Introduction
Hypertension (HTN) is one of the most common chronic
diseases with an uncertain etiology. HTN is associated with
cerebrovascular and cardiovascular diseases and mortality (1,2). It
causes severe myocardial injury, heart failure, chronic kidney
disease, peripheral vascular disease, stroke and derived
complications (3-5).
The complex pathogenesis of HTN is determined by an intricate
combination of various genetic and environmental factors. Although
several genetic factors show a variability of 25-60% in human HTN
(4), the genetic mechanism of HTN
remains uncertain.
Leptin is involved in the regulation of various
cardiac and vascular events such as thrombosis, angiogenesis and
cardiac hypertrophy (6). It also
affects the regulation of metabolism, immunity and reproduction
(7). Leptin has a physiological
function through the leptin receptor (LEPR), a molecule distributed
in various tissues that can mediate important effects of leptin as
a hormone involved in whole-body energy homeostasis (8).
MicroRNAs (miRNAs/miRs) are small-sized noncoding
RNAs consisting of nucleotides that mainly bind to 3'-untranslated
regions (3'-UTRs) (9-11).
They can either inhibit the translation of mRNA or accelerate its
degradation. As biomarkers for several diseases, miRNAs have the
potential to contribute to the understanding of pathogenesis
mechanisms and the development of therapeutic medicine (4). As miRs are involved in various
cellular processes including proliferation, differentiation,
apoptosis and development, polymorphisms within miRs and the
dysregulation of their expression are associated with metabolic
diseases such as cancer, type 2 diabetes mellitus (T2DM),
cardiovascular disease and gestational diabetes mellitus (12,13).
For example, miR-27a shows a higher expression in the plasma of
patients with hypertension and the omenta of obese patients
(14). A study by Ciccacci et
al (15) suggested that the G
allele in miR-27a demonstrates a protective effect in T2DM, as
overexpression of miR-27a in pre-adipocytes suppresses the
expression of the peroxisome proliferator-activated receptor γ and
the differentiation of adipocytes. MiR-155 expression levels in the
aortae of adult spontaneous hypertensive rats are decreased and
negatively correlate with blood pressure (16), suggesting that miR-155 might be
involved in the development and pathological progression of HTN.
Circulating miR-155 is associated with T2DM and the rs767649
polymorphism in the pre-miR155 gene is associated with miR-155
expression (17).
The gene encoding LEPR is located on chromosome
1p31(18). A deletion/insertion
(Del/Ins) polymorphism located in the 3'-UTR of the LEPR gene may
affect the rate of translation or degradation of mRNA by creating a
stem-loop structure (19-21).
Del/Ins polymorphism in the LEPR gene has mainly been reported to
be associated with susceptibility to T2DM (19,22-27),
although this remains controversial. Previous studies have also
suggested that miR-27a and miR-155 play an important role in the
regulation of LEPR expression (28,29).
These miRNAs have also been found to be differentially expressed in
various diseases.
Although the functions and gene expression of LEPR
and miRNAs are known to be involved in susceptibility to various
diseases, the effects of polymorphisms in LEPR and miRNAs genes on
susceptibility to HTN have not yet been sufficiently elucidated.
Thus, the present study performed a case-control study to
investigate potential associations between LEPR 3'UTR Del/Ins,
miR-27aA>G (rs895819) and miR-155T>A (rs767649) polymorphisms
and HTN in a Korean population.
Materials and methods
Study population
The present study included a total of 479
individuals, 232 patients with HTN, including 190 male and 42
female patients (mean ± SD age, 47.35±8.23 years; age range, 31-67
years), and 247 control subjects, including 132 male and 115 female
subjects (mean ± SD age, 48.93±9.98 years; age range, 32-80 years),
recruited from Jeju National University Hospital (Jeju, Korea)
between January 2020 and December 2020. HTN was diagnosed after
repeated measurements with a systolic blood pressure (SBP) >140
mmHg and a diastolic blood pressure (DBP) >90 mmHg. The present
study also included subjects who were currently taking
antihypertensive drugs. Patients who were diagnosed with other
chronic diseases were excluded from the current study. Healthy
control subjects had normal blood pressure and were not on
medication, and were enrolled during the same period. The normal
group was randomly selected from those who did not have chest pain,
diabetes, hypertension or general illnesses as determined through
physical examination. All study participants provided written
informed consent. The study protocol was approved by the
Institutional Review Board of Jeju National University Hospital,
Republic of Korea (approval no. JEJUNUH 2020-07-005). The
biospecimens and data used in this study were collected by the
Biobank of Jeju National University Hospital, a member of the South
Korea Biobank Network supported by the Ministry of Health and
Welfare.
Phenotype measurements
The blood pressure of all participants was measured
in a sitting position, after resting for at least 10 min. Risk
factors for HTN were also measured. Body mass index (BMI) was
calculated after weighing. Waist circumference (WC) was measured
with a non-stretchable fiber measuring tape. All subjects fasted
for >12 h before biochemical measurements. Levels of plasma
fasting blood glucose (FBG) (cat. no. GLU CN R1 99118592, cat. no.
GLU CN R2 99718692; FUJIFILM Wako Pure Chemical Corp.),
triglycerides (TG; cat. no. TG CN R1 99333192, cat. no. TG CN R2
99933292; FUJIFILM Wako Pure Chemical Corp.) and high-density
lipoprotein-cholesterol (HDL-C) (cat. no. ML HDLC S R1 55379, cat.
no. ML HDLC S R2 55380; Minaris Medical Co., Ltd.) were measured
with commercially available enzymatic colorimetric tests using an
automatic biochemical analyzer (TBA 200FR NEO; Toshiba Medical
Systems).
Genotype analysis
Genomic DNA was extracted from white blood cells
using a G-DEX blood extraction kit (cat. No. 172R-1000; Intron
Biotechnology, Inc.). Genotyping of LEPR Del/Ins, miR-27a
(rs895819) and miR-155 (rs767649) polymorphisms were performed
using a PCR and restriction fragment length polymorphism (RFLP)
technique, as described in previous papers (28-30).
Restriction enzyme digestion was performed at 37˚C for 17 h with
RsaI, DraIII and Tsp45I for LEPR Del/Ins,
miR-27a (rs895819) and miR-155 (rs767649) polymorphisms,
respectively (New England BioLabs, Inc.). Detailed PCR conditions
and genotype sizes are summarized in Table I.
 | Table IDetails of the SNPs (LEPR,
miR-27aA>G and miR-155T>A) that were included in the present
study. |
Table I
Details of the SNPs (LEPR,
miR-27aA>G and miR-155T>A) that were included in the present
study.
| SNP | Primer sequence | PCR annealing,
Tm./product (˚C/bp) | Genotype (bp) | RE |
|---|
| LEPR Del/Ins | 5'-ATA ATG GGT AAT
ATA AAG TGT AAT AGA GTA-3' | 55˚C/114 | Del: 114 | RsaI |
| | 5'-AGA GAA CAA ACA
GAC AAC ATT-3' | | Del/Ins: 114, 90,
29 | |
| | | | Ins: 90, 29 | |
| miR-27a A>G | 5'-GAA CTT AGC CAC
TGT GAA CAC GAC TTC G-3' | 55˚C/182 | AA: 155, 29 | DraIII |
| | 5'-TTG CTT CCT GTC
ACA AAT CAC ATT G-3' | | AG: 182, GG: 182 | |
| miR-155 T>A | 5'-CCT GTA TGA CAA
GGT TGT GTT TG-3' | 60˚C/294 | TT: 294 | Tsp45I |
| | 5'-GCT GGC ATA CTA
TTC TAC CCA TAA-3' | | TA: 260, 155,
139 | |
| | | | AA: 155, 138 | |
The following reference sequence shows the
amplification region for analyzing the LEPR polymorphism (where
bases in square brackets indicate the positions of the primers, the
parentheses indicate pentanucleotide insertion and where * is the
RsaI cutting site): 5'
[ATAATGGGTAATATAAAGTGTAATAGATT]*(ACTTT)AAAGTGTAATAGATTATAGTTGTGGGTGGGAGAGAGAAAAGAAACCAGAGTCAAATTTGAAAATAATTGTTCCAAATG[AATGTTGTCTGTTTGTTCTCT]
3'. It is truncated by RsaI only in the case of the
insertion genotype. However, the deletion genotype does not have
pentanucleotide insertion and is not cleaved by RsaI. The
following sequence shows the amplification region for analyzing the
miR-27a polymorphism (where bases in square brackets indicate the
positions of the primers, where * is the DraIII cutting
site):
5'-[GAACTTAGCCACTGTGAACACGACTTGG]*GTGGACCCTGCTCACAAGCAGCTAAGCCCTGCTCCTCAGGCCAGGCACAGGCTTCGGGGCCTCTCTGCCACCCCGTCCCCGGGCAGCATCCTCGGTGGCAGAGCTCAGGGTCGGTTGGAAATCCCTGG[AATGTGATTTGTGACAGGAAGCAA]-3'.
The following sequence shows the amplification region for analyzing
the miR-155 polymorphism (where bases in square brackets indicate
the positions of the primers, where * is the Tsp45I cutting
site):
[5'-CCTGTATGACAAGGTTGTGTTTG]AGAACAAAAGGGGACCTGTGTGACATGTTTTATTTCAATATACATTTAGAGTTTGAACAAATAAAAAAAGCTATTAGAATTTTTAATATATATAACACATTATCAAAAACACTG*CACTTTTCTGAGTGCTCTAATCAGGCAATTCGTATGTATATTTAAAAAGAGGGAAAAAGCTAATTAGTATACTAAATATATTTATTTTAATACCTGTGCTCATATGTTATCTTTAAAGAACAGGAATAAAAT[TTATGGGTAGAATAGTATGCCAGC-3'].
Statistical analysis
P-values were calculated using a unpaired two-sided
Student's t-test or Mann-Whitney U test for continuous variables
and a χ2 test for categorical variables between the
control subjects and the HTN group. Values are presented as mean ±
standard deviation (SD) for clinical profiles of the control
subjects and the HTN group. Logistic regression analysis adjusted
for age and sex was applied to risk assessment with LEPR Del/Ins,
miR-27a (rs895819) and miR-155 (rs767649) polymorphisms for HTN.
Values are presented as adjusted odds ratios (AOR) and 95%
confidence intervals (95% CI). P<0.05 was considered to indicate
a statistically significant difference. All statistical analyses
were performed using GraphPad Prism version 4.0 (GraphPad Software,
Inc.) and MedCalc version 12.7.1.0 (MedCalc Software Ltd.).
Results
The HTN group had significantly higher BMI, WC, SBP,
DBP, FBG and HDL-C values (P<0.0001) compared with the normal
group. By contrast, HDL-C levels were significantly lower in the
HTN group compared with in the normal group (P<0.001). However,
age was not significantly different between the HTN and control
group (P=0.09). The demographics of the HTN patient group and the
control group are presented in Table
II.
 | Table IIBaseline characteristics between
patients and control subjects. |
Table II
Baseline characteristics between
patients and control subjects.
| Characteristic | Controls
(n=247) | HTN (n=232) | P-value |
|---|
| Age, years | 45.9±10.0 | 47.4±8.2 | 0.09 |
| BMI,
kg/m2 | 23.9±3.5 | 26.1±3.4 | <0.0001 |
| WC, cm | 81.3±8.7 | 87.2±9.2 | <0.0001 |
| SBP, mmHg | 118.5±11.2 | 138.4±13.4 | <0.0001 |
| DBP, mmHg | 71.4±8.6 | 88.5±8.8 | <0.0001 |
| FBG, mg/dl | 88.2±8.9 | 95.5±14.2 | <0.0001 |
| TG, mg/dl | 92.1±57.4 | 133.5±96.0 | <0.0001 |
| HDL-C, mg/dl | 57.5±14.3 | 52.2±11.3 | <0.0001 |
For the LEPR gene polymorphism, the genotypes were
assessed as follows: A single 114 bp fragment for the wild-type
homozygous alleles (Del genotype), two fragments of 90 and 24 bp
for the mutated type (Ins genotype) and three fragments of 114, 90
and 24 bp for the heterozygous alleles (Del/Ins genotype) (Fig. 1). The 24 bp fragment is not shown in
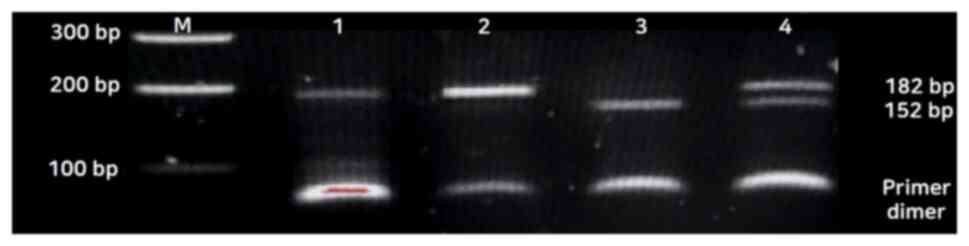
Fig. 1. For the miR-27aA>G
polymorphism, homozygosity of the common allele (AA genotype) was
represented by 155 and 27 bp bands, while the homozygosity of the
variant allele (GG genotype) revealed itself as a 182 bp band. The
heterozygous alleles (AG genotype) were revealed by 182, 155 and 27
bp bands (Fig. 2). The 27 bp
fragment is not shown in Fig. 2.
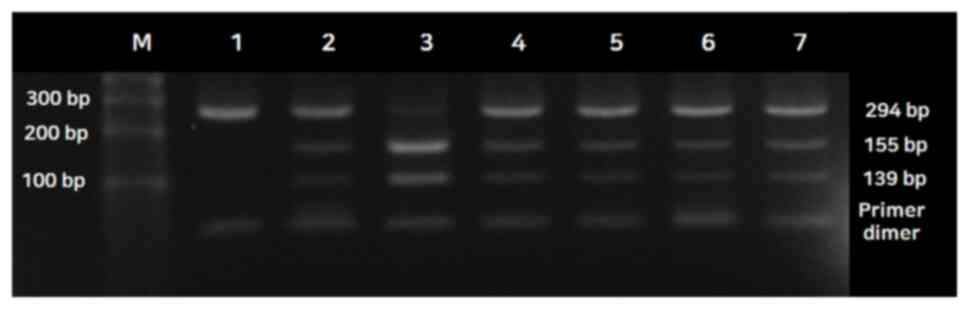
The lowest bands in lanes 1-6 are primer dimers. For the
miR-155T>A gene polymorphism, the genotypes were assessed as
follows: A single 294 bp fragment for the wild-type homozygous
alleles (TT genotype), two fragments of 155 and 139 bp for the
mutated type (AA genotype) and three fragments of 294, 155, and 139
bp for the heterozygous alleles (TA genotype) (Fig. 3).
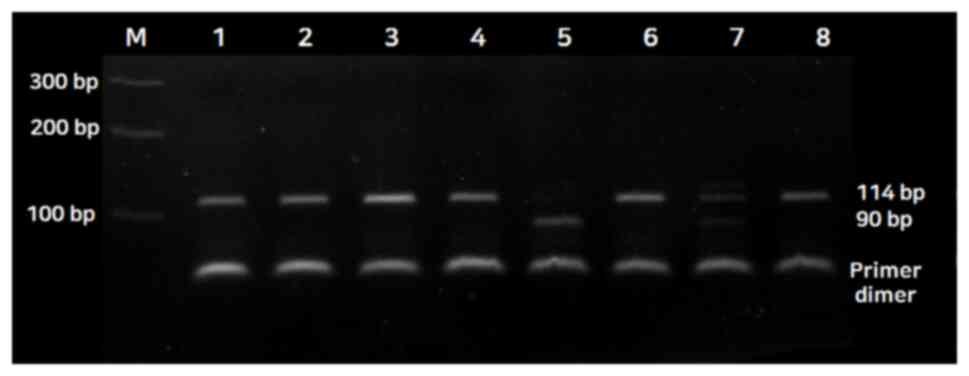 | Figure 1Restriction fragment length
polymorphism results showing the restriction enzyme RsaI
digestion pattern of LEPR Del/Ins. M, 100-bp size marker; lanes 1,
2, 3, 4, 6 and 8, Del homozygous alleles; lane 5, Ins homozygous
allele; lane 7, heterozygous allele (Del/Ins); LEPR, Leptin
receptors; Del/Ins, deletion/insertion. |
Table III presents
the comparison results of the genotype and allele frequencies of
the LEPR Ins/Del, miR-27a A>G and miR-155 T>A polymorphisms
between the HTN and control group. The LEPR Del/Ins and Ins/Ins
genotypes and Ins allele were significantly associated with a
decreased risk of HTN (Del/Del vs. Del/Ins: AOR=0.642, 95% CI
0.417-0.986, P=0.043; Del/Del vs. Ins/Ins: AOR=0.386, 95% CI
0.151-0.987, P=0.047; and Del allele vs. Ins allele: AOR=0.609, 95%
CI 0.430-0.864, P=0.005). The frequency of the dominant model
(Del/Del + Del/Ins vs. Ins/Ins) was also significantly lower in the
case group compared with in the control group (AOR=0.597, 95% CI
0.397-0.898, P=0.013). The miR-27a AG genotype and dominant model
were significantly associated with an increased risk of HTN (AA vs.
AG: AOR=1.719, 95% CI 1.108-2.666, P=0.016; and AA + AG vs. GG:
AOR=1.557, 95% CI 1.027-2.359, P=0.037). By contrast, the miR-155
T>A polymorphism was not significantly different between the
patients with HTN and control subjects.
 | Table IIIComparison of genotype frequencies of
LEPR, miR-27a and miR-155 polymorphisms between patients with HTN
and controls. |
Table III
Comparison of genotype frequencies of
LEPR, miR-27a and miR-155 polymorphisms between patients with HTN
and controls.
| Genotypes | Controls
(n=247) | HTN (n=232) | AOR (95% CI) | P-value |
|---|
| LEPR Del/Ins | | | | |
|
DD | 148 (59.9) | 172 (74.1) | 1.000
(reference) | |
|
DI | 82 (33.2) | 53 (22.8) | 0.642
(0.417-0.986) | 0.043a |
|
II | 17 (6.9) | 7 (3.0) | 0.386
(0.151-0.987) | 0.047a |
| Dominant | | | 0.597
(0.397-0.898) | 0.013a |
| Recessive | | | 0.434
(0.169-1.115) | 0.083 |
| Del allele | 378 (76.5) | 397 (85.6) | 1.000
(reference) | |
| Ins allele | 116 (23.5) | 67 (14.4) | 0.609
(0.430-0.864) | 0.005a |
| miR-27a A>G
(rs895819) | | | | |
|
AA | 84 (34.0) | 62 (26.7) | 1.000
(reference) | |
|
AG | 109 (44.1) | 126 (54.3) | 1.719
(1.108-2.666) | 0.016a |
|
GG | 54 (21.9) | 44 (19.0) | 1.193
(0.681-2.089) | 0.538 |
| Dominant | | | 1.557
(1.027-2.359) | 0.037a |
| Recessive | | | 0.869
(0.543-1.391) | 0.558 |
| A allele | 277 (56.1) | 250 (53.9) | 1.000
(reference) | |
| G allele | 217 (43.9) | 214 (46.1) | 1.152
(0.881-1.508) | 0.302 |
| miR-155 T>A
(rs767649) | | | | |
|
TT | 76 (30.8) | 70 (30.2) | 1.000
(reference) | |
|
TA | 120 (48.6) | 113 (48.7) | 1.050
(0.675-1.633) | 0.828 |
|
AA | 51 (20.6) | 49 (21.1) | 1.177
(0.687-2.017) | 0.553 |
| Dominant | | | 1.086
(0.721-1.635) | 0.693 |
| Recessive | | | 1.157
(0.724-1.851) | 0.542 |
| T allele | 272 (55.1) | 253 (54.5) | 1.000
(reference) | |
| A allele | 222 (44.9) | 211 (45.5) | 1.087
(0.831-1.422) | 0.543 |
To investigate the association between genotype
combination and the risk of HTN, the present study analyzed the
combined genotypes of LEPR Ins/Del, miR-27a A>G and miR-155
T>A polymorphisms from the HTN and control groups. As shown in
Table IV, the Del/Del-AG and
Ins/Ins-AA combined genotypes (AOR=1.704, 95% CI 1.005-2.888,
P=0.048; AOR=0.106, 95% CI 0.013-0.881, P=0.038, respectively) of
the LEPR Del/Ins and miR-27a A>G polymorphisms and the AG-AA
combined genotype (AOR=2.575, 95% CI 1.019-6.507, P=0.046) of the
miR-27a A>G and miR-155 T>A polymorphisms could be linked
with a genetic susceptibility to HTN.
 | Table IVGenotype combination analysis for
SNPs in patients with HTN and controls. |
Table IV
Genotype combination analysis for
SNPs in patients with HTN and controls.
| A, LEPR
Del-Ins/miR-27a A>G |
|---|
| Genotype
combination | |
|---|
| SNP1 | SNP2 | Controls
(n=247) | HTN (n=232) | AOR (95% CI) | P-value |
|---|
| DD | AA | 52 (21.1) | 48 (20.7) | 1.000
(reference) | |
| | AG | 64 (25.9) | 92 (39.7) | 1.704
(1.005-2.888) | 0.048a |
| | GG | 32 (13.0) | 32 (13.8) | 1.084
(0.558-2.106) | 0.812 |
| DI | AA | 21 (8.5) | 13 (5.6) | 0.874
(0.370-2.068) | 0.760 |
| | AG | 39 (15.8) | 31 (13.4) | 1.001
(0.526-1.905) | 0.999 |
| | GG | 22 (8.9) | 9 (3.9) | 0.626
(0.242-1.619) | 0.334 |
| II | AA | 11 (4.5) | 1 (0.4) | 0.106
(0.013-0.881) | 0.038a |
| | AG | 6 (2.4) | 3 (1.3) | 0.797
(0.166-3.831) | 0.777 |
| | GG | 0 (0.0) | 3 (1.3) | N/A | 0.998 |
| B, miR-27a
A>G/miR-155 T>A |
| Genotype
combination | |
| SNP1 | SNP2 | Controls
(n=247) | HTN (n=232) | AOR (95% CI) | P-value |
| AA | TT | 21 (8.5) | 16 (6.9) | 1.000
(reference) | |
| | TA | 41 (16.6) | 34 (14.7) | 1.333
(0.567-3.137) | 0.510 |
| | AA | 22 (8.9) | 12 (5.2) | 0.967
(0.340-2.748) | 0.950 |
| AG | TT | 32 (13.0) | 39 (16.8) | 2.053
(0.851-4.955) | 0.109 |
| | TA | 57 (23.1) | 56 (24.1) | 1.591
(0.721-3.511) | 0.250 |
| | AA | 20 (8.1) | 31 (13.4) | 2.575
(1.019-6.507) | 0.046a |
| GG | TT | 23 (9.3) | 15 (6.5) | 1.274
(0.448-3.620) | 0.650 |
| | TA | 22 (8.9) | 23 (9.9) | 1.774
(0.660-4.770) | 0.256 |
| | AA | 9 (3.6) | 6 (2.6) | 1.452
(0.354-5.963) | 0.605 |
To ascertain the synergistic effect between LEPR
Ins/Del, miR-27a A>G and miR-155 T>A polymorphisms, an allele
combination analysis was conducted (Table V). The results demonstrated that the
Ins-A-A and Ins-G-T allele combination frequencies of the LEPR
Ins/Del, miR-27a A>G and miR-155 T>A polymorphisms were
significantly lower in the HTN group compared with in the control
group (AOR=0.089, 95% CI 0.026-0.301, P<0.0001;
AOR=0.403, 95% CI 0.204-0.800, P=0.038, respectively).
 | Table VComparison of allele combination
between patients with HTN and controls (LEPR Del/Ins-miR-27a
A>G-miR-155 T>A). |
Table V
Comparison of allele combination
between patients with HTN and controls (LEPR Del/Ins-miR-27a
A>G-miR-155 T>A).
| Allele
combination | Overall
(n=958) | Controls
(n=494) | HTN (n=464) | OR (95% CI) | P-value |
|---|
| Del-A-T | 0.2117 | 0.1963 | 0.2204 | 1.000
(reference) | |
| Del-A-A | 0.2307 | 0.2206 | 0.2512 | 1.021
(0.697-1.495) | 0.916 |
| Del-G-T | 0.2209 | 0.2092 | 0.2345 | 1.006
(0.683-1.482) | 0.974 |
| Del-G-A | 0.1457 | 0.139 | 0.1495 | 0.951
(0.616-1.468) | 0.821 |
| Ins-A-T | 0.0656 | 0.0781 | 0.0599 | 0.683
(0.390-1.195) | 0.180 |
| Ins-A-A | 0.0422 | 0.0656 | 0.0073 | 0.089
(0.026-0.301) |
<0.0001a |
| Ins-G-T | 0.0498 | 0.0669 | 0.0305 | 0.403
(0.204-0.800) | 0.008a |
| Ins-G-A | 0.0334 | 0.0242 | 0.0468 | 1.743
(0.818-3.715) | 0.146 |
According to the stratified analysis results shown
in Table VI, LEPR Del/Del vs.
Del/Ins + Ins/Ins induced a decreased risk of HTN based on WC, FBG,
and HDL-C values among the possible risk variables (WC<84.19 cm,
P=0.044; FBG<91.72 mg/dl, P=0.044; HDL-C≥54.91 mg/dl, P=0.045).
Whereas, miR-27a AA vs. AG + GG induced an increased risk of HTN
based on BMI, WC, DBP, FBG, TG and HDL-C values among the possible
risk variables (BMI<25.00 kg/m2, P=0.046; WC<84.19
cm, P=0.021; DBP≥79.68 mmHg, P=0.019; FBG<91.72 mg/dl, P=0.036;
TG<112.19 mg/dl, P=0.019; HDL-C<54.91 mg/dl, P=0.013).
 | Table VIStratified analysis of LEPR, miR-27a
and miR-155 polymorphisms according to risk factors in patients
with HTN. |
Table VI
Stratified analysis of LEPR, miR-27a
and miR-155 polymorphisms according to risk factors in patients
with HTN.
| | LEPR Del/Ins +
Ins/Ins | miR-27a AG +
GG | miR-155 TA +
AA |
|---|
| Variables | AOR (95% CI) | P-value | AOR (95% CI) | P-value | AOR (95% CI) | P-value |
|---|
| BMI,
kg/m2 |
|
<25.00 | 0.557
(0.306-1.012) | 0.055 | 1.921
(1.011-3.650) | 0.046 | 1.062
(0.590-1.910) | 0.842 |
|
≥25.00 | 0.722
(0.393-1.327) | 0.295 | 1.411
(0.787-2.529) | 0.248 | 0.918
(0.493-1.708) | 0.786 |
| WC, cm |
|
<84.19 | 0.547
(0.304-0.984) | 0.044 | 2.034
(1.112-3.720) | 0.021 | 1.192
(0.661-2.151) | 0.559 |
|
≥84.19 | 0.615
(0.339-1.114) | 0.108 | 1.100
(0.594-2.039) | 0.761 | 1.005
(0.553-1.826) | 0.988 |
| SBP, mmHg |
|
<128.16 | 0.726
(0.353-1.491) | 0.383 | 1.436
(0.683-3.019) | 0.340 | 1.132
(0.546-2.346) | 0.739 |
|
≥128.16 | 0.693
(0.335-1.432) | 0.322 | 1.838
(0.895-3.772) | 0.097 | 0.844
(0.397-1.796) | 0.660 |
| DBP, mmHg |
|
<79.68 | 0.541
(0.238-1.230) | 0.143 | 1.214
(0.551-2.671) | 0.631 | 1.228
(0.560-2.692) | 0.609 |
|
≥79.68 | 0.640
(0.318-1.288) | 0.211 | 2.313
(1.148-4.661) | 0.019 | 0.646
(0.296-1.412) | 0.273 |
| FBG, mg/dl |
|
<91.72 | 0.561
(0.321-0.983) | 0.044 | 1.901
(1.043-3.465) | 0.036 | 0.986
(0.567-1.715) | 0.959 |
|
≥91.72 | 0.743
(0.394-1.402) | 0.360 | 1.188
(0.633-2.230) | 0.592 | 1.011
(0.524-1.951) | 0.974 |
| TG, mg/dl |
|
<112.19 | 0.646
(0.391-1.066) | 0.087 | 1.907
(1.114-3.264) | 0.019 | 1.245
(0.746-2.076) | 0.402 |
|
≥112.19 | 0.570
(0.270-1.204) | 0.141 | 1.196
(0.576-2.483) | 0.632 | 0.747
(0.350-1.593) | 0.450 |
| HDL-C, mg/dl |
|
<54.91 | 0.660
(0.378-1.150) | 0.142 | 2.033
(1.161-3.561) | 0.013 | 0.925
(0.527-1.621) | 0.784 |
|
≥54.91 | 0.532
(0.287-0.986) | 0.045 | 1.116
(0.600-2.077) | 0.729 | 1.296
(0.700-2.400) | 0.410 |
Discussion
According to a recent report, HTN affects 34.2 and
61.4% of the Korean population in the age ranges of >30 and 65
years of age, respectively (31).
These facts led the current study to analyze potential associations
of LEPR Del/Ins, miR-27a (rs895819) and miR-155 (rs767649)
polymorphisms with a predisposition to HTN.
Regarding the LEPR 3'UTR Del/Ins polymorphism, the
presence of a pentanucleotide insertion generates a putative
stem-loop structure in the mRNA (19) and could affect the rate of
degradation and/or translation of mRNA. In the present study, the
LEPR Del/Ins and Ins/Ins genotypes as well as the Ins allele were
significantly associated with a decreased risk of HTN. The WC, FBG,
and HDL-C values were significantly decreased in Del/Ins + Ins/Ins
carriers compared with in subjects with Del/Del. Previous studies
have shown that the LEPR Del/Ins polymorphism is mainly associated
with T2DM and its related traits (19,22-27).
The results of the present study were consistent with the findings
of the following studies, although the subjects of the present
study were different. In particular, the results of the present
study were similar to those of a Japanese study (26). In young Japanese men, plasma
HDL-cholesterol and apoA-I levels were significantly lower in
homozygous or heterozygous carriers of the Ins allele compared with
in subjects homozygous for the Del allele. In another study, the
results of LEPR Del/Ins polymorphism remained non-significant in
morbidly obese French Caucasian families; however, subjects having
a Del/Ins genotype showed a slight trend towards lower insulin
values at 30 min after an oral glucose load compared with Del/Del
individuals (24).
A total of three papers regarding the LEPR Del/Ins
polymorphism have been published by a Finnish group (19,23,25).
Serum insulin levels in morbidly obese subjects were found to be
lower in heterozygous carriers of the Ins allele compared with in
subjects homozygous for the Del allele (23). Individuals having a 3'UTR Del/Del
genotype also had a slightly higher body weight throughout the
study compared with those with an Ins allele in Finnish subjects
showing impaired glucose tolerance (19). In addition, carriers of the Ins
allele had a reduced risk of diabetes compared with non-carriers
(25). Similarly, in other groups,
the Ins allele of the LEPR Del/Ins polymorphism is associated with
a reduced risk of development of diabetes in Mexican (27) and Kashmiri (28) populations. These results suggest
that alterations in the leptin signaling system can contribute to
serum insulin levels and the development of T2DM. However, to the
best of our knowledge, there is no study on the relationship
between LEPR Del/Ins polymorphism and HTN. Therefore, the results
of the present study need to be confirmed with larger samples from
other hypertensive groups in the future.
MiRNAs are involved in the pathogenesis of HTN as
well as various diseases such as cancer, coronary artery disease
and diabetes (4). In particular,
circulating miRNAs that target other miRNAs have been proposed to
play a role in HTN in a tissue-based study (32). LEPR can interact with numerous
miRNAs, including miR-27a and miR-155, which were selected in the
present study. Of these two chosen miRNAs polymorphisms, the
miR-27a AG genotype and dominant model of the present study was
associated with an increased risk of HTN. The evidence for the
involvement of miR-27a in hypertension includes that monocyte
miR-27a in extracellular vesicles can decrease Mas receptor
expression and eNOS phosphorylation in the endothelium, impair
Ang-(1-7)-mediated
vasodilation and cause hypertension (33). Another study has shown that a
combination of mitoQ and endurance training has more prominent
effects in improving cardiac health and ameliorating blood pressure
in patients with HTN compared with mitoQ or endurance training
alone through miR-27a modulation and amelioration of mitochondrial
reactive oxygen species production (34). Plasma miRNA-27a also has a higher
expression in the plasma of patients with HTN and the omenta of
obese patients (14). Furthermore,
miR-27a rs895819 polymorphism has been reported to alter disease
susceptibility by interfering with the maturation and expression of
miR-27a. The relationship between miR-27a polymorphism and disease
susceptibility has been studied in gastrointestinal cancer
(35), renal cell cancer (36), breast cancer (37) and myocardial infarction (38). To the best of our knowledge, the
present study is the first to reveal the relationship between
miR-27a rs895819 polymorphism and the risk of HTN.
In a previous study, miR-155 expression levels in
the aortae of adult spontaneous hypertensive rats are decreased and
negatively correlated with blood pressure (16), suggesting that miR-155 might be
involved in the development and pathological progression of HTN.
Nevertheless, in the present study, miR-155 (rs767649) polymorphism
was not associated with HTN risk. However, since susceptibility to
multifactorial diseases such as HTN is often indicated by a
combination of multiple polymorphic variants rather than a single
polymorphism, it cannot necessarily be said that a single
polymorphism is unrelated. In the present study, Ins-A-A and
Ins-G-T allele combinations of LEPR Ins/Del, miR-27a A>G and
miR-155 T>A polymorphisms were associated with a decreased risk
of HTN. In addition, the Del/Del-AG combined genotype of LEPR
Del/Ins and miR-27a A>G polymorphisms and the AG-AA combined
genotype of miR-27a A>G and miR-155 T>A polymorphisms were
also associated with the risk of hypertension. The combined
analysis of alleles and genotypes of these polymorphisms can bring
synergies that are difficult to obtain from a single polymorphism,
which can be an important tool for research related to disease
risk.
The current study had several limitations. The
sample size of the study was relatively small. Hence, for
additional verification, it will be necessary to conduct further
studies on larger sample sizes containing more diverse patient
cohorts. In addition, there was no in vitro evidence to
complement the genetic association study. Therefore, the study
results warrant further functional studies to elucidate the
mechanisms by which polymorphisms of LEPR Del/Ins, miR-27aA>G,
and miR-155T>A affect HTN development.
In conclusion, the present study investigated the
relationship between HTN susceptibility and LEPR Del/Ins,
miR-27aA>G (rs895819) and miR-155T>A (rs767649)
polymorphisms. The present study revealed that the LEPR Del/Ins and
miR-27aA>G polymorphisms were associated with HTN susceptibility
and that the combination of three polymorphisms was synergistic in
analyzing HTN susceptibility. The present study could provide a
basis for epidemiological studies of HTN and effective strategies
for the early prevention of HTN in the Korean population. To the
best of our knowledge, this is the first such study on HTN. The
study results need to be confirmed in more diverse ethnic
populations in the future. In addition, the study results require
validation in laboratory-based functional studies.
Acknowledgements
Not applicable.
Funding
Funding: This research was supported by the Basic Science
Research Program through the National Research Foundation of Korea
funded by the Ministry of Education, Science and Technology (grant
no. NRF-2017R1D1A3B03027985).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
SHH collected the medical data and performed the
genetic analysis. YRK was a major contributor in analyzing and
interpreting the data, and in writing the manuscript. All authors
read and approved the final manuscript. YRK and SHH confirmed the
authenticity of all the raw data.
Ethics approval and consent to
participate
All enrolled subjects provided written informed
consent to participate in the study. This study was approved by the
Institutional Review Board (approval no. JEJUNUH 2020-07-005) of
Jeju National University Hospital (Jeju, Korea).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Oh J, Matkovich SJ, Riek AE, Bindom SM,
Shao JS, Head RD, Barve RA, Sands MS, Carmeliet G, Osei-Owusu P, et
al: Macrophage secretion of miR-106b-5p causes renin-dependent
hypertension. Nat Commun. 11(4798)2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Sun L, Zhang J and Li Y: Chronic central
miR-29b antagonism alleviates angiotensin II-induced hypertension
and vascular endothelial dysfunction. Life Sci.
253(116862)2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Lionakis N, Mendrinos D, Sanidas E,
Favatas G and Georgopoulou M: Hypertension in the elderly. World J
Cardiol. 4:135–147. 2012.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Romaine SPR, Charchar FJ, Samani NJ and
Tomaszewski M: Circulating microRNAs and hypertension-from new
insights into blood pressure regulation to biomarkers of
cardiovascular risk. Curr Opin Pharmacol. 27:1–7. 2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Yin J, Liu H, Huan L, Song S, Han L, Ren
F, Zhang Z, Zang Z, Zhang J and Wang S: Role of miR-128 in
hypertension-induced myocardial injury. Exp Ther Med. 14:2751–2756.
2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Gogiraju R, Hubert A, Fahrer J, Beate K,
Straub BK, Brandt M, Wenzel P, Münzel T, Konstantinides S and
Schäfer GHK: Endothelial leptin receptor deletion promotes cardiac
autophagy and angiogenesis following pressure overload by
suppressing Akt/mTOR signaling. Circ Heart Fail.
12(e005622)2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Al-Hussaniy HA, Ali Hikmate Alburghaif AH
and Naji MA: Leptin hormone and its effectiveness in reproduction,
metabolism, immunity, diabetes, hopes and ambitions. J Med Life.
14:600–605. 2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Houseknecht KL and Portocarrero CP: Leptin
and its receptors: Regulators of whole-body energy homeostasis.
Domest Anim Endocrinol. 15:457–475. 1998.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Aquino-Jarquin G: Emerging role of
CRISPR/Cas9 technology for microRNAs editing in cancer research.
Cancer Res. 77:6812–6817. 2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Cao Q, Lu K, Dai S, Hu Y and Fan W:
Clinicopathological and prognostic implications of the miR-200
family in patients with epithelial ovarian cancer. Int J Clin Exp
Pathol. 7:2392–2401. 2014.PubMed/NCBI
|
|
11
|
Zhao H, Guo Y, Sun Y, Zhang N and Wang X:
miR-181a/b-5p ameliorates inflammatory response in
monocrotaline-induced pulmonary arterial hypertension by targeting
endocan. J Cell Physiol. 235:4422–4433. 2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Ghaedi H, Tabasinezhad M, Alipoor B,
Shokri F, Movafagh A, Mirfakhraie R, Omrani MD and Masotti A: The
pre-mir-27a variant rs895819 may contribute to type 2 diabetes
mellitus susceptibility in an Iranian cohort. J Endocrinol Invest.
39:1187–1193. 2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Pheiffer C, Dias S, Rheeder P and Adam S:
Decreased expression of circulating miR-20a-5p in south African
women with gestational diabetes mellitus. Mol Diagn Ther.
22:345–352. 2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Collares RVA, Salgado W Jr, Pretti da
Cunha Tirapelli D and Santos JS: The expression of LEP, LEPR, IGF1
and IL10 in obesity and the relationship with microRNAs. PLoS One.
9(e93512)2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Ciccacci C, Di Fusco D, Cacciotti L,
Morganti R, D'Amato C, Greco C, Rufini S, Novelli G, Sangiuolo F,
Spallone V and Borgiani P: MicroRNA genetic variations: Association
with type 2 diabetes. Acta Diabetol. 50:867–872. 2013.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Xu CC, Han WQ, Xiao B, Li NN, Zhu DL and
Gao PJ: Differential expression of microRNAs in the aorta of
spontaneously hypertensive rats. Acta Physiologica Sinica.
60:553–560. 2009.PubMed/NCBI
|
|
17
|
Latini A, Spallone V, D'Amato C, Novelli
G, Borgiani P and Ciccacci C: A common polymorphism in miR155 gene
promoter region is associated with a lower risk to develop type 2
diabetes. Acta Diabetologica. 56:717–718. 2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Winick JD, Stoffel M and Friedman JF:
Identification of microsatellites linked to the human leptin
receptor gene on chromosome 1. Genomics. 36:221–222.
1996.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Salopuro T, Pulkkinen L, Lindstrom J,
Eriksson JG, Valle TT, Hämäläinen H, Ilanne-Parikka P,
Keinänen-Kiukaanniemi S, Tuomilehto J, Laakso M, et al: Genetic
variation in leptin receptor gene is associated with type 2
diabetes and body weight: The finish diabetes prevention study. Int
J Obes (Lond). 29:1245–1251. 2005.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Ross J: mRNA stability in mammalian cells.
Microbiol Rev. 59:423–450. 1995.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wang Z and Kiledjian M: The poly
(A)-binding protein and an mrna stability protein jointly regulate
an endoribonuclease activity. Mol Cell Biol. 20:6334–6341.
2000.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Thompson DB, Sutherland J, Appel W and
Ossowski V: A physical map at 1p31 encompassing the acute insulin
response locus and the leptin receptor. Genomics. 39:227–230.
1997.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Oksanen L, Kaprio J, Mustajoki P and
Kontula K: A common pentanucleotide polymorphism of the
3'-untranslated part of the leptin receptor gene generates a
putative stem-loop motif in the mRNA and is associated with serum
insulin levels in obese individuals. Int J Obes Relat Metab Disord.
22:634–640. 1998.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Francke S, Clement K, Dina C, Inoue H,
Behn P, Vatin V, Basdevant A, Guy-Grand B, Permutt MA, Froguel P,
et al: Genetic studies of the leptin receptor gene in morbidly
obese French Caucasian families. Hum Genet. 100:491–496.
1997.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Lakka HM, Oksanen L, Tuomainen TP, Kontula
K and Salonen JT: The common pentanucleotide polymorphism of the
3'-untranslated region of the leptin receptor gene is associated
with serum insulin levels and the risk of type 2 diabetes in
non-diabetic men: A prospective case-control study. J Intern Med.
248:77–83. 2000.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Nishikai K, Hirose H, Ishii T, Hayashi M,
Saito I and Saruta T: Effects of leptin receptor gene
3'-untranslated region polymorphism on metabolic profiles in young
Japanese men. J Atheroscler Thromb. 11:73–78. 2004.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Nannipieri M, Posadas R, Bonotti A,
Bonotti A, Williams K, Gonzalez-Villalpando C, Stern MP and
Ferrannini E: Polymorphism of the 3'-untranslated region of the
leptin receptor gene, but not the adiponectin SNP45 polymorphism,
predicts type 2 diabetes: A population-based study. Diabetes Care.
29:2509–2511. 2006.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Hameed I, Masoodi SR, Afroze D, Bhat RA,
Naykoo NA, Mir SA, Mubarik I and Ganai BA: CTTTA Deletion/Insertion
polymorphism in 3'-UTR of LEPR gene in type 2 diabetes subjects
belonging to Kashmiri population. J Diabetes Metab Disord.
13(124)2014.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Rah HC, Chung KW, Ko KH, Kim ES, Kim JO,
Sakong JH, Kim JH, Lee WS and Kim NK: miR-27a and miR-449b
polymorphisms associated with a risk of idiopathic recurrent
pregnancy loss. PLoS One. 12(e0177160)2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Choi GH, Ko KH, Kim JO, Kim J, Oh SH, Han
IB, Cho KG, Kim OJ, Bae J and Kim NK: Association of miR-34a,
miR-130a, miR-150 and miR-155 polymorphisms with the risk of
ischemic stroke. Int J Mol Med. 38:345–356. 2016.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Korea Centers for Disease Control &
Prevention: Korea Health Statistics 2020: Korea national health and
nutrition examination survey (KNHANES Ⅷ-2). Retrieved from
https://knhanes.kdca.go.kr/knhanes/sub04/sub04_04_01.do,
2020.
|
|
32
|
Marques FZ and Charchar FJ: microRNAs in
essential hypertension and blood pressure regulation. Adv Exp Med
Biol. 888:215–235. 2015.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Zou X, Jialiang Wang J, Chen C, Xiaorong
Tan X, Huang Y, Jose PA, Yang J and Zeng C: Secreted monocyte
miR-27a, via mesenteric arterial mas receptor-eNOS pathway, causes
hypertension. Am J Hypertens. 33:31–42. 2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Masoumi-Ardakani Y, Najafipour H, Nasri
HR, Aminizadeh S, Jafari S and Moflehi D: Effect of combined
endurance training and mitoQ on cardiac function and serum level of
antioxidants, NO, miR-126, and miR-27a in hypertensive individuals.
Biomed Res Int. 2022(8720661)2022.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Shankaran ZS, Walter CEJ, Prakash N,
Ramachandiran K, Doss GP and Johnson T: Investigating the role of
microRNA-27a gene polymorphisms and its interactive effect with
risk factors in gastrointestinal cancers. Heliyon.
6(e03565)2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Shi D, Li P, Ma L, Zhong D, Chu H, Yan F,
Lv Q, Qin C, Wang W, Wang M, et al: A genetic variant in
pre-miR-27a is associated with a reduced renal cell cancer risk in
a Chinese population. PLoS One. 7(e46566)2012.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Wang B, Ma N and Wang Y: Association
between the hsa-mir-27a variant and breast cancer risk: A
meta-analysis. Asian Pacific J Cancer Prev. 13:6207–6210.
2012.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Yang Y, Shi X, Du Z, Zhou G and Zhang X:
Associations between genetic variations in microRNA and myocardial
infarction susceptibility: A meta-analysis and systematic review.
Herz. 47:524–535. 2022.PubMed/NCBI View Article : Google Scholar
|