Introduction
Alzheimer's disease (AD) is a dementia with a
complex pathogenesis. It is a slowly progressing neurodegenerative
disease, with clinical manifestations including progressive memory
loss, language and intellectual impairment, and the inability to
take care of oneself. It is estimated that the number of patients
with AD will exceed 78 million worldwide, in 2030 (World Alzheimer
Report 2021; https://www.alzint.org/resource/world-alzheimer-report-2021/).
The presence of extracellular β-amyloid (Aβ) deposits and the
intracellular accumulation of hyperphosphorylated tau are still the
main neuropathological criteria for the diagnosis of AD (1,2).
Although significant progress has been made in elucidating the key
pathogenetic mechanisms, the etiopathogenesis of AD is far from
clear. Alternative medicines based on natural herbs have garnered
attention in recent years due to their abundance, diversity, low
toxicity, effectiveness, and low cost. Chinese herbal medicines
with a wide range of pharmacological activities have become
prospective interventions for the prevention and treatment of human
diseases (3).
Tianma [Gastrodia elata Blume, (GEB)] is an
exceptional traditional herb. The GEB dry tuber is featured in the
2015 edition of the Chinese Pharmacopoeia (4). Recent studies have revealed that GEB
and its active components have diverse pharmacological effects on
Parkinson's disease (5), AD
(6,7), and improving ischemia-reperfusion
injury (8). The water extract of
GEB can be used as a food additive to promote the neuroregenerative
process (9). It inhibits the
production of stress-related proteins and activates the
neuroprotective genes in humans (10). GEB also controls
chaperone/proteasome degradation pathways in SH-SY5Y cells, such as
the production of CALR and HSP70/90 proteins, to enhance synapse
regeneration, thereby playing a role in neuroprotection (11). Gastrodin, the major bioactive
component in GEB, was demonstrated to ameliorate memory deficits in
mice by inhibiting the expression of β-site APP-cleaving enzyme 1
(BACE1) protein and reducing the levels of eIF2α and phosphorylated
PKR in cells (12). Furthermore,
gastrodin significantly improved cell viability in a cell culture
model of AD induced by Aβ25-35 and reduced the release of lactate
dehydrogenase (LDH), thereby protecting neurons from Aβ toxicity
(13). p-Hydroxybenzyl alcohol is
another key component in GEB. p-Hydroxybenzyl alcohol was revealed
to reduce the accumulation of reactive oxygen species in
Caenorhabditis elegans (C. elegans) and decrease Aβ
plaque deposition (14). There are
numerous studies on GEB and its active components (15,16),
however the underlying mechanisms of action remain unclear due to
the complexity of bioactive compounds in traditional Chinese
medicine (TCM). It is therefore necessary to further study the
therapeutic potential of GEB in AD.
Network pharmacology is a useful tool for
identifying the bioactive substances in TCM formulations. It can
also reveal the mechanisms that underlie the effects of TCM
(17) and provide insight into the
overall regulatory mechanism of the ‘component-target-pathway’.
Using the network pharmacology approach, numerous studies have
successfully investigated the mechanisms of action of active
components in various Chinese herbal medicines, and have identified
drugs with the potential to treat diseases. For example,
Yizhiqingxin formula may exert its neuroprotective effect in AD via
the PI3K/Akt signaling pathway. Polygonatine A, polygonatine C and
4',5-dihydroxyflavone were demonstrated to significantly increase
the survival rate and calcium levels in a PC12 cell model of
amyloid β toxicity, and exhibited potential in preventing and
treating AD (18,19). Network pharmacology has been
gradually applied to the study of TCM components. Whole-formula,
multi-component and multi-target analyses can help optimize
compound prescription, according to the tenets of TCM (20). The Gene Expression Omnibus (GEO) is
a global database of high-quality gene expression data, with
expansive information on gene expression profiles, including
significant differences in expression between diseased tissues and
normal tissues (18). Therefore, in
the present study, network pharmacology was combined with the GEO
database to investigate the mechanism of action of GEB in the
treatment of AD.
A component-target-gene network was first
constructed to explore the underlying molecular mechanisms. The
core network and targets of GEB were then identified using a
protein-protein interaction (PPI) network, and the key pathways
were clarified using Gene Ontology (GO) and Kyoto Encyclopedia of
Genes and Genomes (KEGG) enrichment analyses. Finally, molecular
docking and reverse transcription-quantitative polymerase chain
reaction (RT-qPCR) experiments were used to verify the association
between key targets and components.
Materials and methods
Network pharmacology analysis.
Analysis of active components
The active components of GEB were obtained by
searching for ‘TIAN MA’ or ‘Gastrodia elata BI’ in the
Encyclopedia of Traditional Chinese Medicine (ETCM; http://www.tcmip.cn/ETCM/index.php/Home/Index/),
Bioinformatics Analysis Tool for Molecular mechANisms of
Traditional Chinese Medicine (BATMAN-TCM; http://bionet.ncpsb.org.cn/batman-tcm/) and literature
from China National Knowledge Infrastucture (CNKI). Subsequently,
using the PubChem database (http://pubchem.ncbi.nlm.nih.gov/), the obtained active
components were converted to Canonical SMILES format and imported
into the SwissTargetPrediction database (http://swisstargetprediction.ch/) (21) with species set to ‘Homo sapiens’ to
acquire the targets of the active components of GEB.
Obtaining gene targets related to
AD
GeneCards (https://www.genecards.org/), OMIM (https://omim.org/), CTD (http://ctdbase.org/) and DisGeNET (http://www.disgenet.org/) databases (22,23)
were used to screen potential targets associated with AD by
searching for the keyword ‘Alzheimer disease’. The results from the
four databases were integrated and deduplicated for the following
analyses.
Construction of the drug-active
component-target-AD network
Limma (version 3.44.3) (24) R package was used to screen for
differential genes (DEGs) between 10 AD and 13 control samples in
GSE5281(18) with |log2FC|>1 and
adjusted P-value <0.05. The potential therapeutic targets for
the anti-AD effects of GEB were obtained by overlapping targets of
active components, AD-related targets and DEGs. The drug-active
component-target-AD network was then constructed and visualized
using Cytoscape software (version 3.2.1) (25).
Construction of a PPI network
Next, potential therapeutic targets were input into
STRING database (https://string-db.org). In order to build a PPI
network with a confidence level >0.4, Cytoscape software
(version 3.2.1) was used to visualize the PPI network and calculate
the degree of each node in the PPI network to obtain the core
target. Core targets were defined as the top five nodes with the
highest degrees. Heat maps were created using the Pheatmap (version
0.7.7) (26) package in R for
potential targets.
GO and KEGG enrichment analyses
ClusterProfiler (version 3.18.0) (27) R package was used for GO and KEGG
enrichment analyses of GEB-AD-related core targets. The GO system
contains three main categories including biological process (BP),
cellular component (CC) and molecular function (MF). An adjusted
P-value of <0.05 was considered to indicate significant
enrichment.
Molecular docking
Core targets acting as large molecular receptors,
and the corresponding active compounds, acting as small molecule
ligands, were selected for molecular docking analysis. The 3D
structures of the core targets were downloaded from the PDB
database (https://www.rcsb.org/). The 2D
structures of the corresponding active ingredients were downloaded
from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). The raw files of
core targets and the corresponding active components were processed
with the AutoDock (version 4.2) (28) tool and converted to PDBQT format for
molecular docking. Binding activity was expressed as binding
energy; the lower the bonding energy, the more stable the docking
module (29). In general, a docking
energy <-5 kcal/mol indicates good docking (30).
Experimental verification.
Materials
4,4'-Dihydroxydiphenyl methane (DM) and
protocatechuic aldehyde (PA) were purchased from Chengdu Alfa
Biotechnology Co., Ltd. (purity ≥98%). Other reagents included
RNAsimple Total RNA Kit (cat. no. DP419; Tiangen Biotech Co.,
Ltd.), GoScript™ Reverse Transcription System (cat. no. A5000;
Promega Corporation), GoTaq® qPCR Master Mix (cat. no.
A6001; Promega Corporation). Transgenic C. elegans strains
CL4176 [dvIs27 (myo-3p::Aβ1-42::let-851 3'UTR)
+ rol-6 (su1006)] and CL2006 [dvIs2 (pCL12
(unc-54/human Aβ1-42 minigene) + pRF4)], and Escherichia
coli OP50 were obtained from the Caenorhabditis Genetics Center
(CGC) of the University of Minnesota (Minneapolis, USA). The
network pharmacology workflow is presented in Fig. 1.
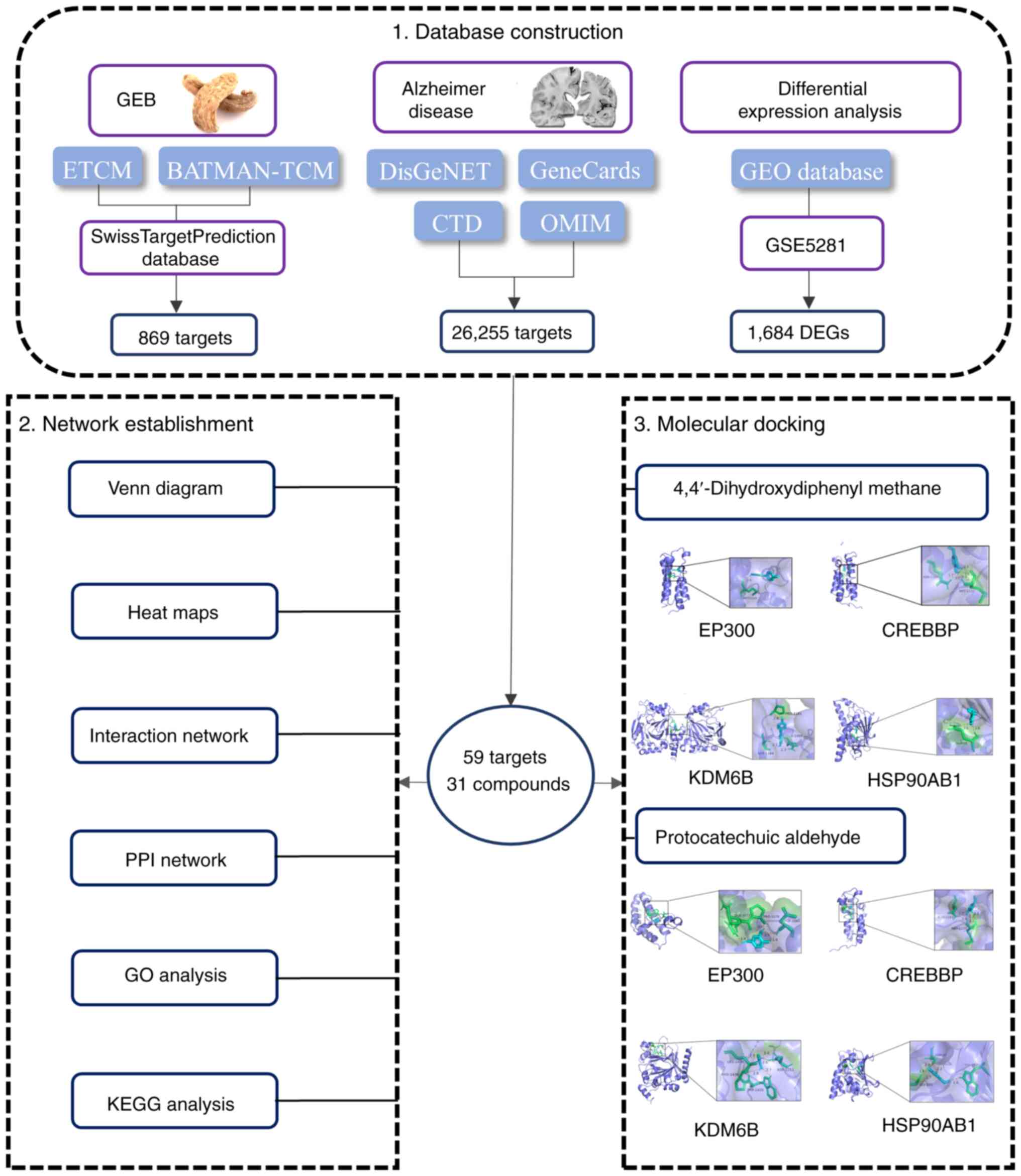 | Figure 1Flowchart of network pharmacology and
molecular docking. GEB, Gastrodia elata Blume; GEO, Gene
Expression Omnibus; ETCM, Εncyclopedia of Τraditional Chinese
Μedicine; BATMAN-TCM, Bioinformatics Analysis Tool for Molecular
mechANisms of Traditional Chinese Medicine; CTD, Comparative
Toxicogenomics Database; OMIM, Online Mendelian Inheritance in Man;
DEGs, differentially expressed genes; PPI, protein-protein
interaction; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes
and Genomes. |
C. elegans strains and drug
treatment
The strain CL2006 was propagated at 20˚C, and the
strain CL4176 was propagated at 16˚C. All C. elegans strains
were routinely propagated on solid nematode growth medium (NGM;
consisting of 300 ml deionized water including 0.82 g peptone,
Oxoid Limited; Thermo Fisher Scientific; 1.2 g NaCl, Tianjin
Fengchuan Chemical Reagent Co., Ltd.; 5.15 g agar, Beijing Solarbio
Science & Technology Co., Ltd.; autoclaved and supplemented 1
mM MgSO4, 1 mM CaCl2, Tianjin Fengchuan Chemical Reagent Co., Ltd.;
12.9 mM cholesterol solution, MACKLIN) with E. coli OP50 as
a food source. C. elegans was treated with sodium
hypochlorite, and synchronized larvae (L1 stage) were obtained
after eggs were incubated overnight in M9 buffer (containing 22 mM
KH2PO4, 42 mM Na2HPO4, autoclaved and supplemented with 1 mM MgSO4;
all the reagents were from Tianjin Fengchuan Chemical Reagent Co.,
Ltd.) for experiment. Transgenic C. elegans strains CL2006
and CL4176 were used as AD animal models. C. elegans were
randomly divided into control and drug pretreatment groups. The
preparation was dissolved in 0.1% DMSO and diluted with ultrapure
water to the appropriate concentration. Based on a previous study
(31), various concentrations of DM
(0.125, 0.25, 0.5, 1, 2 mM) and PA (0.125, 0.25, 0.5, 1 mM) were
used to assess safety and efficacy in C. elegans.
Paralysis assay
CL4176, a temperature-sensitive transgenic model,
has been demonstrated to inactivate the smg-1 system when the
temperature increases from 16˚C to 25˚C, which leads to
overexpression of Aβ in muscles and a paralysis phenotype. CL4176
nematodes synchronized to the L1 stage were transferred to 35 mm
culture plates with or without OP50 and drugs, cultured at 16˚C for
36 h and then at 25˚C for transgene induction. After 24 h of
culture at 25˚C, the paralyzed worms were observed every 2 h. When
they could not move or respond to platinum wire stimulation, they
were considered paralyzed (32).
Fluorescence staining of Aβ
deposits
Transgenic CL2006 worms synchronized to the L4 stage
(early adult stage) were inoculated on NGM plates with or without
drugs, and N2 was used as a negative control for Aβ deposition
(32). Following incubation at 20˚C
for 48 h, the worms were collected with M9 buffer and fixed in 4%
paraformaldehyde/PBS (pH 7.4) (cat. no. BL539A; Biosharp Life
Sciences) at 4˚C for 24 h. The worms were then treated with 5%
β-mercaptoethanol (cat. no. M828395; Macklin, Inc.), 1% Triton
X-100 (cat. no. MB2486; Dalian Meilun Biology Technology Co., Ltd.)
and 125 mM Tris (pH 7.4) (cat. no. T8060; Beijing Solarbio Science
& Technology Co., Ltd.) at 37˚C for 24 h. The samples were
stained with 0.125% thioflavin S (cat. no. S19293; Shanghai Yuanye
Bio-Technology Co., Ltd.) in 50% ethanol at room temperature for 2
min, and then with 50% ethanol. The samples were thereafter rinsed
and transferred to a glass slide for observation under a laser
scanning confocal microscope (LSM900; Carl Zeiss AG). The number of
thioflavin S-reactive deposits in the area anterior to the
pharyngeal bulb was counted.
RNA extraction and quantitation
Synchronized L1 stage CL4176 nematodes were
transferred to NGM medium with or without drugs (~500 nematodes in
each plate) and cultured at 15˚C for 36 h, and then at 25˚C for a
further 36 h. The worms were collected in an EP tube with M9
buffer, rinsed, and the supernatant was discarded (33). Total RNA was extracted using
RNAsimple Total RNA Kit, and the concentration and purity were
measured with an ultra-micro spectrophotometer (SMA6000; Merinton
Instrument, Inc.). cDNA was obtained by reverse transcription using
GoScript™ Reverse Transcription System and a thermocycler (Veriti
Applied Biosystems; Thermo Fisher Scientific, Inc.), and target
mRNA was quantified using GoTaq® qPCR Master Mix and
real-time PCR (C1000 Touch PCR; Bio-Rad Laboratories, Inc.).
Reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) was performed under the following operating conditions:
95˚C for 10 min, followed by 40 cycles of 95˚C for 15 sec, 55˚C for
30 sec and 72˚C for 30 sec. The reaction was then cooled and
maintained at 4˚C. β-actin was used as a housekeeping gene.
Relative gene expression was calculated using the 2-ΔΔCq
method (34), and each reaction was
repeated three times. The primers used in this assay are listed in
Table SI.
Statistical analysis
GraphPad Prism software 8.0 (GraphPad Software,
Inc.) was used for statistical analyses. All data are presented as
the means ± SD. All experiments were repeated three times. Curves
were plotted using Kaplan-Meier survival curves and statistical
analyses of differences between groups in the paralysis assays were
performed using the log-rank test. In addition, unpaired Student's
t-test was used to compare differences between two groups.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Prediction of components and targets
based on network pharmacology. Analysis of active components
A total of 21 and 23 active components in GEB were
obtained from the ETCM and BATMAN-TCM databases, respectively.
After integrating and de-duplicating the data, 34 active components
were obtained. After removing three active components
(bis(4-hydroxybenzyl)ether mono-β'-d-glucopyranoside,
tris-[4-(β'-d-glucopyranosyloxy)benzyl]citrate and vanillyl
alcohol) with no canonical SMILES format, the remaining 31 active
components (Table I) were imported
into the SwissTargetPrediction database. A total of 869 targets of
the active components of GEB were obtained. By mining the
GeneCards, OMIM, CTD and DisGeNET databases, and after removing
duplicate data, a total of 26,255 AD-related targets were
obtained.
 | Table IComponents of GEB for analysis. |
Table I
Components of GEB for analysis.
| No. | Compounds | Molecular
weight | Molecular
formula |
|---|
| 1 | Sitosterol | 414.707 | C29H50O |
| 2 | Vanillin | 152.147 | C8H8O3 |
| 3 |
20-Hexadecanoylingenol | 586.94 | C36H58O6 |
| 4 |
4-(4'-Hydroxybenzyloxy)benzyl methyl
ether | 244.286 | C15H16O3 |
| 5 |
4,4'-Dihydroxydiphenyl methane | 200.233 | C13H12O2 |
| 6 |
4-Ethoxymethylphenyl-4'-hydroxybenzylether | 258.312 | C16H18O3 |
| 7 | 4-Hydroxybenzyl
alcohol | 124.137 | C7H8O2 |
| 8 | 4-Hydroxybenzyl
methyl ether | 138.164 | C8H10O2 |
| 9 |
4-Hydroxybenzylamine | 123.152 | C7H9NO |
| 10 |
7-Hydroxybiopterin | 253.22 | C9H11N5O4 |
| 11 | Vanillin
acetate | 212.2 | C10H12O5 |
| 12 |
Bis(4-Hydroxybenzyl)ether | 230.259 | C14H14O |
| 13 | Cetylic Acid | 256.42 | C16H32O2 |
| 14 | Citric Acid | 192.124 | C6H8O7 |
| 15 | Citronellal | 154.249 | C10H18O |
| 16 | Daucosterol | 576.847 | C35H60O6 |
| 17 | Dauricine | 624.766 | C38H44N2O6 |
| 18 |
Ethoxysanguinarine | 377.390 | C22H19NO5 |
| 19 | γ-Sitosterol | 414.7 | C29H50O |
| 20 | Gastrodamine | 245.274 | C14H15NO3 |
| 21 | Gastrodin | 286.278 | C13H18O7 |
| 22 | Gaultheroside
A | 552.6 | C27H36O12 |
| 23 | M-Hydroxybenzoic
acid | 138.121 | C7H6O3 |
| 24 |
P-Hydroxybenzaldehyde | 122.121 | C7H6O2 |
| 25 | P-Hydroxybenzyl
alcohol | 124.137 | C7H8O2 |
| 26 | P-Hydroxybenzyl
ethyl ether | 152.190 | C9H12O2 |
| 27 | Protocatechuic
aldehyde | 138.120 | C7H6O3 |
| 28 | Succinic Acid | 118.088 | C4H6O4 |
| 29 | Suchilactone | 368.380 | C21H20O6 |
| 30 | Sucrose | 342.296 | C12H22O11 |
| 31 | Suffruticoside
A | 612.5 | C27H32O16 |
Obtaining gene targets related to
AD
To obtain more robust AD-related targets, a total of
1,684 DEGs between AD and control samples were identified, among
which 975 were upregulated and 709 were downregulated. The red dots
represent upregulated genes, the green dots represent downregulated
genes, and the gray dots represent genes with no significant
difference (Fig. S1). Thereafter,
by overlapping 869 targets of the active components of GEB (26,255
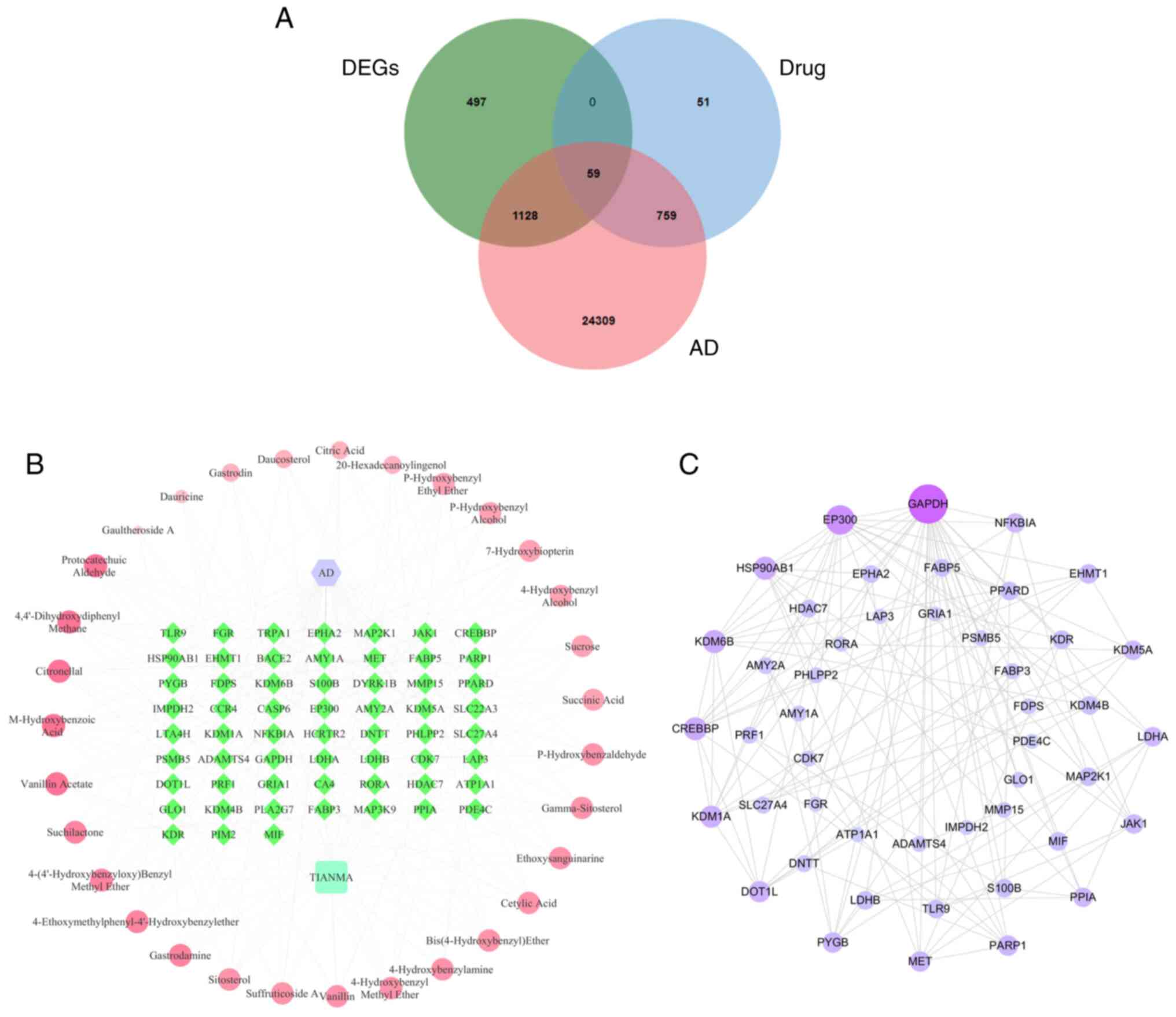
AD-related targets and 1,684 DEGs), a total of 59 potential targets
of the anti-AD therapeutic effects of GEB were obtained (Fig. 2A and Table SII). The heat map shows the
expression of 59 potential therapeutic targets in AD and control
samples (Fig. S2). Each square
represents a gene. The greater the expression, the darker the color
(red, upregulated; blue, downregulated). Each row represents the
expression of each gene in different samples; the column represents
expression of all genes in each sample.
Construction of the drug-active
component-target-AD network
Based on these results, a drug-active
component-target-AD network with 92 nodes (AD, TIANMA, 31 active
compounds, and 59 target genes) and 269 edges (Fig. 2B) was constructed. Furthermore, to
investigate the interactions of the 59 therapeutic targets of GEB,
a PPI network was constructed using the STRING database. The PPI
network was composed of 47 nodes and 119 edges, revealing that
therapeutic targets had a strong and complex association with each
other. The color of the nodes, ranging from blue to purple,
indicates the degree, ranging from small to large (Fig. 2C). GAPDH, EP300, HSP90AB1, KDM6B and
CREBBP had the highest degrees, and where therefore considered to
be core targets and were used for molecular docking analysis.
GO and KEGG enrichment analyses
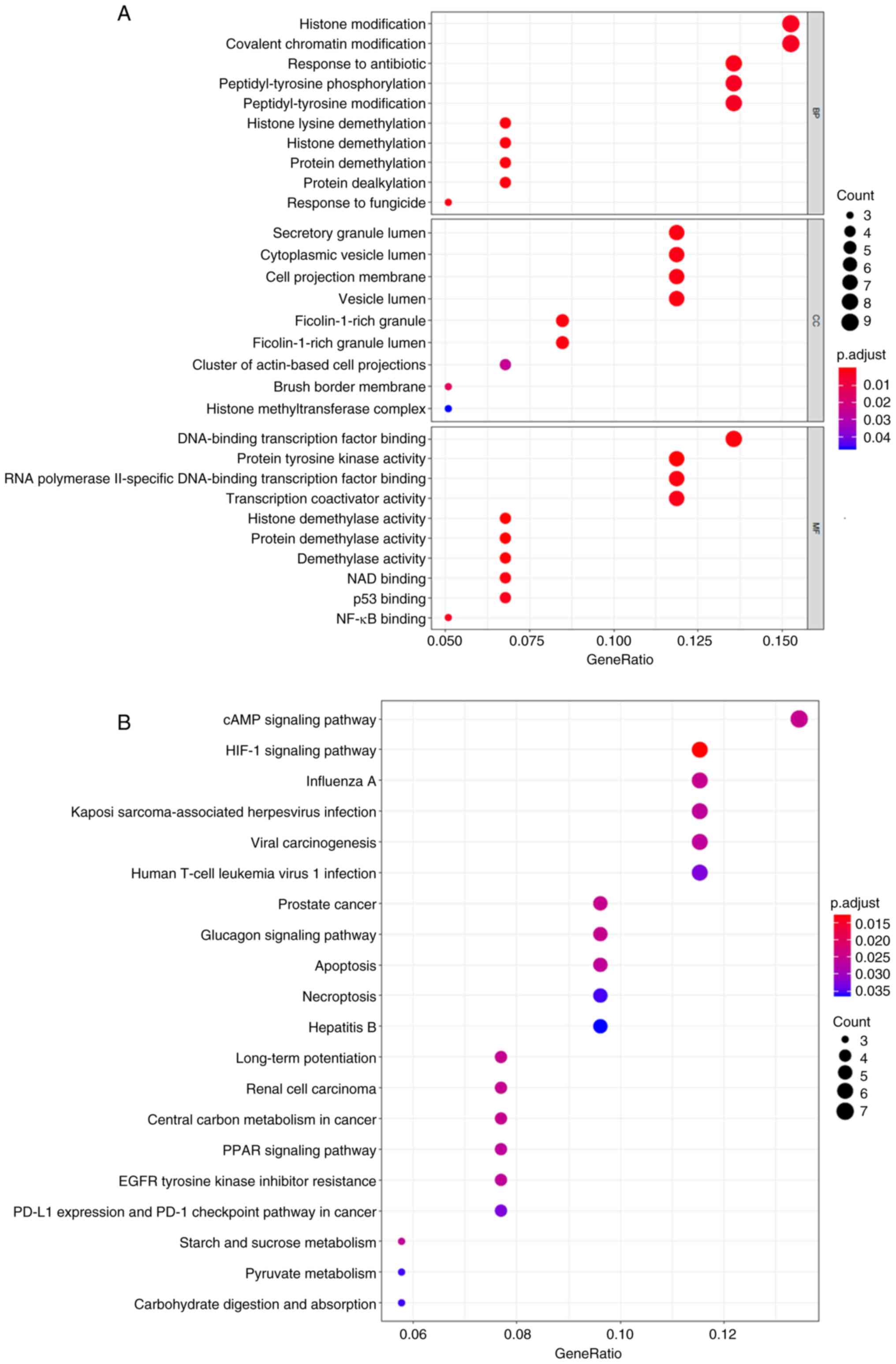
Next, the biological functions of the 59 potential
therapeutic targets were explored, which were significantly
enriched in the 163 BP, 9 CC, 38 MF and 21 KEGG pathways. The top
10 BP, MF and CC pathways were involved in gene transcription, such
as ‘histone demethylation’, ‘histone modification’, ‘covalent
chromatin modification’, ‘DNA-binding transcription factor
binding’, ‘transcription coactivator activity’ and ‘NAD binding’
(Fig. 3A). The top 10 enriched KEGG
pathways were closely associated with neurodegeneration and
neuroendocrinology, including the ‘cAMP signaling pathway’, ‘HIF-1
signaling pathway’ and ‘Glucagon signaling pathway’ (Fig. 3B and Table SIII).
Results of molecular docking
As mentioned above, GAPDH, EP300, HSP90AB1, KDM6B
and CREBBP may be the core potential targets of GEB in the
treatment of AD. GAPDH is an internal reference, and was therefore
not considered further. DM and PA, two components which are at the
forefront in the network diagram were investigated. These two
components, with high moderate values in the network diagram, were
matched with four core protein targets, and their docking potential
was studied. The binding energies were between -8.12 and -5.04
kcal/mol (Table II), exhibiting
good docking activity (the more negative the value, the stronger
the molecular docking). Both compounds interact with the four
target proteins mainly through hydrogen bonds.
 | Table IIAffinity score of potential targets
and compounds by molecular docking. |
Table II
Affinity score of potential targets
and compounds by molecular docking.
| Target | Compound | Affinity
(kcal/mol) |
|---|
| EP300 |
4,4'-Dihydroxydiphenyl methane | -7.69 |
| KDM6B |
4,4'-Dihydroxydiphenyl methane | -8.04 |
| HSP90AB1 |
4,4'-Dihydroxydiphenyl methane | -8.83 |
| CREBBP |
4,4'-Dihydroxydiphenyl methane | -8.12 |
| EP300 | Protocatechuic
aldehyde | -5.49 |
| KDM6B | Protocatechuic
aldehyde | -5.04 |
| HSP90AB1 | Protocatechuic
aldehyde | -5.93 |
| CREBBP | Protocatechuic
aldehyde | -5.41 |
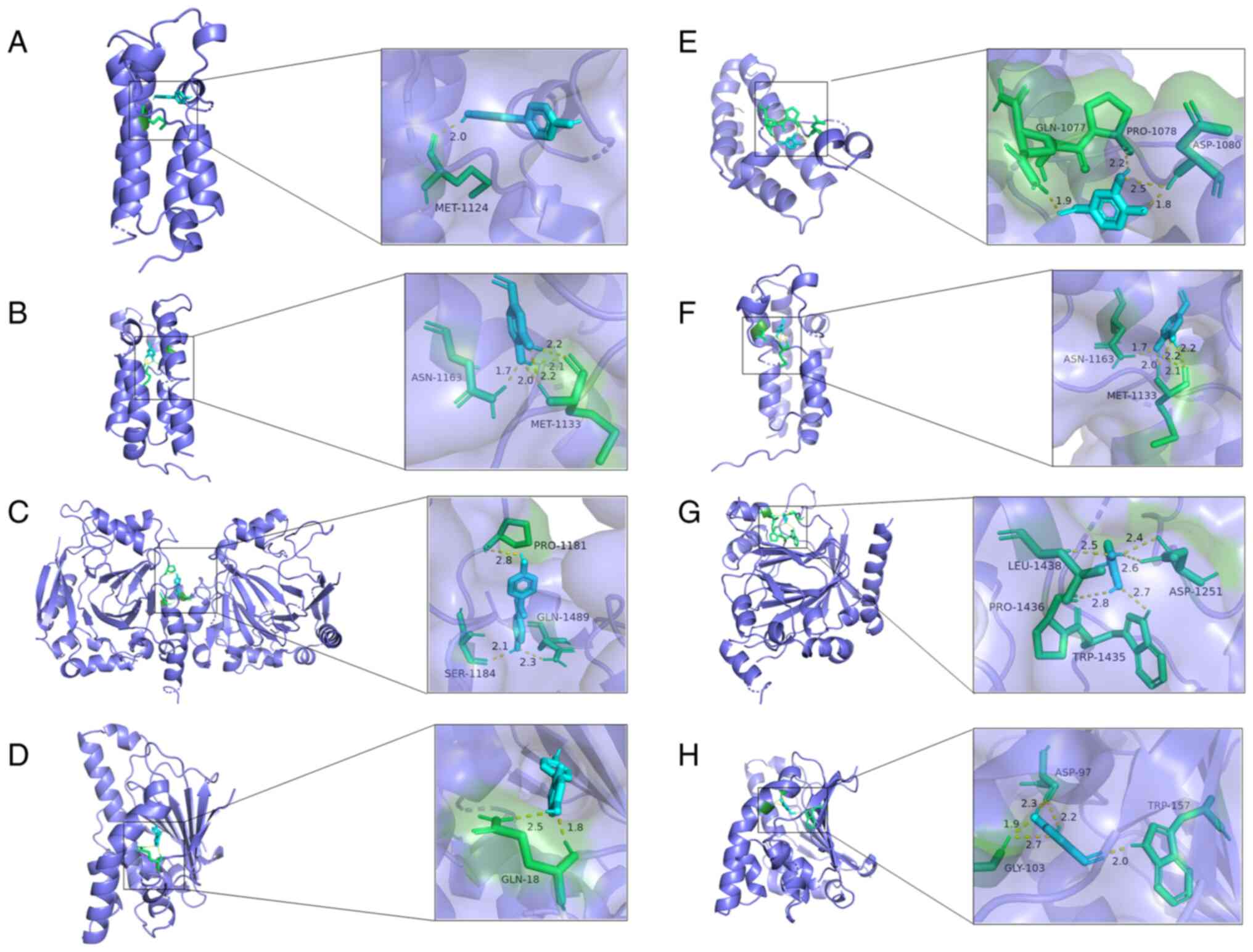
The results revealed that DM forms a hydrogen bond
with MET-1124 on EP300 (Fig. 4A).
DM forms five hydrogen bonds with ASN-1163 and MET-1133 on CREBBP
(Fig. 4B). DM forms three hydrogen
bonds, with GLN-1489, SER-1184 and PRO-1181, on KDM6B (Fig. 4C). DM forms two hydrogen bonds, with
GLN-18, on HSP90AB1 (Fig. 4D). PA
forms four hydrogen bonds, with GLN-1077, ASP-1080 and PRO-1078, on
EP300 (Fig. 4E). PA forms five
hydrogen bonds, with ASN-1163 and MET-1133, on CREBBP (Fig. 4F). PA forms five hydrogen bonds,
with LEU-1438, ASP-1251, TRP-1435 and PRO-1436, on KDM6B (Fig. 4G). PA forms five hydrogen bonds,
with GLY-103, ASP-97 and TRP-157, on HSP90AB1 (Fig. 4H).
Effect of GEB on transgenic C.
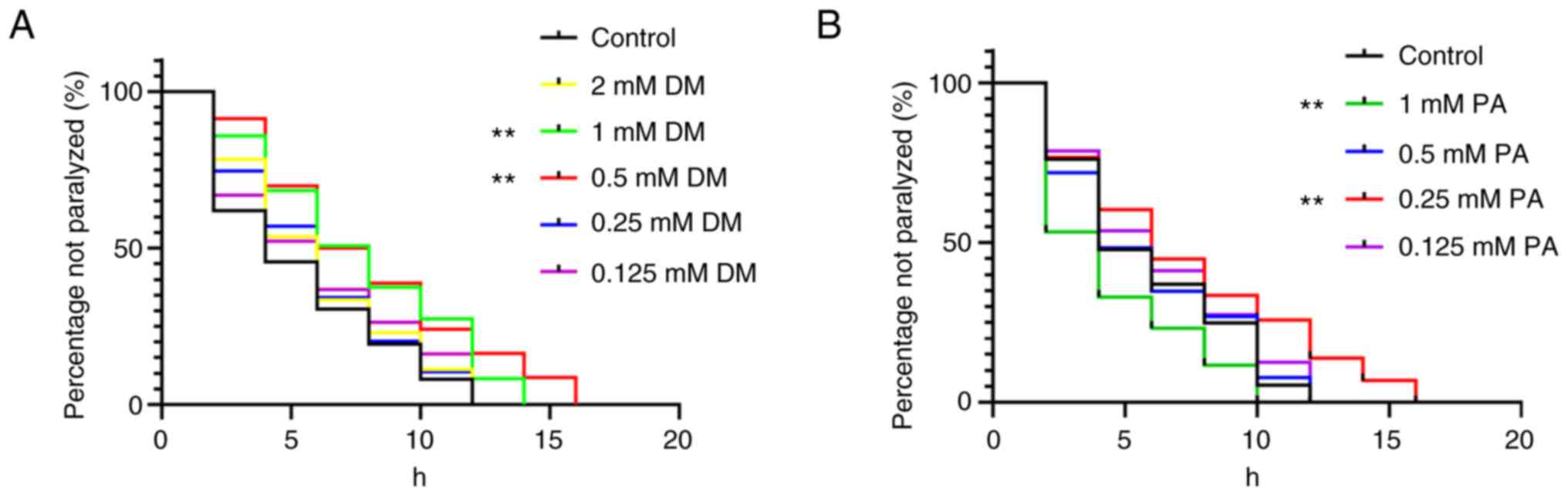
elegans. Effects of DM and PA on paralysis in C. elegans
DM (0, 0.125, 0.25, 0.5, 1, 2 mM) and PA (0, 0.125,
0.25, 0.5 mM) had no obvious toxicity. When the concentration of DM
exceeded 0.5 mM, the effect of delaying C. elegans paralysis
exhibited a downward trend, and 0.5 mM DM exerted the most marked
effect (P<0.01) (Fig. 5A). When
the concentration of PA exceeded 0.5 mM, it displayed a toxic
effect; a PA concentration of 0.25 mM exhibited the most marked
therapeutic effect (P<0.01) (Fig.
5B). Therefore, DM and PA have potential anti-AD activity.
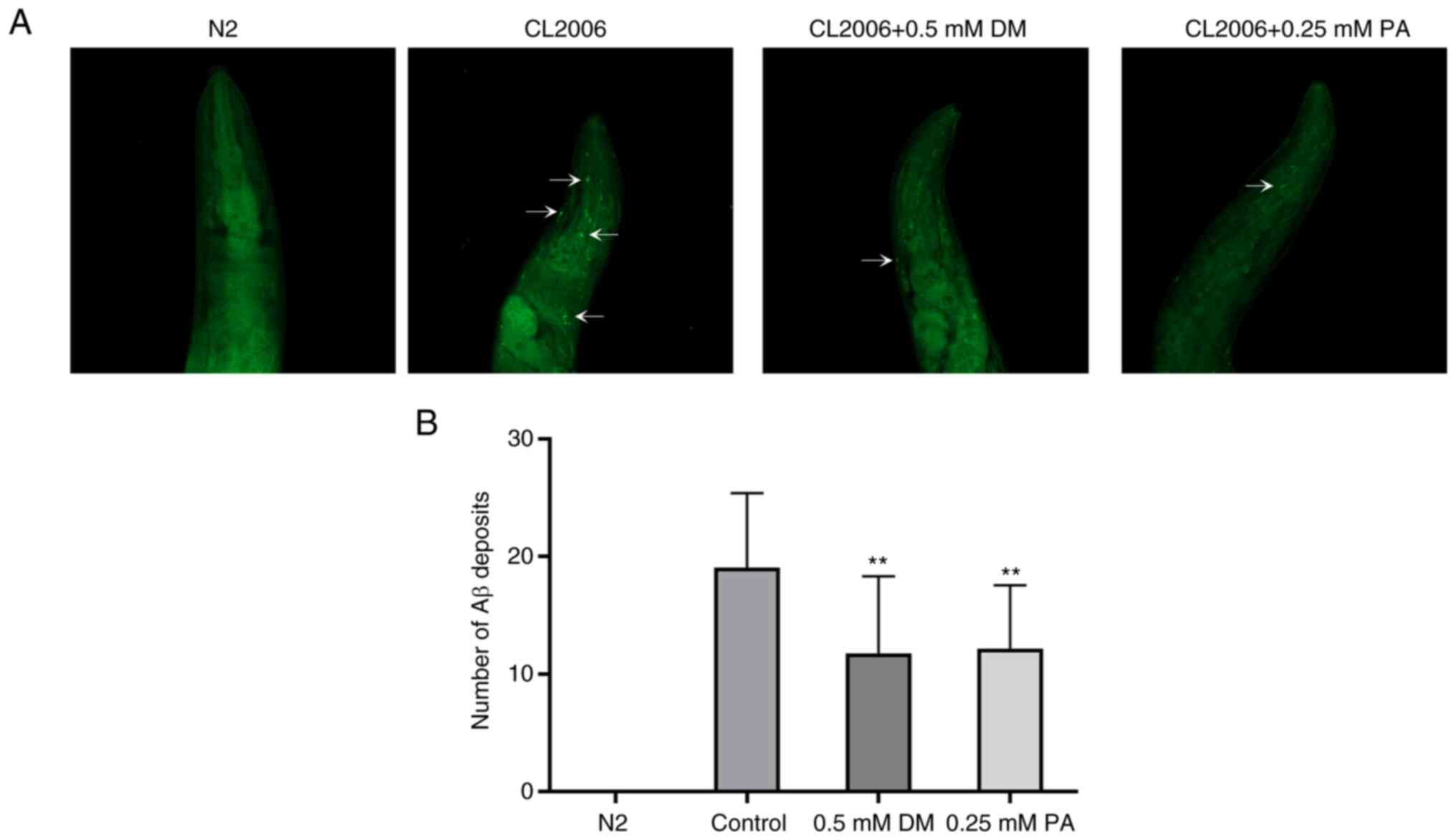
DM and PA inhibit the deposition of Aβ
in transgenic C. elegans
In order to study the effects of DM and PA on the
aggregation of Aβ, the formation of amyloid fibrils was observed
using the thioflavin-S fluorescence method. The transgenic strain
CL2006 used in this experiment expressed the Aβ protein fragment
related to the development of AD. These worms showed expression and
aggregation of Aβ in muscle, leading to progressive paralysis.
CL2006 was stained with triterpenes for Aβ at the end of DM and PA
treatment (green fluorescent labeling). The fluorescence image of
the head area of CL2006 nematodes showed that compared with
untreated worms (negative control), Aβ deposition in those treated
with 0.5 mM DM or 0.25 mM PA significantly decreased. The wild N2
strain exhibited no Aβ deposition in any part of the animal
(Fig. 6A). These results indicated
that both DM and PA inhibit the aggregation and deposition of Aβ in
muscle cells of transgenic worms (Fig.
6B).
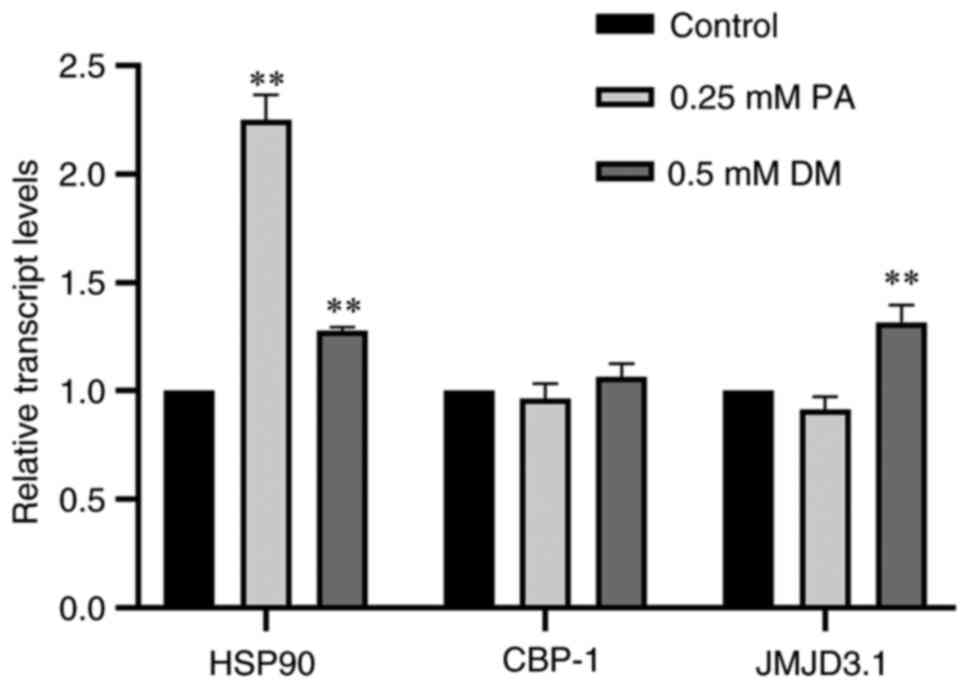
Effects of DM and PA on gene
expression in C. elegans
Based on the results of network pharmacology and
molecular docking, the regulatory effects of two active components
of DM and PA with the core targets EP300, HSP90AB1, KDM6B and
CREBBP were analyzed and verified using RT-qPCR. In C.
elegans, the gene homologue of EP300 and CREBBP is CBP-1, that
for KDM6B is JMJD3.1, and that for HSP90AB1 is HSP90 (35-37).
Therefore, CBP-1, JMJD3.1 and HSP90 were
examined. Compared with the control group, the expression levels of
HSP90 in the DM and PA groups were significantly increased
(P<0.01) (Fig. 7). There was no
significant effect on the expression levels of CBP-1 in the
DM or PA group. JMJD3.1 was upregulated in the DM group
(P<0.01).
Discussion
In the present study, the combined power of network
pharmacology and GEO were utilized to analyze the effects of GEB in
AD. Firstly, a regulatory network of 31 active constituents and 59
key targets was constructed, and it was revealed that most
compounds in GEB have overlapping targets and synergistic effects,
suggesting that GEB has the capacity for multi-component and
multi-target treatment of diseases, which is consistent with the
concept of diseases having multiple pathogenic factors and complex
pathogeneses. In the network diagram ‘drug-compound-target-AD’, the
two components with the highest degree values were DM and PA,
strongly indicating that they have potential anti-AD activity. Most
previous studies on the anti-AD effects of GEB focused on extracts
and gastrodin, while DM and PA have not been investigated. New
components in GEB for the treatment of AD may be identified
(38,39).
GO analysis revealed that GEB may be associated with
the biological process of regulating epigenetic histone
modification (40). KEGG analysis
showed that GEB may be associated with the cAMP signaling pathway
that regulates the accumulation of Aβ and neuronal apoptosis
(41). Among the 59 key targets,
five core targets were obtained through PPI: GAPDH, EP300,
HSP90AB1, KDM6B and CREBBP. GAPDH was excluded as it was used as
the internal reference. Among the four remaining proteins, HSP90AB1
is a stress-induced heat shock protein (HSP). HSPs can protect
neurons by preventing protein misfolding to ensure proteome
homeostasis and play a role in resisting oxidative stress and
inhibiting apoptosis (42,43). HSP90AB1 is significantly
downregulated in AD patients (44).
Some studies suggest that HSP90AB1 is closely linked to astrocytes.
Aβ and tau proteins may be degraded by HSP90AB1 through autophagy
(45,46). KDM6B is a specific histone 3 lysine
27 (H3K27) demethylase. KDM6B regulates the deposition of zebrafish
glial cells by regulating the expression of chemokine, Wnt and FGF
signaling pathway genes (47). In
addition, KDM6B serves as an effective target for black chokeberry
ethanolic extract to inhibit Aβ-induced neuronal cell death
(48). EP300 is a major acetylase
involved in tau protein aggregation and neurodegeneration in AD
(49). A previous study revealed
that histone acetyltransferase, P300, can prevent and treat AD by
regulating acetylation of AD-related gene promoters such as PS1 and
BACE1(50). CREBBP is an acetylated
histone that affects age-related memory loss (51). The inhibition of CREBBP exerts an
anti-inflammatory effect. Furthermore, studies have shown that CCC
protects nerve cells and ameliorates cognitive impairment in an AD
mouse model by regulating CREBBP (52). CREBBP is therefore considered an
attractive target in drug design and development (53). In future studies, the regulatory
effects of DM and PA on core targets will be examined through
molecular docking and other experiments.
Among the pathological mechanisms of AD, that of Aβ
deposition is a hot research topic, and inhibiting Aβ plaque
aggregation is an effective strategy for preventing and treating AD
(54). C. elegans is a
classic model organism for studying AD and plays an important role
in drug screening (55). There are
numerous AD transgenic C. elegans lines (56,57),
which exhibit a number of advantages such as a short life cycle,
easy culture and high gene homology (58). Transgenic C. elegans can
directly produce Aβ plaques, which renders the observation and
research of Aβ convenient (59). In
the present study, it was determined that both DM and PA
effectively delayed the paralysis phenotype and inhibited Aβ
deposition in C. elegans. DM and PA exhibited similar
efficacies, indicating that they directly inhibit Aβ aggregation.
Both DM and PA belong to the phenolic components of GEB. Consistent
with a previous study, including p-hydroxybenzyl alcohol, they can
inhibit the deposition of Aβ in C. elegans, exert anti-aging
and antioxidative effects in C. elegans, as well as exert
anti-AD biological activities (14). Notably, RT-qPCR revealed that both
DM and PA regulate the expression of the heat shock protein HSP90,
but have no impact on CBP-1. Only DM positively regulated JMJD3.1.
Collectively, these findings suggest that DM and PA may have
therapeutic potential for AD, and can serve as focal points of
future studies of GEB.
There are some limitations to the present study. The
network pharmacology approach is based on reported components and
targets. There may be more disease targets and components that have
yet to be discovered and the composition of GEB is complex, and
therefore, network pharmacology may not fully reflect the effect of
GEB in AD. Furthermore, studies using other animals, such as mice
and rats, are required to better model the therapeutic actions of
DM and PA in AD. In future studies, the anti-AD effect of DM and
PA, two active components of GEB, will be further explored and the
mechanism in combination with animal models such as rats and mice
will be studied, in order to obtain data that may be clinically
used.
In conclusion, network pharmacology identified two
core components in GEB, DM and PA, and the core targets, KDM6B,
HSP90AB1, EP300 and CREBBP. Molecular docking revealed that DM and
PA stably bound to the four core targets. DM (5 mM) and PA (0.25
mM) significantly delayed paralysis and inhibited the accumulation
and deposition of Aβ plaques. Both DM and PA upregulated the core
target gene HSP90AB1, while DM upregulated KDM6B. DM and PA may be
therapeutic candidates for AD.
Supplementary Material
Volcano map of differentially
expressed genes. The abscissa log2foldchange represents the
difference (Alzheimer's disease/normal) and the ordinate represents
the confidence level [-log10(adj. P. value)]. Each point in the
figure represents a gene. The dashed lines on the horizontal and
vertical axes represent the log2foldchange absolute threshold of
0.5 and P-value threshold of 0.05, respectively.
Heat map of 59 key targets in control
and AD samples. Each square represents a gene. The greater the
expression, the darker the color (red, upregulated; blue,
downregulated expression). Each row represents the expression of
each gene in different samples; each column represents the
expression of all genes in each sample.
Primer sequences used for PCR.
Potential targets associated with AD
and GEB.
Results of KEGG analysis (top
20).
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the National Natural
Science Foundation of China (grant no. 81960733), the Open Project
of Yunnan Key Laboratory of Dai and Yi Medicines (grant no.
202210ZD2206), the Xingdian Talent Support Program-Special for
Young Talent (2022), the National Administration of Traditional
Chinese Medicine High-level Key Discipline Construction Project
‘Dai Medicine’.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XS, YL and XD made considerable contributions to the
experimental design, statistical data analysis and English language
editing. XD and LY are responsible for drafting the manuscript and
revising it for important intellectual content. XS and LY confirm
the authenticity of all the raw data. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Solanki I, Parihar P and Parihar MS:
Neurodegenerative diseases: From available treatments to
prospective herbal therapy. Neurochem Int. 95:100–108.
2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Kent SA, Spires-Jones TL and Durrant CS:
The physiological roles of tau and Aβ: implications for Alzheimer's
disease pathology and therapeutics. Acta Neuropathol. 140:417–447.
2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
de Caires S and Steenkamp V: Use of
Yokukansan (TJ-54) in the treatment of neurological disorders: A
review. Phytother Res. 24:1265–1270. 2010.PubMed/NCBI View
Article : Google Scholar
|
|
4
|
The Pharmacopoeia Commission of PRC.
Pharmacopoeia of the People's Republic of China. Volume 1. 10th
Edition. China Medical Science and Technology Press, Beijing,
pp58-59, 2015.
|
|
5
|
Lin YE, Lin CH, Ho EP, Ke YC, Petridi S,
Elliott CJ, Sheen LY and Chien CT: Glial Nrf2 signaling mediates
the neuroprotection exerted by Gastrodia elata Blume in
Lrrk2-G2019S Parkinson's disease. Elife. 10(e73753)2021.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Kim HJ, Moon KD, Lee DS and Lee SH: Ethyl
ether fraction of Gastrodia elata Blume protects amyloid beta
peptide-induced cell death. J Ethnopharmacol. 84:95–98.
2003.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Park YM, Lee BG, Park SH, Oh HG, Kang YG,
Kim OJ, Kwon LS, Kim YP, Choi MH, Jeong YS, et al: Prolonged oral
administration of Gastrodia elata extract improves spatial learning
and memory of scopolamine-treated rats. Lab Anim Res. 31:69–77.
2015.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Wang D, Wang Q, Chen R, Yang S, Li Z and
Feng Y: Exploring the effects of Gastrodia elata Blume on the
treatment of cerebral ischemia-reperfusion injury using
UPLC-Q/TOF-MS-based plasma metabolomics. Food Funct. 10:7204–7215.
2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Lu KH, Ou GL, Chang HP, Chen WC, Liu SH
and Sheen LY: Safety evaluation of water extract of Gastrodia elata
Blume: Genotoxicity and 28-day oral toxicity studies. Regul Toxicol
Pharmacol. 114(104657)2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Manavalan A, Ramachandran U, Sundaramurthi
H, Mishra M, Sze SK, Hu JM, Feng ZW and Heese K: Gastrodia elata
Blume (tianma) mobilizes neuro-protective capacities. Int J Biochem
Mol Biol. 3:219–241. 2012.PubMed/NCBI
|
|
11
|
Ramachandran U, Manavalan A, Sundaramurthi
H, Sze SK, Feng ZW, Hu JM and Heese K: Tianma modulates proteins
with various neuro-regenerative modalities in differentiated human
neuronal SH-SY5Y cells. Neurochem Int. 60:827–836. 2012.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Zhang JS, Zhou SF, Wang Q, Guo JN, Liang
HM, Deng JB and He WY: Gastrodin suppresses BACE1 expression under
oxidative stress condition via inhibition of the PKR/eIF2α pathway
in Alzheimer's disease. Neuroscience. 325:1–9. 2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Liu ZH, Hu HT, Feng GF, Zhao ZY and Mao
NY: Protective effects of gastrodin on the cellular model of
Alzheimer's disease induced by Abeta25-35. Sichuan Da Xue Xue Bao
Yi Xue Ban. 36:537–540. 2005.PubMed/NCBI(In Chinese).
|
|
14
|
Liu Y, Lu YY, Huang L, Shi L, Zheng ZY,
Chen JN, Qu Y, Xiao HT, Luo HR and Wu GS: Para-hydroxybenzyl
alcohol delays the progression of neurodegenerative diseases in
models of caenorhabditis elegans through activating multiple
cellular protective pathways. Oxid Med Cell Longev.
2022(8986287)2022.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Ding Y, Bao X, Lao L, Ling Y, Wang Q and
Xu S: p-Hydroxybenzyl alcohol prevents memory deficits by
increasing neurotrophic factors and decreasing inflammatory factors
in a mice model of Alzheimer's disease. J Alzheimers Dis.
67:1007–1019. 2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Mishra M, Huang J, Lee YY, Chua DSK, Lin
X, Hu JM and Heese K: Gastrodia elata modulates amyloid precursor
protein cleavage and cognitive functions in mice. Biosci Trends.
5:129–138. 2011.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Hopkins AL: Network pharmacology. Nat
Biotechnol. 25:1110–1111. 2007.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Liang WS, Dunckley T, Beach TG, Grover A,
Mastroeni D, Walker DG, Caselli RJ, Kukull WA, McKeel D, Morris JC,
et al: Gene expression profiles in anatomically and functionally
distinct regions of the normal aged human brain. Physiol Genomics.
28:311–322. 2007.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Yang X, Guan Y, Yan B, Xie Y, Zhou M, Wu
Y, Yao L, Qiu X, Yan F, Chen Y and Huang L: Evidence-based
complementary and alternative medicine bioinformatics approach
through network pharmacology and molecular docking to determine the
molecular mechanisms of Erjing pill in Alzheimer's disease. Exp
Ther Med. 22(1252)2021.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Li S and Zhang B: Traditional Chinese
medicine network pharmacology: Theory, methodology and application.
Chin J Nat Med. 11:110–120. 2013.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Daina A, Michielin O and Zoete V:
SwissTargetPrediction: Updated data and new features for efficient
prediction of protein targets of small molecules. Nucleic Acids
Res. 47:W357–W364. 2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Piñero J, Ramírez-Anguita JM,
Saüch-Pitarch J, Ronzano F, Centeno E, Sanz F and Furlong LI: The
DisGeNET knowledge platform for disease genomics: 2019 update.
Nucleic Acids Res. 48:D845–D855. 2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Piñero J, Bravo À, Queralt-Rosinach N,
Gutiérrez-Sacristán A, Deu-Pons J, Centeno E, García-García J, Sanz
F and Furlong LI: DisGeNET: A comprehensive platform integrating
information on human disease-associated genes and variants. Nucleic
Acids Res. 45:D833–D839. 2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43(e47)2015.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Otasek D, Morris JH, Bouças J, Pico AR and
Demchak B: Cytoscape Automation: Empowering workflow-based network
analysis. Genome Biol. 20(185)2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Kolde R: Pheatmap: Pretty Heatmaps. R
Package Version. CRAN Repository, 2012.
|
|
27
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Morris GM, Huey R, Lindstrom W, Sanner MF,
Belew RK, Goodsell DS and Olson AJ: AutoDock4 and AutoDockTools4:
Automated docking with selective receptor flexibility. J Comput
Chem. 30:2785–2791. 2009.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Morris GM, Huey R and Olson AJ: Using
AutoDock for ligand-receptor docking. Curr Protoc Bioinformatics.
8(Unit 8.14)2008.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Yang Y, He Y, Wei X, Wan H, Ding Z, Yang J
and Zhou H: Network pharmacology and molecular docking-based
mechanism study to reveal the protective effect of salvianolic acid
C in a rat model of ischemic Stroke. Front Pharmacol.
12(799448)2021.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Chen XY, Liao DC, Sun ML, Cui XH and Wang
HB: Essential oil of acorus tatarinowii Schott Ameliorates
Aβ-Induced toxicity in caenorhabditis elegans through an autophagy
pathway. Oxid Med Cell Longev. 2020(3515609)2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Zhi D, Wang D, Yang W, Duan Z, Zhu S, Dong
J, Wang N, Wang N, Fei D, Zhang Z, et al: Dianxianning improved
amyloid β-induced pathological characteristics partially through
DAF-2/DAF-16 insulin like pathway in transgenic C. elegans. Sci
Rep. 7(11408)2017.PubMed/NCBI View Article : Google Scholar
|
|
33
|
DanQing L, YuJie G, ChengPeng Z, HongZhi
D, Yi H, BiSheng H and Yan C: N-butanol extract of Hedyotis diffusa
protects transgenic Caenorhabditis elegans from Aβ-induced
toxicity. Phytother Res. 35:1048–1061. 2021.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Williams K, Christensen J, Rappsilber J,
Nielsen AL, Johansen JV and Helin K: The histone lysine demethylase
JMJD3/KDM6B is recruited to p53 bound promoters and enhancer
elements in a p53 dependent manner. PLoS One.
9(e96545)2014.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Mathies LD, Lindsay JH, Handal AP,
Blackwell GG, Davies AG and Bettinger JC: SWI/SNF complexes act
through CBP-1 histone acetyltransferase to regulate acute
functional tolerance to alcohol. BMC Genomics.
21(646)2020.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Eckl J, Sima S, Marcus K, Lindemann C and
Richter K: Hsp90-downregulation influences the heat-shock response,
innate immune response and onset of oocyte development in
nematodes. PLoS One. 12(e0186386)2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Ng CF, Ko CH, Koon CM, Xian JW, Leung PC,
Fung KP, Chan HYE and Lau CBS: The aqueous extract of rhizome of
gastrodia elata protected drosophila and PC12 cells against
beta-amyloid-induced neurotoxicity. Evid Based Complement Alternat
Med. 2013(516741)2013.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Fasina OB, Wang J, Mo J, Osada H, Ohno H,
Pan W, Xiang L and Qi J: Gastrodin from gastrodia elata enhances
cognitive function and neuroprotection of AD mice via the
regulation of gut microbiota composition and inhibition of neuron
inflammation. Front Pharmacol. 13(814271)2022.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Narayan P and Dragunow M: Alzheimer's
disease and histone code alterations. Adv Exp Med Biol.
978:321–336. 2017.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Li H, Yang S, Wu J, Ji L, Zhu L, Cao L,
Huang J, Jiang Q, Wei J, Liu M, et al: cAMP/PKA signaling pathway
contributes to neuronal apoptosis via regulating IDE expression in
a mixed model of type 2 diabetes and Alzheimer's disease. J Cell
Biochem. 119:1616–1626. 2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Gorenberg EL and Chandra SS: The role of
Co-chaperones in synaptic proteostasis and neurodegenerative
disease. Front Neurosci. 11(248)2017.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Ramos E, Romero A, Marco-Contelles J,
López-Muñoz F and Del Pino J: Modulation of heat shock response
proteins by ASS234, targeted for neurodegenerative diseases
therapy. Chem Res Toxicol. 31:839–842. 2018.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Li J, Xu C, Zhang J, Jin C, Shi X, Zhang
C, Jia S, Xu J, Gui X, Xing L, et al: Identification of
miRNA-target gene pairs in the parietal and frontal lobes of the
brain in patients with alzheimer's disease using bioinformatic
analyses. Neurochem Res. 46:964–979. 2021.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Gonzalez-Rodriguez M, Villar-Conde S,
Astillero-Lopez V, Villanueva-Anguita P, Ubeda-Banon I,
Flores-Cuadrado A, Martinez-Marcos A and Saiz-Sanchez D:
Neurodegeneration and astrogliosis in the human CA1 hippocampal
subfield are related to hsp90ab1 and bag3 in Alzheimer's disease.
Int J Mol Sci. 23(165)2021.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Jeong HO, Park D, Im E, Lee J, Im DS and
Chung HY: Determination of the mechanisms that cause sarcopenia
through cDNA microarray. J Frailty Aging. 6:97–102. 2017.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Tang D, Lu Y, Zuo N, Yan R, Wu C, Wu L,
Liu S and He Y: The H3K27 demethylase controls the lateral line
embryogenesis of zebrafish. Cell Biol Toxicol.
29:10.1007/s10565-021-09669-y. 2021.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Kim J, Lee KP, Beak S, Kang HR, Kim YK and
Lim K: Effect of black chokeberry on skeletal muscle damage and
neuronal cell death. J Exerc Nutrition Biochem. 23:26–31.
2019.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Min SW, Chen X, Tracy TE, Li Y, Zhou Y,
Wang C, Shirakawa K, Minami SS, Defensor E, Mok SA, et al: Critical
role of acetylation in tau-mediated neurodegeneration and cognitive
deficits. Nat Med. 21:1154–1162. 2015.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Lu X, Deng Y, Yu D, Cao H, Wang L, Liu L,
Yu C, Zhang Y, Guo X and Yu G: Histone acetyltransferase p300
mediates histone acetylation of PS1 and BACE1 in a cellular model
of Alzheimer's disease. PLoS One. 9(e103067)2014.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Barral S, Reitz C, Small SA and Mayeux R:
Genetic variants in a ‘cAMP element binding protein’
(CREB)-dependent histone acetylation pathway influence memory
performance in cognitively healthy elderly individuals. Neurobiol
Aging. 35:2881.e2887–2881.e2810. 2014.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Kim MJ, Park SY, Kim Y, Jeon S, Cha MS,
Kim YJ and Yoon HG: Beneficial effects of a combination of curcuma
longa L. and Citrus junos against beta-amyloid peptide-induced
neurodegeneration in mice. J Med Food. 25:12–23. 2022.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Akinsiku OE, Soremekun OS and Soliman MES:
Update and potential opportunities in CBP [Cyclic Adenosine
Monophosphate (cAMP) Response Element-Binding Protein
(CREB)-Binding Protein] research using computational techniques.
Protein J. 40:19–27. 2021.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Scheltens P, De Strooper B, Kivipelto M,
Holstege H, Chételat G, Teunissen CE, Cummings J and van der Flier
WM: Alzheimer's disease. Lancet. 397:1577–1590. 2021.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Paul D, Chipurupalli S, Justin A, Raja K
and Mohankumar SK: Caenorhabditis elegans as a possible model to
screen anti-Alzheimer's therapeutics. J Pharmacol Toxicol Methods.
106(106932)2020.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Link CD: Expression of human beta-amyloid
peptide in transgenic Caenorhabditis elegans. Proc Natl Acad Sci
USA. 92:9368–9372. 1995.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Fay DS, Fluet A, Johnson CJ and Link CD:
In vivo aggregation of beta-amyloid peptide variants. J Neurochem.
71:1616–1625. 1998.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Morcos M and Hutter H: The model
Caenorhabditis elegans in diabetes mellitus and Alzheimer's
disease. J Alzheimers Dis. 16:897–908. 2009.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Zhu S, Li H, Dong J, Yang W, Liu T, Wang
Y, Wang X, Wang M and Zhi D: Rose essential oil delayed Alzheimer's
disease-like symptoms by SKN-1 pathway in C. elegans. J Agric Food
Chem. 65:8855–8865. 2017.PubMed/NCBI View Article : Google Scholar
|