Introduction
In mammalian cells, the only pathway for inositol
breakdown is via myo-Inositol oxygenase (MIOX) (1). MIOX is a non-heme iron enzyme, which
performs the conversion of myo-Inositol to glucuronic acid, the
first committed step in myo-Inositol catabolism (2,3). The
reaction involves oxidative cleavage of the bond between C6 and C1
to produce D-glucuronic acid. D-glucuronic acid is successively
converted to L-gulonate, 3-keto-L-gulonate, L-xylulose, xylitol,
D-xylulose and D-xylulose-5-phosphate, which then enters the
pentose phosphate cycle (4). MIOX is
predominantly expressed in the renal tubules, but is also observed
at lower levels in extra-renal tissues, where diabetic
complications are widely shown (3).
Altered inositol metabolism is implicated in a number of diabetic
complications, including cataracts, nephropathy, retinopathy and
neuropathy. MIOX enzymatic activity is proportional to its serum
glucose concentration (5). In
addition, there is evidence that a polymorphism (rs761745) in the
promoter region of the MIOX gene is associated with the
progress of type 1 diabetes mellitus (1,6).
The structure of the MIOX gene has been
characterized in humans and other mammals, including mice and rats,
and MIOX sequences for cows, wild boars, dogs, orangutans, rhesus
monkeys, common marmosets, bonobos, northern greater galagos, and
Tasmanian devils are reported in GenBank (2,4,7).
In humans, the MIOX gene is located on
chromosome 22 in band 22q13.3, and consists of a DNA coding
sequence (CDS) of 858 nucleotides (nt) split by 11 introns (exon
1,15 nt; exon 2,81 nt; exon 3,81 nt; exon 4,163 nt; exon 5,68 nt;
exon 6,110 nt; exon 7,68 nt, exon 8,50 nt; exon 9,113 nt; and exon
10,109 nt). The open reading frame (ORF) encodes a polypeptide of
285 amino acid residues with a mass of 33.01 kDa (8).
Although the function of the MIOX enzyme in human
physiology has been determined, there are few MIOX-associated
studies (8). Currently, there are gaps
in the genomic information of MIOX in experimental animal models;
thus, characterizing the physiological differences between humans
and other MIOX proteins is difficult. An animal model closely
associated with humans was selected in the current study. The
current study describes the gene encoding MIOX in the baboon
(Papio hamadryas). It is hypothesized that the baboon is an
ideal animal model due to the fact that baboons spontaneously
develop type 1 and 2 diabetes, which is considered to be a
physiological research advantage.
Materials and methods
Animal specimens and nucleic acid
isolation from biological samples
Samples from three male baboons were collected from
the following frozen archived tissues: Testis, kidney, heart,
omental fat, skeletal muscle, pancreas, mononuclear cells, liver
and hypothalamus. The samples were collected according to ethical
guidelines reviewed by the Institutional Animal Care and Use
Committee of the Texas Institute of Biomedical Research (TIBR) (San
Antonio, TX, USA). The animals had been housed at the Southwest
National Primate Research Center at TIBR and were sacrificed under
humanitarian conditions. All of the baboons were housed together
and fed ad libitum on standard low-fat chow (15% Monkey
Diet, 8715; Harlan Teklad Labs, Houston, TX, USA).
Total RNA was isolated from the tissue samples using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), according to the manufacturer's instructions.
The RNA samples were treated with DNaseI (Invitrogen; Thermo Fisher
Scientific, Inc.) for 10 min at 37°C to remove any remainder of
genomic DNA. The quality and integrity of the RNA were evaluated
using standard spectrophotometric and electrophoretic methods,
respectively.
Reverse transcription (RT) and
polymerase chain reaction (PCR) amplification
The RT reactions were developed with 1 µg total RNA
using random primers and a high-capacity cDNA RT kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.), according to the
manufacturer's instructions. To amplify the baboon MIOX cDNA with
PCR, 2X Taq PCR Master Mix (Qiagen, Inc., Valencia, CA, USA) was
used, 5 µl synthesized cDNA served as the template, and 10 pM of
each consensus primer (forward, 5′-TATAGGAGCCTGGCTGAGGA-3′ and
reverse, 5′-CAGGCCTAGGTCCAGCAG-3′), designed based on previously
reported, highly conserved primate MIOX sequences with the online
UniPrime2 tool http://uniprime.batlab.eu (9). The complete MIOX CDS was amplified (based
on the human MIOX gene sequence). The PCR thermal cycling program
included initial denaturation at 94°C for 3 min, followed by 28
cycles of denaturation at 94°C for 30 sec, annealing at 59°C for 35
sec, and elongation at 72°C for 45 sec, with a final elongation at
72°C for 6 min. Universal β-actin gene primers (Ambion; Thermo
Fisher Scientific, Inc.) were used to amplify the positive control.
All PCR amplification products were confirmed by electrophoresis in
agarose gel (1%) at 75 V for 35 min, stained with ethidium bromide
and visualized under UV light. To visualize ethidium
bromide-stained DNA gels we used a transilluminator (an ultraviolet
lightbox).
Molecular cloning and sequencing
The PCR products were cloned with the
TOPO® XL PCR Cloning kit with the
pCR®-XL-TOPO® 3.5 kb vector (Invitrogen;
Thermo Fisher Scientific, Inc.). Electrocompetent Top 10
Escherichia coli cells (Invitrogen; Thermo Fisher
Scientific, Inc.) were transformed with the ligation reactions,
according to the manufacturer's instructions. The cloning products
were sequenced with an ABI PRISM 3100 Genetic Analyzer (Applied
Biosystems Life Technologies) using the universal M13 primers and
BigDye® Terminator Cycle Sequencing Reagents (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The electropherograms
were analyzed with the GeneStudio Pro tool (GeneStudio, Inc.,
Suwanee, GA, USA). The DNA sequences of the MIOX transcripts were
used to deduce the amino acid sequences using the Transeq online
softwa http://www.ebi.ac.uk/Tools/st/emboss_transeq/
(10), and these were subsequently
aligned with other MIOX sequences using the ClustalW program
(11). All procedures were performed
with three clones of each amplicon to exclude artifacts.
Phylogenetic analysis
The data obtained from the sequencing analysis were
used as the target sequence in a Basic Local Alignment Search Tool
search to determine the identity. The sequences were subsequently
aligned using the ClustalW program http://www.ebi.ac.uk/Tools/msa/clustalw2 (11). The GenBank accession numbers of the
sequences used in the current study are presented in Table I. A phylogenetic tree was constructed
from the mRNA sequences using MEGA version 6 software (12) with the neighbor-joining method
(13) with 1,000 bootstrap
replications (14). To identify the
evolutionary forces underlying the divergence in the primate MIOX
genes, the synonymous (dS; encoded amino acid does not change) and
non-synonymous (dN; the encoded amino acid changes) substitutions
were quantified using the modified Nei-Gojobori method (15), and the MIOX CDs from apes, Old World
monkeys (OWMs) and New World monkeys (NWMs), with the lemur
counterpart sequence as the out-group. The hypothesis of positive
or adaptive evolution (dN>dS), purifying selection (dN<dS)
and neutrality (dN=dS) was then tested using a codon-based Z test
of selection (16), which is included
in the MEGA version 6 software (12).
P<0.05 was considered to indicate a statistically significant
difference.
 | Table I.Primates MIO-X sequences from NCBI
GenBank. |
Table I.
Primates MIO-X sequences from NCBI
GenBank.
| Species (scientific
name) | mRNA accession
no. |
|---|
| Apes |
|
|---|
| Human
(Homo sapiens) | NM_017584.5 |
| Pygmy
chimpanzee (Pan paniscus) | XM_003811021.1 |
| Gorilla
(Gorilla gorilla) | XM_004063692.1 |
| Sumatran
orangutan (Pongoa belii) | NM_001131282.2 |
| Old World
monkeys |
|
| Hamadryas
baboon (Papio hamadryas) | KP299165 |
| Olive
baboon (Papioanubis) | XM_003905754.1 |
| Gibbon
(Noma scusleucogenys) | XM_003281467.1 |
| Rhesus
monkey (Macaca mulatta) | XM_001116334.2 |
|
Crab-eating macaque | XM_005566896.1 |
| (Macaca
fascicularis) |
|
| New World
monkeys |
|
| Squirrel
monkey | XM_003932782.1 |
|
(Saimiri boliviensis
boliviensis) |
|
| Lemur
(out-group) |
|
| Galago
(Otolemur garnettii) | XM_003783084.1 |
Results
Molecular biology
A single band of the expected size (858 bp) was
amplified by RT-PCR from renal tissue cDNA. This amplicon was not
observed in the other tissue samples (testis, heart, omental fat,
skeletal muscle, pancreas, mononuclear cells, liver or
hypothalamus) that were analyzed. Thus, the MIOX transcript was
only observed in the kidney. The novel sequence was deposited in
the GenBank database (accession no. KP299165).
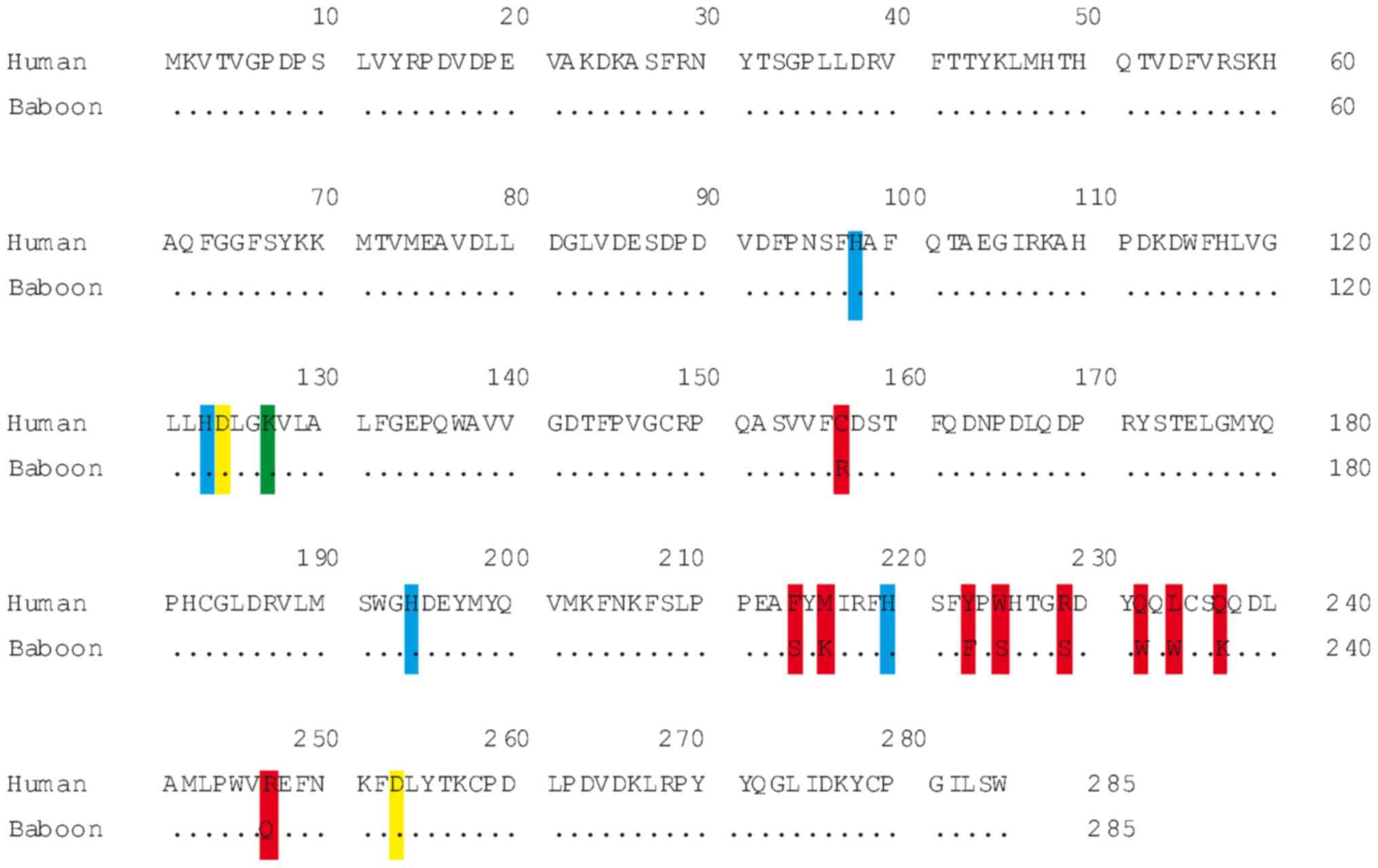
The nucleotide and amino acidic sequences of baboon
MIOX were compared with their human counterparts. The percentage
nucleotide similarity between the baboon and human genes was 95%
and the percentage amino acid identity between the proteins was
96%. In total, 10 amino acid changes were identified between the
two primate species. Thorsell et al (1) proposed a novel role for Lys127 (K127) in
the crystallized human MIOX protein in controlling access to the
diiron cluster. That position is conserved in the baboon enzyme as
predicted from the ORF. In our previous study, it was found that,
in the active site of the baboon MIOX enzyme, the diiron cluster is
bound in a conserved crevice formed between α4, α5, α6, α7 and α8
(17). This represents a strictly
conserved diiron cluster with a first coordination sphere of four
histidines (H98, H123, H194 and H220) and two aspartates (D124 and
D253) (1). All these positions are
conserved in the baboon MIOX protein (Fig.
1).
Phylogenetic analysis
Apes, OWMs and NWMs demonstrated a clear tendency
toward purifying selection (dN<dS; P=0.0001). A screening test
was performed using Fisher's exact test selection with the models
of Nei-Gojobori, Li-Wu-Luo, Pamilo-Bianchi-Li and Kumar. The same
results were obtained with all the models used (Table II).
 | Table II.Evolutionary forces that underlie the
process of divergence in the MIO-X primate genes. |
Table II.
Evolutionary forces that underlie the
process of divergence in the MIO-X primate genes.
| Species | dN | dS | P-valuea |
|---|
| Human | 0.0339 | 0.5298 | 0.0001 |
| Pygmy chimpanzee | 0.0347 | 0.5633 | 0.0001 |
| Gorilla | 0.0365 | 0.4986 | 0.0001 |
| Sumatran
orangutan | 0.0348 | 0.4868 | 0.0001 |
| Hamadryas baboon | 0.0390 | 0.5404 | 0.0001 |
| Olive baboon | 0.0348 | 0.5344 | 0.0001 |
| Gibbon | 0.0330 | 0.4781 | 0.0001 |
| Rhesus monkey | 0.0366 | 0.5402 | 0.0001 |
| Crab eating
macaque | 0.0339 | 0.5144 | 0.0001 |
| Squirrel monkey | 0.0434 | 0.4354 | 0.0001 |
Statistical analysis
Statistical and mathematical calculations made in
this work were performed using different algorithms of the MEGA
software. Although there are many mathematical algorithms, none is
better than another, they simply start from different assumptions.
These algorithms are known and accepted among the expert community
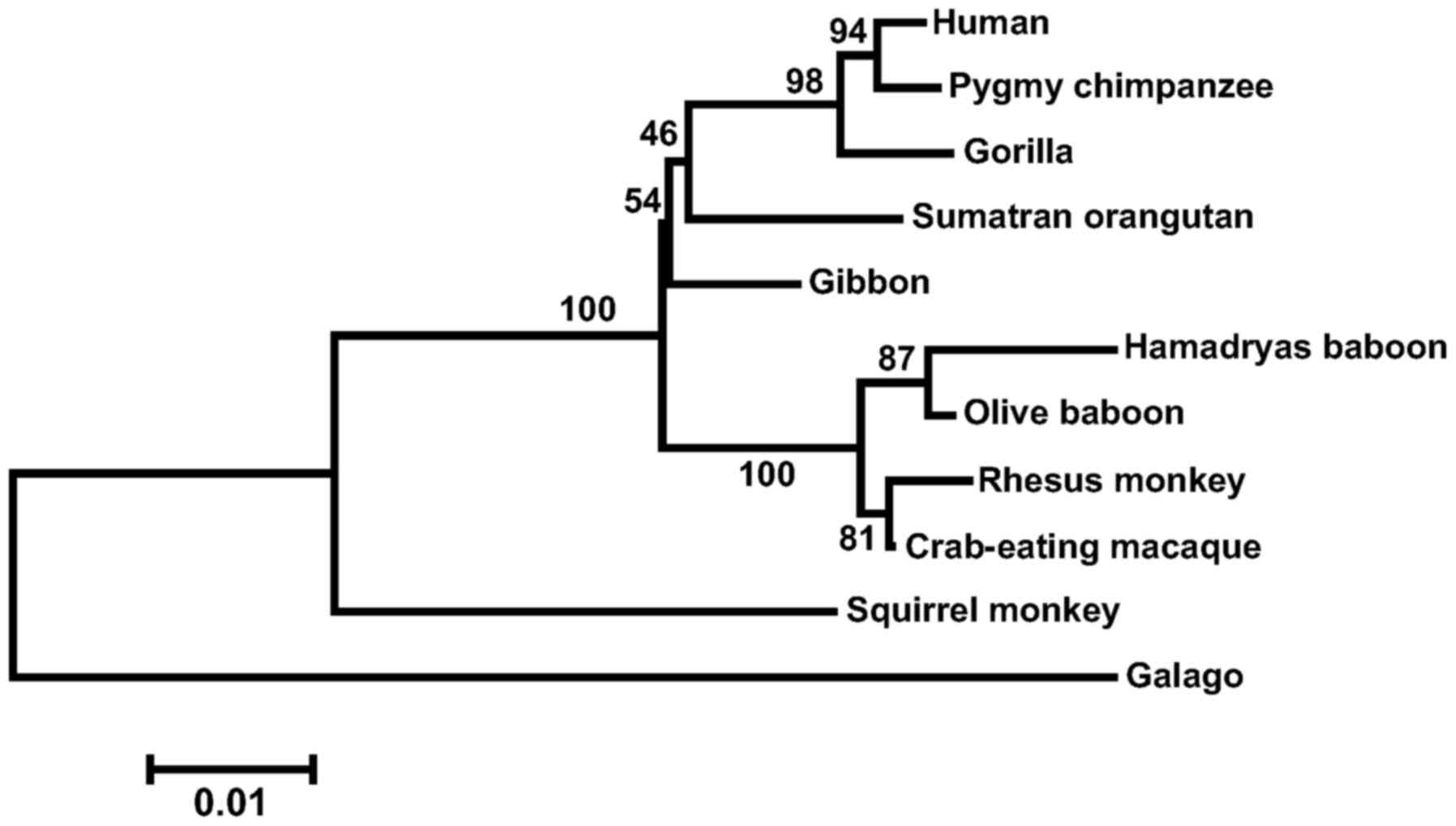
in the subject. The phylogenetic tree constructed from primate MIOX
sequences (Fig. 2) shows four
lineage-specific clades, corresponding to apes, OWMs, NWMs and
lemurs (out-group). It supports the orthology within the primate
MIOX genes. Bootstrap values are shown on the branches of the tree.
Approaching results were obtained with other phylogenetic
methods.
Discussion
Molecular biology
The MIOX sequence in baboons was observed to be
highly similar to that of other apes. The preservation of
exon-intron limits and the lack of evident mutations indicate that
the baboon MIOX gene is functional. To the best of our
knowledge, this is the first study that reports the expression
profile of the novel MIOX gene in different tissues from
baboons. The results of the current study indicate that, in
baboons, the MIOX gene is only expressed in the kidney,
while in humans, MIOX gene expression is observed in kidney
and extrarenal tissue (3). To
elucidate this difference, further studies are required, which
include promoter alignment, bioinformatics analyses (in order to
predict transcriptional factor binding sites) and searching CpG
islands that are susceptible to methylation. Conservation of the
structural amino acid (lysine 127), histidines (98, 123, 194 and
220) and aspartates (124 and 253) (1),
suggests that the three-dimensional shape may be identical in
baboon and human MIOX protein (Fig.
1). Kidney expression in the two species suggests that the
machinery, which regulates gene expression is similar in humans and
baboons. In addition, it indicates that the MIOX gene may
exert a similar physiological impact. However, further studies
under different physiological conditions, such as insulin
resistance, diabetes, obesity, fasting or other conditions
associated with metabolic syndrome, are required.
Phylogenetic analysis
Positive selection (dN>dS) means that
non-synonymous substitutions are functional and profitable to the
organism, and provide an evolutionary benefit. While purifying
selection (dN<dS) implies that evolutionary pressure eases off.
The dN and dS rates were estimated in the present study and it was
found that the evolutionary force that acts is purifying selection
(Table II). Thus, it was hypothesized
that purifying selection is an indication that the MIOX gene
is functional in the studied species, as functional family genes
fit this hypothesis. In conclusion, only one MIOX gene was
found for baboons and splicing variants were not detected.
Furthermore, MIOX gene expression was only identified in the
kidney. The dN and dS rates conclusively demonstrated that
MIOX genes are included in the hypothesis of purifying
selection.
Acknowledgements
The present study was financed by grants from the
Mexican Council of Science and Technology (CONACYT; grant nos.
167697 and 157965). The authors gratefully acknowledge the critical
reading of the manuscript by Sergio Lozano-Rodríguez.
References
|
1
|
Thorsell AG, Persson C, Voevodskaya N,
Busam RD, Hammarström M, Gräslund S, Gräslund A and Hallberg BM:
Structural and biophysical characterization of human myo-inositol
oxygenase. J Biol Chem. 283:15209–15216. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Arner RJ, Prabhu KS, Thompson JT,
Hildenbrandt GR, Liken AD and Reddy CC: Myo-Inositol oxygenase:
Molecular cloning and expression of a unique enzyme that oxidizes
myo-inositol and D-chiro-inositol. Biochem J. 360:313–320. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Arner RJ, Prabhu KS, Krishnan V, Johnson
MC and Reddy CC: Expression of myo-inositol oxygenase in tissues
susceptible to diabetic complications. Biochem Biophys Res Commun.
339:816–820. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Arner RJ, Prabhu KS and Reddy CC:
Molecular cloning, expression, and characterization of myo-inositol
oxygenase from mouse, rat, and human kidney. Biochem Biophys Res
Commun. 324:1386–1392. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nayak B, Xie P, Akagi S, Yang Q, Sun L,
Wada J, Thakur A, Danesh FR, Chugh SS and Kanwar YS: Modulation of
renal-specific oxidoreductase/myo-inositol oxygenase by
high-glucose ambience. Proc Natl Acad Sci USA. 102:17952–17957.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang B, Hodgkinson A, Millward BA and
Demaine AG: Polymorphisms of myo-inositol oxygenase gene are
associated with Type 1 diabetes mellitus. J Diabetes Complications.
24:404–408. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yang Q, Dixit B, Wada J, Tian Y, Wallner
EI, Srivastva SK and Kanwar YS: Identification of a renal-specific
oxido-reductase in newborn diabetic mice. Proc Natl Acad Sci USA.
97:9896–9901. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zeng LC, Liu F, Zhang X, Zhu ZD, Wang ZQ,
Han ZG and Ma WJ: hOLF44, a secreted glycoprotein with distinct
expression pattern, belongs to an uncharacterized olfactomedin-like
subfamily newly identified by phylogenetic analysis. FEBS Lett.
571:74–80. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Boutros R, Stokes N, Bekaert M and Teeling
EC: UniPrime2: A web service providing easier Universal Primer
design. Nucleic Acids Res. 37:W209–W213. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rice P, Longden I and Bleasby A: EMBOSS:
The European Molecular Biology Open Software Suite. Trends Genet.
16:276–277. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Thompson JD, Gibson TJ, Plewniak F,
Jeanmougin F and Higgins DG: The CLUSTAL_X windows interface:
Flexible strategies for multiple sequence alignment aided by
quality analysis tools. Nucleic Acids Res. 25:4876–4882. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tamura K, Stecher G, Peterson D, Filipski
A and Kumar S: MEGA6: Molecular Evolutionary Genetics Analysis
version 6.0. Mol Biol Evol. 30:2725–2729. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Saitou N and Nei M: The neighbor-joining
method: A new method for reconstructing phylogenetic trees. Mol
Biol Evol. 4:406–425. 1987.PubMed/NCBI
|
|
14
|
Efron B, Halloran E and Holmes S:
Bootstrap confidence levels for phylogenetic trees. Proc Natl Acad
Sci USA. 93:13429–13434. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nei M and Gojobori T: Simple methods for
estimating the numbers of synonymous and nonsynonymous nucleotide
substitutions. Mol Biol Evol. 3:418–426. 1986.PubMed/NCBI
|
|
16
|
Zhang J, Rosenberg HF and Nei M: Positive
Darwinian selection after gene duplication in primate ribonuclease
genes. Proc Natl Acad Sci USA. 95:3708–3713. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Thorsell AG, Persson C, Voevodskaya N,
Busam RD, Hammarström M, Gräslund S, Gräslund A and Hallberg BM:
Structural and biophysical characterization of human myo-inositol
oxygenase. J Biol Chem. 283:15209–15216. 2008. View Article : Google Scholar : PubMed/NCBI
|