Introduction
Multiple myeloma (MM) is a clonal plasma cell
malignancy characterized by bone, renal, hematological and often
neurological complications (1).
Although the response rate of MM patients has significantly
improved over the last decade due to the broad use of novel agents,
the majority of patients will eventually succumb due to
complications associated with the development of resistant disease
(2). There remains a requirement for
new therapeutic options for patients with this B-cell malignancy.
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) has
become a potential therapeutic drug for MM, as it selectively
induces apoptosis in various types of tumor cells, including
myeloma cells, while it shows no significant untoward effects on
normal cells (3,4). Recombinant mutant human TRAIL (rmhTRAIL)
is optimized from wild-type TRAIL. It has significant improvements
in terms of stability, solubility and biological activity (5).
It has been found that the sensitivity of myeloma
cells to TRAIL varies considerably in vitro and in
vivo. However, the exact targets and resistance mechanisms of
TRAIL on MM cells are controversial. Certain studies have addressed
that TRAIL has a specific apoptosis-inducing effect in tumor cells
by combining with TRAIL receptors on cell membranes (6–8). By
contrast, certain studies argue that the sensitivity of MM cells to
TRAIL has no reference to the level of TRAIL receptors (9,10). To
determine whether there are other targets and resistance mechanisms
of TRAIL on MM cells, the differentially expressed proteins were
compared between TRAIL-sensitive and TRAIL-resistant cell lines
prior and subsequent to rmhTRAIL administration by a global
proteomic-based approach, and the promising target and
resistance-related proteins were analyzed.
Materials and methods
Reagents and cell lines
RmhTRAIL freeze-dried powder (Beijing Sunbio Biotech
Co., Ltd., Beijing, China) was diluted in distilled water to a 1
mg/ml rmhTRAIL solution, and was preserved and protected from air
at −20°C in aliquots, and diluted to a working concentration in
RPMI-1640 prior to use. The human myeloma cell line RPMI 8226
(Beijing Sunbio Biotech Co., Ltd.) and U266 (Cancer Institute and
Hospital, Chinese Academy of Medical Sciences, Beijing, China) were
cultured in RPMI-1640 supplemented with 1% penicillin/streptomycin,
1 mmol/l L-glutamine and 10% fetal bovine serum at 37°C, 5%
CO2 in air.
TRAIL treatment
RPMI 8226 and U266 cells (2×107 cells each) were
treated with 15.625 and 1,000 ng/ml rmhTRAIL, respectively, for 24
h (8226TRAIL and U266TRAIL), and untreated cells were used as the
control (8226CON and U266CON). The concentrations were selected as
the apoptosis ratios were 4.29 and 0.54%, respectively, according
to our previous study (unpublished data).
One-dimensional sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (1D-SDS-PAGE)
Following treatment, the cells were washed three
times with phosphate-buffered saline. Total proteins from the four
groups of cells were extracted from cells using the Total Protein
Extraction kit (Beijing Biosynthesis Biotechnology Co., Ltd.,
Beijing, China). Cellular debris was removed by centrifugation for
30 min at 13,000 × g and at 4°C. Protein concentrations were
determined using the bicinchonininc acid (Beijing Biosynthesis
Biotechnology Co., Ltd.) assay. Protein samples were separated by
12% 1D-SDS-PAGE and stained with Coomassie Brilliant Blue R250
solution (Beijing Biosynthesis Biotechnology Co., Ltd.). Protein
zones were manually excised from the gels. Subsequently, gel
sections were destained and dehydrated with acetonitrile.
Sample preparation
Subsequent to the samples being destained and
dehydrated, the proteins in the gel sections were reduced with
dithiothreitol, alkylated with iodoacetamide and incubated with
12.5 µg/µl sequencing grade trypsin (Promega, Madison, WI, USA) at
37°C for 12 h. For protein quantification, peptides of the four
groups of cells were labeled with disparate TMT6 reagents (Pierce
Biotechnology, Thermo Fisher Scientific, Inc., Rockford, IL, USA).
In detail, the U266TRAIL group was labeled with TMT6-128, U266CON
group with TMT6-129, 8226TRAIL group with TMT6-130 and 8226CON
group with TMT6-131 respectively, according to the manufacturer's
protocol. The labeling reaction was carried out by incubation of
tryptic peptides with the TMT reagents for 1 h at room temperature,
and was quenched by hydroxylamine. The TMT-labeled peptides were
desalted using the stage tips. Following the labeling, the peptides
were extracted with 0.1% formic acid, and dried in a vacuum
centrifuge. The volumes of the extraction were adjusted to 25 µl
with 0.1% trilfluoroacetic acid, of which 20 µl was analyzed by
liquid chromatography-tandem mass spectrometry (LC-MS/MS).
LC-MS/MS
For LC-MS/MS analysis, each digestion product was
separated by a 65-min gradient elution at a flow rate 0.25 µl/min
with the EASY-nLCII™ integrated nano-HPLC system (Proxeon
Biosystems A/S, Odense C, Denmark), which is directly interfaced
with the Thermo LTQ-Orbitrap mass spectrometer. The LTQ-Orbitrap
mass spectrometer was operated in the data-dependent acquisition
mode using the Xcalibur software (Thermo Fisher Scientific, Inc.).
The experiment consisted of a single full-scan mass spectrum in the
Orbitrap (400–1,800 m/z, 30,000 resolutions) followed by 20
data-dependent MS/MS scans in the ion trap at 35% normalized
collision energy. The MS/MS spectra from each LC-MS/MS run were
searched against the selected database using an in-house Proteome
Discovery searching algorithm (10).
Bioinformatics analysis of
proteins
The MS/MS peak lists were searched against the IPI
human database using SEQUEST software (http://fields.scripps.edu/sequest/). The search
criteria were as follows: Full tryptic specificity was required;
one missed cleavages were allowed; carbamidomethylation was set as
fixed modification; the oxidation was set as the variable
modification; precursor ion mass tolerances were set at 10 ppm for
all MS acquired in the Orbitrap mass analyzer; and the fragment ion
mass tolerance was set at 0.8 Da for all MS2 spectra acquired in
the linear ion trap (11). The
peptides data were further filtered by selecting proteins with
ProtScore >2 and at least two unique peptides. Cut-off TMT
ratios of fold-change for protein expression were >1.5 for
upregulation and <0.67 for downregulation.
Differentially expressed proteins were classified
based on the Protein ANalysis THrough Evolutionary Relationships
(PANTHER) system (http://www.pantherdb.org), which is a unique resource
that classifies genes and proteins by their functions (12,13). Certain
proteins were annotated manually based on literature searches and
closely associated homologues.
The differentially expressed protein interaction
networks were built automatically by the Search Tool for the
Retrieval of Interacting Genes/Proteins (STRING) system (http://string-db.org) with the default setting, except
that organism, confidence (score) and interactors shown were set to
‘human’, ‘0.20’, and ‘no more than 10 interactors’, respectively
(14,15). The gene name list of these proteins was
input to search against the database, which contains known and
predicted protein-protein interactions. The retrieve included a
detailed network, which highlights several hub proteins.
Results
Proteome profiles of rmhTRAIL-treated
and control myeloma cells
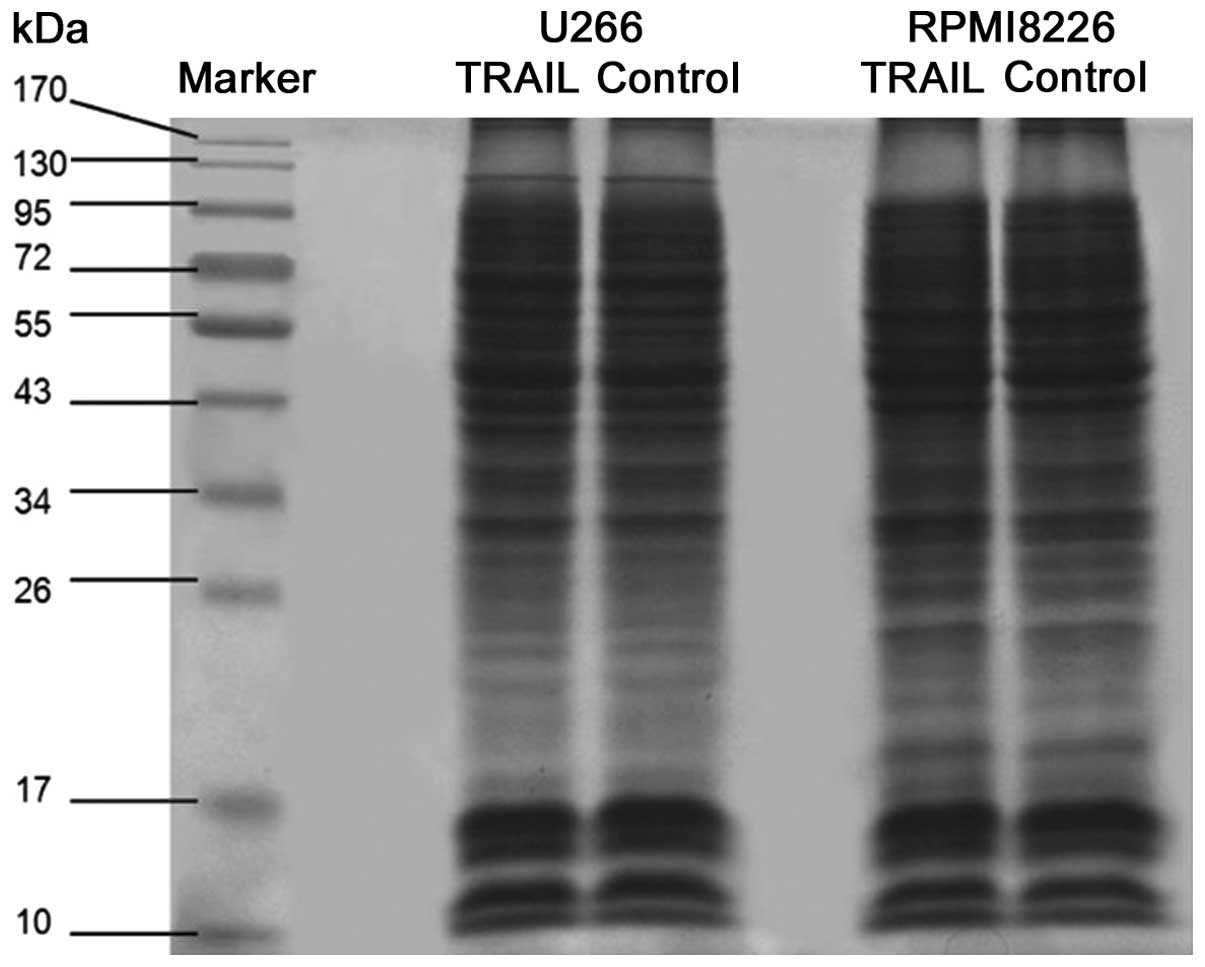
Four groups of proteins were separated in SDS-PAGE
gels, which were subsequently stained with Coomassie Brilliant Blue
R250. Each of the four stained polyacrylamide gels was divided into
10 sections (Fig. 1).
LC-MS/MS identification revealed that 1,594 proteins
were identified, which had a ProtScore >2 and at least two
unique peptides. 42% (675 of 1,594) proteins were identified by
>5 peptides, 12% (190 of 1,594) by 4 peptides, 17% (278 of
1,594) by 3 peptides and 28% (451 of 1,594) by 2 peptides.
Target proteins of rmhTRAIL on RPMI
8226 cells
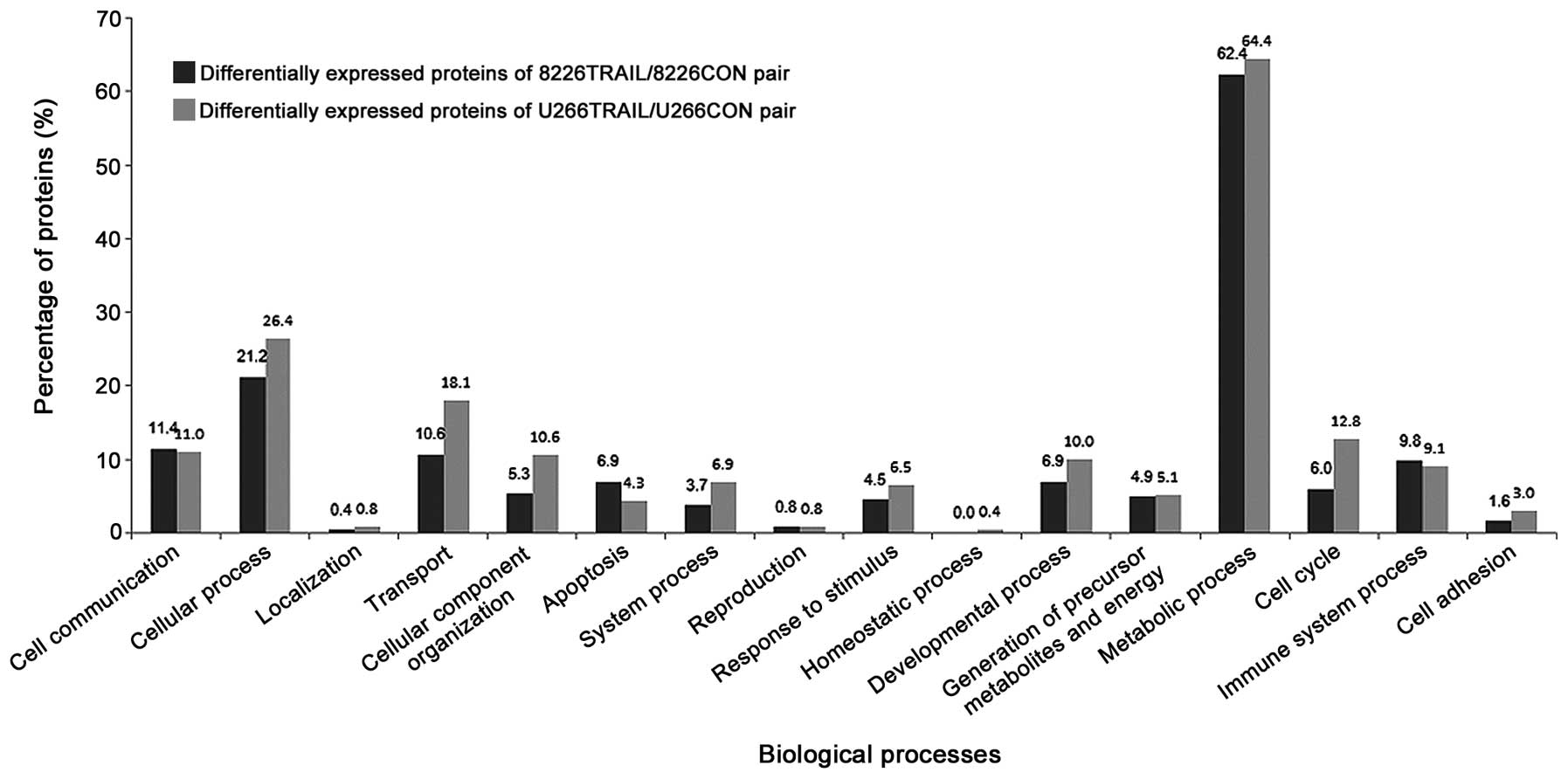
Among the total 1,594 proteins, 337 differentially
expressed proteins were screened in 8226TRAIL/8226CON pair, while
757 differentially expressed proteins in U266TRAIL/U266CON pair,
among which 245 and 492 proteins could be recognized and analyzed
by PANTHER. These proteins were involved in various biological
processes (Fig. 2).
Differentially expressed proteins of the two pairs
were further analyzed, and the proteins that participated in
apoptosis were screened by PANTHER. Among them, 6 pro-apoptotic
proteins were screened as possible target proteins of rmhTRAIL on
RPMI 8226 cells (Table I), which were
upregulated in the 8226TRAIL/8226CON pair, while no change was
observed in the U266TRAIL/U266CON pair. These were calpain small
subunit 1 (CPNS1), peflin (PEF1), B-cell receptor-associated
protein 31 (BAP31), apoptosis-associated speck-like protein
containing CARD (ASC), BAG family molecular chaperone regulator 2
(BAG2) and chromobox protein homolog 3 (CBX3), respectively.
 | Table I.Target proteins of rmhTRAIL on
RPMI8226 cells according to liquid chromatography-tandem mass
spectrometry identification. |
Table I.
Target proteins of rmhTRAIL on
RPMI8226 cells according to liquid chromatography-tandem mass
spectrometry identification.
| Namea | Function |
Accessionb | Scorec |
8226TRAIL/8226CONd |
U266TRAIL/U266CONe |
|---|
| Calpain small subunit
1 (CPNS1) | Induction of
apoptosis; immune system process; proteolysis | IPI00025084 | 37.23 | 1.941 | 0.800 |
| Peflin (PEF1) | Induction of
apoptosis; immune system process; proteolysis | IPI00018235 | 30.84 | 1.646 | 1.021 |
| B-cell
receptor-associated protein 31 (BAP31) | Apoptosis;
intracellular protein transport | IPI00218200 | 18.53 | 1.958 | 0.831 |
| Isoform 2 of
apoptosis-associated speck-like protein containing a CARD
(ASC) | Apoptosis;
proteolysis | IPI00221360 | 17.16 | 1.925 | 0.731 |
| BAG family molecular
chaperone regulator 2 (BAG2) | Apoptosis; protein
folding | IPI00000643 | 11.29 | 1.599 | 0.822 |
| Chromobox protein
homolog 3 (CBX3) | Apoptosis; regulation
of transcription from RNA polymerase II promoter; establishment or
maintenance of chromatin | IPI00297579 | 7.47 | 2.303 | 0.929 |
Resistance-related proteins of
rmhTRAIL on U266 cells
Among the total 1,594 proteins, 1,081 differentially
expressed proteins were screened in the U266CON/8226CON pair, while
662 differentially expressed proteins were in the
U266TRAIL/8226TRAIL pair, among which 702 and 450 proteins could be
recognized and analyzed by PANTHER. The differentially expressed
proteins of the two pairs were further analyzed, and the proteins
that participated in apoptosis or proliferation were screened by
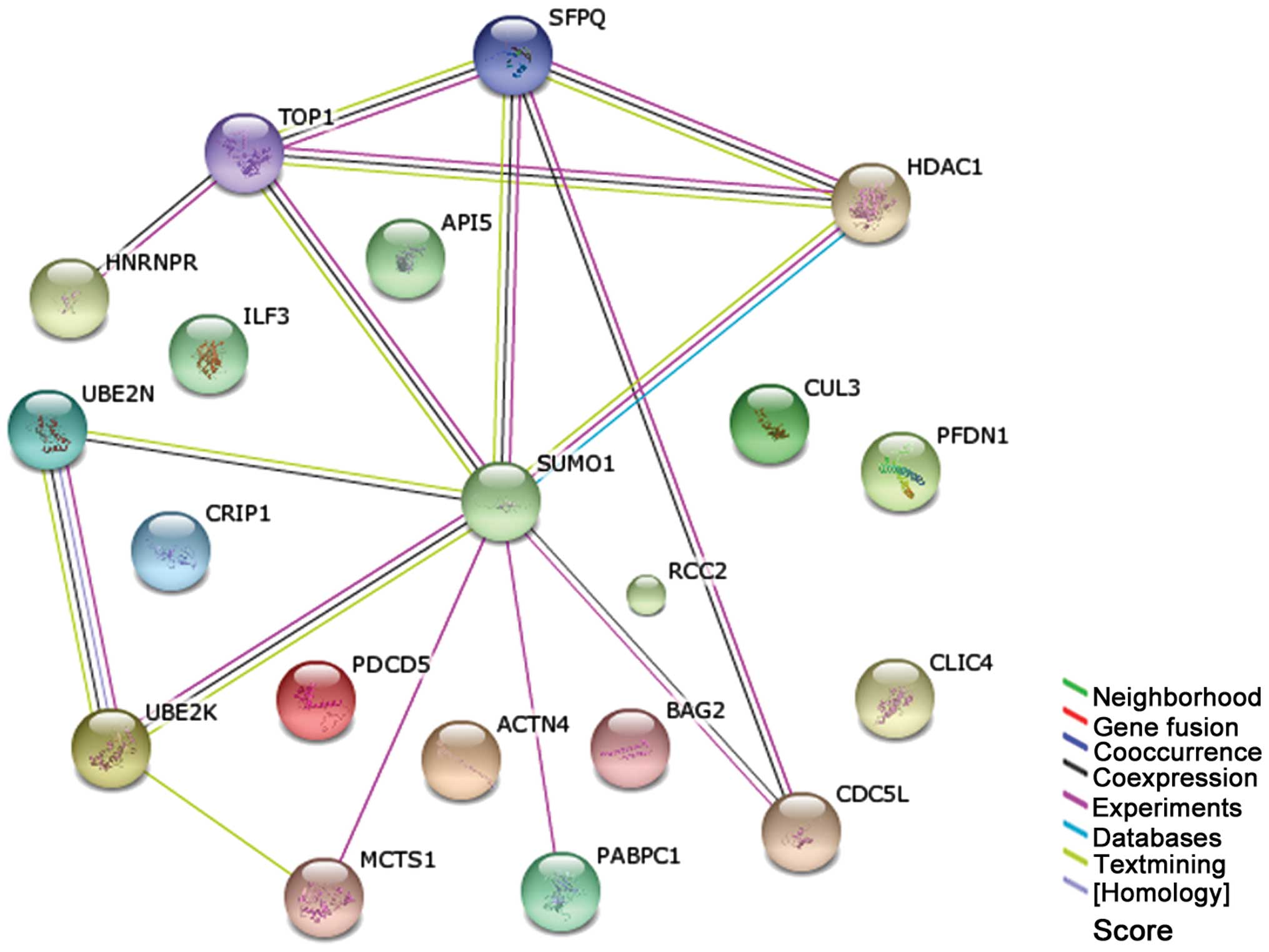
PANTHER. Data showed that there were 31 proliferation- or
anti-apoptosis-related proteins expressed at higher levels in the
TRAIL-resistant cells U266 compared to RPMI 8226 cells prior and
subsequent to rmhTRAIL treatment. Among the 31 identified proteins,
20 can be linked through direct interaction into a protein-protein
interaction network based on the prediction results of STRING
system (Fig. 3). Notably, small
ubiquitin-related modifier 1 (SUMO1) and several other proteins,
which participated in the ubiquitin proteasome pathway (UPP), were
hubs in this network. Of the 20 proteins, 12 are known SUMO1
interactors. These results showed that UPP proteins were
overexpressed in U266 cells and were independent of rmhTRAIL
treatment, suggesting that UPP may have a vital role in mediating
TRAIL-resistance in U266 cells.
Discussion
The present study analyzed the TRAIL-targeted
proteins and the resistance-related proteins by a global
proteomic-based approach. According to the results, the possible
target proteins of rmhTRAIL on RPMI 8226 cells were CPNS1, PEF1,
BAP31, ASC, BAG2 and CBX3. The possible resistance mechanism of
rmhTRAIL on U266 cells was the overexpression of UPP-related
proteins.
To date, studies in MM show that TRAIL induces cell
apoptosis via the extrinsic and intrinsic pathways (16). According to this view, TRAIL receptors
are considered essential in these two approaches, which is
inconsistent with certain studies that argue the sensitivity of MM
cells to TRAIL has no reference to the level of TRAIL receptors
(9,10).
With the present data, the pro-apoptosis proteins CPNS1, PEF1,
BAP31, ASC, BAG2 and CBX3 may be the apoptosis-induced mechanisms
in addition to the activation of death receptors.
Furthermore, a previous study showed that the
resistance mechanisms of myeloma cells to TRAIL contain various
pathways, including variable levels of TRAIL receptors, variable
expression levels of molecules participating extrinsic pathway or
intrinsic pathway, and drug resistance mediated by bone marrow
stromal cells (17). UPP is an
important mechanism in the degradation of proteins (18,19), which
has not been reported in previous studies of TRAIL-resistance
mechanism in myeloma cells. Recent studies showed that the disorder
of tumor cell regulatory function can be due to the inactivation of
key regulators. Therefore, the proteasome inhibitor has become an
effective therapeutic approach of tumor treatment, particularly for
the treatment of hematological malignancies (18). For myeloma, the proteasome inhibitor
bortezomib has shown significant effectiveness and tolerance, while
the synergistic effect with various drugs has been shown (20). According to the present result, the
combination of TRAIL with the proteasome inhibitor may show a
synergistic effect in the TRAIL-resistant cell line U266, and the
resistance of U266 to TRAIL may be reversed theoretically.
As several pro- and anti-apoptosis proteins have
been screened by LC-MS/MS, their functions could be identified in
the following step to a more extensive verification of the proteins
screened by the present study. In addition, TRAIL receptors were
not identified by LC-MS/MS, which could be due to the limitation of
the mass spectrum, as the proteins with a low concentration could
not be identified (21).
In conclusion, the present study provides a feasible
method to explore targets and resistance mechanisms for drugs and
new information regarding rmhTRAIL on myeloma cells. According to
the results from the global proteomic-based approach, the possible
target proteins of rmhTRAIL on RPMI 8226 cells are CPNS1, PEF1,
BAP31, ASC, BAG2 and CBX3. While the possible resistance mechanism
of rmhTRAIL on U266 cells is the overexpression of UPP-related
proteins.
References
|
1
|
Pratt G, Jenner M, Owen R, Snowden JA,
Ashcroft J, Yong K, Feyler S, Morgan G, Cavenagh J, Cook G, et al:
Updates to the guidelines for the diagnosis and management of
multiple myeloma. Br J Haematol. 167:131–133. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Harousseau JL, Shaughnessy J Jr and
Richardson P: Multiple myeloma. Hematology (Am Soc Hematol Educ
Program). 2004:237–256. 2004.
|
|
3
|
Gazitt Y: TRAIL is a potent inducer of
apoptosis in myeloma cells derived from multiple myeloma patients
and is not cytotoxic to hematopoietic stem cells. Leukemia.
13:1817–1824. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Geng C, Hou J, Zhao Y, Ke X, Wang Z, Qiu
L, Xi H, Wang F, Wei N, Liu Y, et al: A multicenter, open-label
phase II study of recombinant CPT (Circularly Permuted TRAIL) plus
thalidomide in patients with relapsed and refractory multiple
myeloma. Am J Hematol. 89:1037–1042. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fang F, Wang AP and Yang SF: Antitumor
activity of a novel recombinant mutant human tumor necrosis
factor-related apoptosis-inducing ligand. Acta Pharmacol Sin.
26:1373–1381. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pan G, O'Rourke K, Chinnaiyan AM, Gentz R,
Ebner R, Ni J and Dixit VM: The receptor for the cytotoxic ligand
TRAIL. Science. 276:111–113. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pan G, Ni J, Yu G, Wei YF and Dixit VM:
TRUNDD, a new member of the TRAIL receptor family that antagonizes
TRAIL signalling. FEBS Lett. 424:41–45. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Marsters SA, Sheridan JP, Pitti RM, Huang
A, Skubatch M, Baldwin D, Yuan J, Gurney A, Goddard AD, Godowski P,
et al: A novel receptor for Apo2L/TRAIL contains a truncated death
domain. Curr Biol. 7:1003–1006. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mitsiades CS, Treon SP, Mitsiades N, Shima
Y, Richardson P, Schlossman R, Hideshima T and Anderson KC:
TRAIL/Apo2L ligand selectively induces apoptosis and overcomes drug
resistance in multiple myeloma: Therapeutic applications. Blood.
98:795–804. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gómez-Benito M, Martinez-Lorenzo MJ, Anel
A, Marzo I and Naval J: Membrane expression of DR4, DR5 and
caspase-8 levels, but not Mcl-1, determine sensitivity of human
myeloma cells to Apo2L/TRAIL. Exp Cell Res. 313:2378–2388. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hong D, Chen HX, Yu HQ, Wang C, Deng HT,
Lian QQ and Ge RS: Quantitative proteomic analysis of
dexamethasone-induced effects on osteoblast differentiation,
proliferation, and apoptosis in MC3T3-E1 cells using SILAC.
Osteoporos Int. 22:2175–2186. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Thomas PD, Campbell MJ, Kejariwal A, Mi H,
Karlak B, Daverman R, Diemer K, Muruganujan A and Narechania A:
PANTHER: A library of protein families and subfamilies indexed by
function. Genome Res. 13:2129–2141. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mi H, Guo N, Kejariwal A and Thomas PD:
PANTHER version: 6 Protein sequence and function evolution data
with expanded representation of biological pathways. Nucleic Acids
Res. 35(Database): D247–D252. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
von Mering C, Jensen LJ, Snel B, Hooper
SD, Krupp M, Foglierini M, Jouffre N, Huynen MA and Bork P: STRING:
Known and predicted protein-protein associations, integrated and
transferred across organisms. Nucleic Acids Res. 33:D433–D437.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
von Mering C, Jensen LJ, Kuhn M, Chaffron
S, Doerks T, Krüger B, Snel B and Bork P: STRING 7 - recent
developments in the integration and prediction of protein
interactions. Nucleic Acids Res. 35(Database): D358–D362. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Duiker EW, Mom CH, de Jong S, Willemse PH,
Gietema JA, van der Zee AG and de Vries EG: The clinical trail of
TRAIL. Eur J Cancer. 42:2233–2240. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mellier G, Huang S, Shenoy K and Pervaiz
S: TRAILing death in cancer. Mol Aspects Med. 31:93–112. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Adams J: Development of the proteasome
inhibitor PS-341. Oncologist. 7:9–16. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Melvin AT, Woss GS, Park JH, Waters ML and
Allbritton NL: Measuring activity in the ubiquitin-proteasome
system: From large scale discoveries to single cells analysis. Cell
Biochem Biophys. 67:75–89. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kubiczková L, Matějíková J, Sedlaříková L,
Kryukov F, Hájek R and Sevčíková S: Proteasome inhibitors in
treatment of multiple myeloma. Klin Onkol. 26:11–18. 2013.(In
Czech). View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Plowman JE: The proteomics of keratin
proteins. J Chromatogr B Analyt Technol Biomed Life Sci.
849:181–189. 2007. View Article : Google Scholar : PubMed/NCBI
|