sequences, evidence exists that at least
APO-L1 is secreted in the plasma (25,27).
It is believed that TNF-α plays a key role in the inflammation
induced by LDH (28–30). However, Brisby et al
(31) found that the concentration
of TNF-α in the serum of LDH patients is not significantly
different compared to healthy patients. Therefore, the level of
APO-L1 may be a reflection of the TNF-α secreted into the serum and
may serve as a biomarker for LDH. Additionally, APO-L1 is
associated with the lysis of lysosomes and the appearance of an
APO-L version with a signal peptide represents a novel component of
innate immunity (32,33). Herniation of the intervertebral disc
is associated with human autoimmunity. In addition, APO-L1 is a
novel Bcl-2 homology domain 3-only protein, which when
overexpressed and accumulated intracellularly, induces autophagic
cell death in cells as characterized by the increasing formation of
autophagic vacuoles (34). Thus,
APO-L1 may be the signal peptide of LDH.
TN is a plasminogen-binding protein, that can be
encountered in the plasma and in the extracellular matrix (ECM)
(35,36). In addition to its
plasminogen-binding properties, TN can bind to heparin, APO-A,
tissue plasminogen activator, hepatocyte growth factor and
angiostatin (37–39). TN expression has been detected in
various endocrine tissues, as well as in epithelial and mesenchymal
cells, including fibroblasts, monocytes and neutrophils (40–42).
TN also constitutes a crucial component of the ECM in osteogenesis,
muscle development and regeneration, suggesting its important
physiological role in tissue remodeling (43,44).
Serum TN is decreased following trauma or acute myocardial
infarction, during pregnancy and in patients with liver cirrhosis,
rheumatoid arthritis or malignant tumors (37,45–47).
Although the exact biological function of TN has yet to be
established, certain evidence suggests that TN plays an important
role in tissue remodeling. TN increases the tissue-type plasminogen
activator-catalyzed activation of plasminogen in the presence of
poly-d-lysine (35) and activated
plasminogen is considered to play a key role in the degradation of
the ECM. In this study, serum TN was also decreased. As the
majority of the LDH cases are caused by degeneration of the lumbar
disc, in the process of which apoptosis of the nucleus pulposus
cell is first observed and is subsequently followed by the
imbalance of lumbar disc remodeling. In this way, the serum TN
level is reduced in patients with LDH, which is considered to be
associated with regeneration of the lumbar disc, leading to a
decrease in the synthesis and secretion of TN.
Human APO-M was found and initially isolated from
chylomicrons in the 1999 study by Xu and Dahlbäck (48). Xu et al (49) have reported that transforming growth
factor-β can also downregulate APO-M expression and secretion in
HepG2 cells. These data indicate that APO-M may be associated with
the host defense response as the APO-M gene is located in the
histocompatibility complex III region on chromosome 6 (50). A number of genes in this region are
associated with the immune responses and the APO-M gene is
extremely close to the TNF-α gene and lymphotoxin genes (50). Therefore, APO-M may also be
associated with the immune system, or regulated by cytokines or
other inflammatory factors. The detailed association between APO-M
and LDH remains unclear and requires further investigation. The
function of IGL in LDH patients also remains unclear. Thus, it can
be used clinically for screening or monitoring LDH requires further
studies.
Previously, no direct association between the
pathogenesis of LDH and the 4 proteins that were differentially
expressed in the LDH group has been found. To the best of our
knowledge, the present study is the first to report on this
association and is unique as the serum molecular analysis of LDH
was accomplished using a proteomics approach. Thus, proteomic
analysis is an efficient technique for identifying the diagnostic
biomarkers of LDH. In conclusion, the 4 proteins that were
differentially expressed in LDH patients compared to healthy
subjects were recognized. These findings contribute to the
advancement of knowledge in this area of study.
References
|
1
|
Macfarlane GJ, Thomas E, Croft PR, et al:
Predictors of early improvement in low back pain amongst consulters
to general practice: the influence of pre-morbid and
episode-related factors. Pain. 80:113–119. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Frymoyer JW: Lumbar disc disease:
epidemiology. Instr Course Lect. 41:217–223. 1992.
|
|
3
|
Long DM: Decision making in lumbar disc
disease. Clin Neurosurg. 39:36–51. 1992.PubMed/NCBI
|
|
4
|
Balagué F, Nordin M, Shelkhzadeh A, et al:
Recovery of severe sciatica. Spine (Phila Pa 1976). 24:2516–2524.
1999.
|
|
5
|
Igarashi T, Kikuchi S, Shubayev V, et al:
2000 Volvo Award winner in basic science studies: Exogenous tumor
necrosis factor-alpha mimics nucleus pulposus-induced
neuropathology. Molecular, histologic, and behavioral comparisons
in rats. Spine (Phila Pa 1976). 25:2975–2980. 2000.
|
|
6
|
Yoon ST and Patel NM: Molecular therapy of
the intervertebral disc. Eur Spine J. 15(Suppl 3): S379–S388. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kuh SU, Zhu Y, Li J, et al: A comparison
of three cell types as potential candidates for intervertebral disc
therapy: annulus fibrosus cells, chondrocytes, and bone marrow
derived cells. Joint Bone Spine. 76:70–74. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhao CQ, Wang LM, Jiang LS and Dai LY: The
cell biology of intervertebral disc aging and degeneration. Ageing
Res Rev. 6:247–261. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Diefenderfer DL and Brighton CT:
Microvascular pericytes express aggrecan message which is regulated
by BMP-2. Biochem Biophys Res Commun. 269:172–178. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tim Yoon S, Su Kim K, Li J, et al: The
effect of bone morphogenetic protein-2 on rat intervertebral disc
cells in vitro. Spine (Phila Pa 1976). 28:1773–1780.
2003.PubMed/NCBI
|
|
11
|
Fassett DR, Kurd MF and Vaccaro AR:
Biologic solutions for degenerative disc disease. J Spinal Disord
Tech. 22:297–308. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bogduk N: What’s in a name? The labelling
of back pain. Med J Aust. 173:400–401. 2000.
|
|
13
|
Wattiez R, Hermans C, Bernard A, Lesur O
and Falmagne P: Human bronchoalveolar lavage fluid: two-dimensional
gel electrophoresis, amino acid microsequencing and identification
of major proteins. Electrophoresis. 20:1634–1645. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Saintigny G, Schmidt R, Shroot B, Juhlin
L, Reichert U and Michel S: Differential expression of calgranulin
A and B in various epithelia cell lines and reconstructed
epidermis. J Invest Dermatol. 99:639–644. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ruebelt MC, Lipp M, Reynolds TL, et al:
Application of two-dimensional gel electrophoresis to interrogate
alterations in the proteome of genetically modified crops. 3.
Assessing unintended effects. J Agric Food Chem. 54:2169–2177.
2006. View Article : Google Scholar
|
|
16
|
Lee EG, Kim JH, Shin YS, et al:
Application of proteomics for comparison of proteome of Neospora
caninum and Toxoplasma gondii tachyzoites. J Chromatogr
B Analyt Technol Biomed Life Sci. 815:305–314. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tan Y, Lv ZP and Zhang XF: Proteome and
its application in liver disease research. Zhonghua Gan Zang Bing
Za Zhi. 12:700–702. 2004.(In Chinese).
|
|
18
|
Nomura F, Tomonaga T, Sogawa K, et al:
Identification of novel and downregulated biomarkers for alcoholism
by surface enhanced laser desorption/ionization-mass spectrometry.
Proteomics. 4:1187–1194. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tilleman K, Van Beneden K, Dhondt A, et
al: Chronically inflamed synovium from spondyloarthropathy and
rheumatoid arthritis investigated by protein expression profiling
followed by tandem mass spectrometry. Proteomics. 5:2247–2257.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tan X, Cai D, Wu Y, et al: Comparative
analysis of serum proteomes: discovery of proteins associated with
osteonecrosis of the femoral head. Transl Res. 148:114–119. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu XD, Zeng BF, Xu JG, et al: Proteomic
analysis of the cerebrospinal fluid of patients with lumbar disc
herniation. Proteomics. 6:1019–1028. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bradford MM: A rapid and sensitive method
for the quantitation of microgram quantities of protein utilizing
the principle of protein-dye binding. Anal Biochem. 72:248–254.
1976. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Görg A, Obermaier C, Boguth G, et al: The
current state of two-dimensional electrophoresis with immobilized
pH gradients. Electrophoresis. 21:1037–1053. 2000.
|
|
24
|
Wilkins MR, Williams KL, Appel RD and
Hochstrasser DF: Proteome Research: New Frontiers in Functional
Genomics. Springer; Heidelberg: pp. 188–219. 1997
|
|
25
|
Duchateau PN, Pullinger CR, Orellana RE,
et al: Apolipoprotein L, a new human high density lipoprotein
apolipoprotein expressed by the pancreas. Identification, cloning,
characterization, and plasma distribution of apolipoprotein L. J
Biol Chem. 272:25576–25582. 1997. View Article : Google Scholar
|
|
26
|
Horrevoets AJ, Fontijn RD, van Zonneveld
AJ, et al: Vascular endothelial genes that are responsive to tumor
necrosis factor-alpha in vitro are expressed in atherosclerotic
lesions, including inhibitor of apoptosis protein-1, stannin, and
two novel genes. Blood. 93:3418–3431. 1999.
|
|
27
|
Duchateau PN, Movsesyan I, Yamashita S, et
al: Plasma apolipoprotein L concentrations correlate with plasma
triglycerides and cholesterol levels in normolipidemic,
hyperlipidemic, and diabetic subjects. J Lipid Res. 41:1231–1236.
2000.
|
|
28
|
Cuellar JM, Montesano PX and Carstens E:
Role of TNF-alpha in sensitization of nociceptive dorsal horn
neurons induced by application of nucleus pulposus to L5 dorsal
root ganglion in rats. Pain. 110:578–587. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Murata Y, Nannmark U, Rydevik B, et al:
The role of tumor necrosis factor-alpha in apoptosis of dorsal root
ganglion cells induced by herniated nucleus pulposus in rats. Spine
(Phila Pa 1976). 33:155–162. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bachmeier BE, Nerlich AG, Weiler C, et al:
Analysis of tissue distribution of TNF-alpha, TNF-alpha-receptors,
and the activating TNF-alpha-converting enzyme suggests activation
of the TNF-alpha system in the aging intervertebral disc. Ann NY
Acad Sci. 1096:44–54. 2007. View Article : Google Scholar
|
|
31
|
Brisby H, Olmarker K, Larsson K, et al:
Proinflammatory cytokines in cerebrospinal fluid and serum in
patients with disc herniation and sciatica. Eur Spine J. 11:62–66.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pérez-Morga D, Vanhollebeke B,
Paturiaux-Hanocq F, et al: Apolipoprotein L-I promotes trypanosome
lysis by forming pores in lysosomal membranes. Science.
309:469–472. 2005.PubMed/NCBI
|
|
33
|
Vanhamme L, Paturiaux-Hanocq F, Poelvoorde
P, et al: Apolipoprotein L-I is the trypanosome lytic factor of
human serum. Nature. 422:83–87. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wan G, Zhaoriqetu S, Liu Z, et al:
Apolipoprotein L1, a novel Bcl-2 homology domain 3-only
lipid-binding protein, induces autophagic cell death. J Biol Chem.
283:21540–21549. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Clemmensen I, Petersen LC and Kluft C:
Purification and characterization of a novel, oligomeric,
plasminogen kringle 4 binding protein from human plasma:
tetranectin. Eur J Biochem. 156:327–333. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Caterer NR, Graversen JH, Jacobsen C, et
al: Specificity determinants in the interaction of
apolipoprotein(a) kringles with tetranectin and LDL. Biol Chem.
383:1743–1750. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kluft C, Jie AF, Los P, et al: Functional
analogy between lipoprotein(a) and plasminogen in the binding to
the kringle 4 binding protein, tetranectin. Biochem Biophys Res
Commun. 161:427–433. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Mogues T, Etzerodt M, Hall C, et al:
Tetranectin binds to the kringle 1–4 form of angiostatin and
modifies its functional activity. J Biomed Biotechnol. 2004.73–78.
2004.
|
|
39
|
Westergaard UB, Andersen MH, Heegaard CW,
et al: Tetranectin binds hepatocyte growth factor and tissue-type
plasminogen activator. Eur J Biochem. 270:1850–1854. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Borregaard N, Christensen L, Bejerrum OW,
et al: Identification of a highly mobilizable subset of human
neutrophil intracellular vesicles that contains tetranectin and
latent alkaline phosphatase. J Clin Invest. 85:408–416. 1990.
View Article : Google Scholar
|
|
41
|
Clemmensen I, Lund LR, Christensen L and
Andreasen PA: A tetranectin-related protein is produced and
deposited in extracellular matrix by human embryonal fibroblasts.
Eur J Biochem. 195:735–741. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Nielsen H, Clemmensen I and Kharazmi A:
Tetranectin: a novel secretory protein from human monocytes. Scand
J Immunol. 37:39–42. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wewer UM, Iba K, Durkin ME, et al:
Tetranectin is a novel marker for myogenesis during embryonic
development, muscle regeneration, and muscle cell differentiation
in vitro. Dev Biol. 200:247–259. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wewer UM, Ibaraki K, Schjørring P, et al:
A potential role for tetranectin in mineralization during
osteogenesis. J Cell Biol. 127:1767–1775. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Christensen L: The distribution of
fibronectin, laminin and tetranectin in human breast cancer with
special attention to the extracellular matrix. APMIS Suppl.
26:1–39. 1992.PubMed/NCBI
|
|
46
|
Kamper EF, Kopeikina LT, Trontzas P, et
al: Comparative study of tetranectin levels in serum and synovial
fluid of patients with rheumatoid arthritis, seronegative
spondylarthritis and osteoarthritis. Clin Rheumatol. 17:318–324.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kluft C, Los P, Clemmensen I, et al:
Quantitation of plasma levels of tetranectin - effects of oral
contraceptives, pregnancy, treatment with L-asparaginase and liver
cirrhosis. Thromb Haemost. 62:792–796. 1989.PubMed/NCBI
|
|
48
|
Xu N and Dahlbäck B: A novel human
apolipoprotein (apoM). J Biol Chem. 274:31286–31290. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Xu N, Hurtig M, Zhang XY, Ye Q and
Nilsson-Ehle P: Transforming growth factor-beta down-regulates
apolipoprotein M in HepG2 cells. Biochim Biophys Acta. 1683:33–37.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Luo G, Zhang X, Nilsson-Ehle P and Xu N:
Apolipoprotein M. Lipids Health Dis. 3:212004. View Article : Google Scholar
|