Introduction
Polycystic ovary syndrome (PCOS) is commonly
associated with anovulation, hyperandrogenism (clinical and/or
biochemical), and polycystic ovaries (excluding other androgenic,
pituitary, or adrenal causes). It is estimated that 5-10% of all
women of reproductive age suffer from PCOS (1,2). In
addition to its association with multifactorial endocrine
disorders, PCOS is also associated with an increased lifetime risk
of obesity and infertility, which are present in ~74% of PCOS
patients (3). Therefore, timely
diagnosis and treatment of PCOS are not only important for
reproductive health, but also for monitoring and intervention of
complications. These factors are also prerequisites for gaining a
clearer understanding of the pathogenesis of PCOS. Serum
biochemical markers, such as luteinizing hormone (LH),
follicle-stimulating hormone (FSH), and androgen concentration,
have been long used as biochemical indices for the diagnosis of
PCOS. However, serum biochemical marker levels are affected by
different physiological cycles diminishing their utility for
diagnosing PCOS.
The role of exosomal microRNAs (miRNAs/miRs) in the
occurrence, progression, and outcomes of tumors has been reported
(4-6),
while the study of miRNAs related to PCOS in exosomes is in the
early stages (7,8). The etiology of PCOS cases is highly
heterogeneous with genetically complex and individual differences
that are not only related to the endocrine system but are also
greatly influenced by the environment and gene expression factors,
such as miRNAs (9).
miRNAs are short non-coding RNAs that regulate gene
expression at the post-transcriptional level (10,11).
miRNAs have been confirmed to be encapsulated in microvesicles
(12-16),
and exist in most body fluids such as saliva, serum, plasma, urine,
milk, and follicular fluid (17).
Studies comparing the differences in miRNA expression between
follicular fluid exosomes and plasma in women of reproductive age
have shown that, among the 37 miRNAs with upregulated expression in
follicular fluid, 32 were expressed in exosomes and may be involved
in several key signaling pathways of follicular development and egg
cell maturation, such as the WNT, mitogen-activated protein kinase
(MAPK), ErbB, and transforming growth factor (TGF)-β1 signaling
pathways (18). miRNAs in serum are
rich, stable, and easy to detect, and may thus serve as
non-invasive diagnostic markers of PCOS.
The present study aimed to identify novel miRNAs
that are differentially expressed in PCOS patients by screening
exosomal miRNAs in a PCOS cohort. The identified differentially
expressed miRNAs may have potential value as diagnostic biomarkers
for sensitive and accurate diagnosis of PCOS.
Materials and methods
Participants and selection
criteria
Between October 2016 and June 2020, 122 patients
with PCOS (aged 18-35 years) were recruited from Hangzhou Women's
Hospital (Hangzhou, China). Written informed consent was provided
by the patients or their legal guardians according to the
Declaration of Helsinki (19). All
experimental protocols were approved by the Medical Ethics
Committee of the hospital and were performed in accordance with the
relevant guidelines and regulations. All women were diagnosed
according to the Rotterdam criteria and did not have Cushing's
syndrome, late-onset congenital adrenal hyperplasia, thyroid
dysfunction, hyperprolactinemia, or androgen-secreting tumors.
Other exclusion criteria included diabetes, hypertension, chronic
renal disease, smoking, and the use of alcohol or medications. A
group of 112 age-matched first-trimester individuals (aged 18-36
years) with no previous history of reproductive system diseases or
appendicitis served as the control. The control group had normal
and regularly cycling menstrual periods, and their ovaries appeared
normal on ultrasonography. The exclusion criteria for the control
women in the study were the use of drugs, including oral
contraceptives or other hormonal drugs, intrauterine device
placement, and smoking within the past 3 months (20).
We selected the common gynecological diseases,
menopausal syndrome (MPS) and abnormal uterine bleeding (AUB), as
control disease groups to improve the experimental study. The 10
women in the MPS group were diagnosed according to the
International Clinical Practice Guideline of Chinese Medicine
Climacteric Syndrome (21). The 10
women in the AUB group were diagnosed according to the Guidelines
on the Diagnosis and Treatment of Abnormal Uterine Bleeding
(22).
In the first stage of the study, five pairs of serum
samples from five patients with PCOS and five healthy individuals
were combined into two group of samples as the screening group.
Exosomes and total RNA were extracted, and differentially expressed
miRNAs were screened by RNA sequencing (RNA-seq) using a commercial
service (Guangzhou RiboBio Co., Ltd.). Following preliminary
screening, two miRNAs with significantly upregulated expression
(miR-151a-5p and miR-223-3p) and one miRNA with significantly
decreased expression (miR-4488a) were identified and validated.
Reverse transcription-quantitative PCR (RT-qPCR) was performed to
validate the expression of these three miRNAs for the next
stage.
Serum collection, blood glycolipid
assay, hormone assessment, and exosomal purification
After fasting for 8-12 h, 5 ml blood from controls
and patients with PCOS was drawn in the morning. Whole blood was
separated by centrifugation at 1,000 x g for 10 min at room
temperature, and the isolated serum was centrifuged at 10,000 x g
for another 10 min at room temperature to completely remove cells
and debris. Prepared serum samples were stored at -80˚C until
required.
The serum anti-Müllerian hormone (AMH) concentration
was determined using a Roche Cobas e411 automated
electrochemiluminescence immunoassay analyzer (Roche GmbH). Serum
fasting plasma insulin, testosterone, dehydroepiandrosterone
sulphate (DHEA-S), and sex hormone-binding globulin levels were
measured using an automated chemiluminescence immunoassay performed
on a UniCel DxI 800 analyzer (Beckman Coulter, Inc.). Serum glucose
and lipid levels were measured using a Beckman Coulter AU5800
automatic biochemical analyzer (Beckman Coulter, Inc.). All tests
were performed in strict accordance with the manufacturer's
recommended protocols and reagent instructions. The free androgen
index (FAI) was calculated using the equation: FAI=(total
testosterone nmol/lx100)/(sex hormone-binding globulin nmol/l)
(23). Insulin resistance was
calculated using the HOMA method [insulin resistance=(insulin x
glucose)/22.5] (24).
Exosomal RNA extraction and the
RT-qPCR
Plasma exosomes were separated according to the
manufacturer's instructions (Invitrogen; Thermo Fisher Scientific,
Inc.) with minor modifications. Briefly, 500 µl serum was added to
a new tube, and 100 µl total exosome isolation reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) was added. The mixture was then
vortexed until a homogeneous solution was obtained. The sample was
incubated at 4˚C for 30 min, followed by centrifugation at 10,000 x
g for 10 min at room temperature. The supernatant was aspirated and
discarded. The exosomes were present in the pellet that was
re-suspended in 200 µl PBS. Due to the failure of exosome
extraction in some samples, the samples for subsequent RT-qPCR were
reduced to 107 samples in the PCOS group and 101 samples in the
control group.
miRNAs were extracted and purified using a
commercial kit (BioTeke Corporation) and 50 µl miRNA was acquired.
cDNA synthesis and RT-qPCR of miR151a, miR223a, and miR4488a were
performed as recommended by the manufacturer. Briefly, miRNAs were
reverse transcribed into DNA using the RT™ All-in-One Master Mix
(Herogen Biotech). The bulge-loop miRNATM RT-qPCR
Primers Sets (one RT primer and a pair of qPCR primers for each
set) specific for miR-151a-5p, miR-223-3p, and miR-4488 were
designed and synthesized by Guangzhou RiboBio Co., Ltd. The primer
sequences are proprietary and are not disclosed.
Synthetic U6 was added routinely to a final
concentration of 1,024 pmol/ml in all samples to control for
variations during RNA extraction and/or purification due to the
absence of homologous sequences in humans (25,26).
Furthermore, all study participants were recruited during the same
period, and the specimens were stored under the same conditions and
processed in equal volumes at each experimental step to control for
potential bias.
The RT-PCR solution contained 2.0 µl 5 RT™ Mix, 6.5
µl mRNA template, 0.5 µl each primer, and 1.0 µl nuclease-free
water in a total volume of 10 µl (Herogen Biotech). qPCR was
performed in an ABI Prism 7500 Sequence Detection System (Applied
Biosystems; Thermo Fisher Scientific, Inc.) with the following
thermocycling conditions: 25˚C for 10 min, 42˚C for 50 min;
followed by 45 cycles of 95˚C for 15 sec, and 60˚C for 40 sec. U6
mRNA was used as the internal control, and a no-template control
was used as a negative control.
The qPCR mixture contained 10.0 µl EvaGreen qPCR
Master Mix, 2.0 µl cDNA template, 0.5 µl forward and reverse primer
each, and 7.0 µl nuclease-free water in a total volume of 20 µl
(Herogen Biotech). qPCR was performed in an ABI Prism 7500 Sequence
Detection System with the following thermocycling conditions: 95˚C
for 10 min; followed by 40 cycles of 95˚C for 15 sec and 60˚C for
30 sec. For quantitative results, the relative expression levels of
each miRNA are presented as a fold change using the
2-ΔΔCq method (27).
Each sample was assayed in duplicate, and the average was used for
the analysis.
Functional enrichment analyses
The functions of the differentially expressed miRNAs
were analyzed using bioinformatics analysis. Related biological
pathways were analyzed using the Kyoto Encyclopedia of Genes and
Genomes (KEGG, http://www.genome.jp/kegg) and gene set enrichment
using Gene Ontology (GO) (http://www.geneontology.org/). The-log (P-value) was
used as the enrichment score, which indicated the significance of
the correlation.
Statistical analysis
All data were analyzed using SPSS version 17.0 (IBM
Corp.). The normality of the distribution of continuous variables
was assessed using the Kolmogorov-Smirnov test. Data are presented
as the mean ± standard deviation, or median (range). Differences in
the means of two groups of data were compared using a Student's
t-test, and the medians of two groups were compared using a
Mann-Whitney U test. Spearman's correlation coefficients were
calculated to evaluate the relationship between miRNA levels and
other variables in both groups. The optimal cut-off points for the
miRNA and AMH levels to distinguish between the two groups were
evaluated using receiver operating characteristic (ROC) analyses
after calculating the area under the curve (AUC), given the maximum
sum of the sensitivity and specificity (the Youden Index) for the
significance test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Patient characteristics
The metabolic characteristics based on the samples
of the two groups are provided in Table
I. There were no significant differences observed in age,
testosterone levels, and high-density lipid cholesterol levels
(P>0.05). The levels of AMH, DHEA-S, triglyceride, serum total
cholesterol, low-density lipoprotein cholesterol, fasting blood
glucose, and insulin in the serum of the PCOS group were
significantly higher than those of the control subjects (P<0.001
and P<0.05). In contrast, PCOS patients showed lower levels of
sex hormone-binding globulin than controls (P<0.001). PCOS
patients also showed significantly higher FAI and HOMA insulin
resistance values compared to those of the control group
(P<0.001). FSH and LH levels of first-trimester women that
served as control individuals were very low or undetectable and are
not listed. High glucose and insulin levels are important causes of
type 2 diabetes mellitus and insulin resistance in PCOS patients,
and high androgen levels are one of the characteristics of PCOS.
Overall, abnormalities of the metabolic characteristics in PCOS
patients contributed gradually to the development of the disease,
which affected their health and quality of life.
 | Table IHormonal and metabolic variables in
the controls and patients with PCOS. |
Table I
Hormonal and metabolic variables in
the controls and patients with PCOS.
| Variable | PCOS,
n=122c | Control,
n=112c | P-value |
|---|
| Age, years | 26.0
(23.0-28.25) | 28.0
(23.0-30.0) | 0.102 |
| AMH, ng/ml | 9.26
(7.55-12.99) | 2.72
(1.90-4.45) |
<0.001b |
| Testosterone,
nmol/l | 2.43
(1.92-2.84) | 2.41
(1.80-3.15) | 0.971 |
| DHEA-S, µmol/l | 8.99
(6.44-11.51) | 5.09
(3.66-6.82) |
<0.001b |
| SHBG, nmol/l | 38.10
(23.70-65.83) | 91.50
(66.10-141.50) |
<0.001b |
| FAI | 6.21
(2.80-11.57) | 2.32
(1.57-3.44) |
<0.001b |
| TG, mmol/l | 4.85
(4.37-5.45) | 3.89
(3.48-4.31) |
<0.001b |
| TC, mmol/l | 0.98
(0.69-1.51) | 0.73
(0.56-0.94) |
<0.001b |
| HDLC, mmol/l | 1.65±0.40 | 1.57±0.32 | 0.058 |
| LDLC, mmol/l | 2.75
(2.31-3.21) | 2.01
(1.74-2.34) |
<0.001b |
| FBG, mmol/l | 4.76
(4.48-5.22) | 4.64
(4.41-4.88) | 0.019a |
| FINS, µIU/ml | 6.80
(4.68-11.78) | 5.30
(3.70-6.60) |
<0.001b |
| HOMA-IR | 1.47
(0.97-2.69) | 1.05
(0.78-1.36) |
<0.001b |
| LH/FSH, IU/l | 2.24
(1.21-3.94) | - | - |
Expression of miRNA in controls and
PCOS patients
During the screening stage, two miRNAs (miR-151a-5p
and miR-4488) with upregulated expression and one miRNA
(miR-223-3p) with downregulated expression were identified. Next,
the expression levels of these miRNAs in the validation groups were
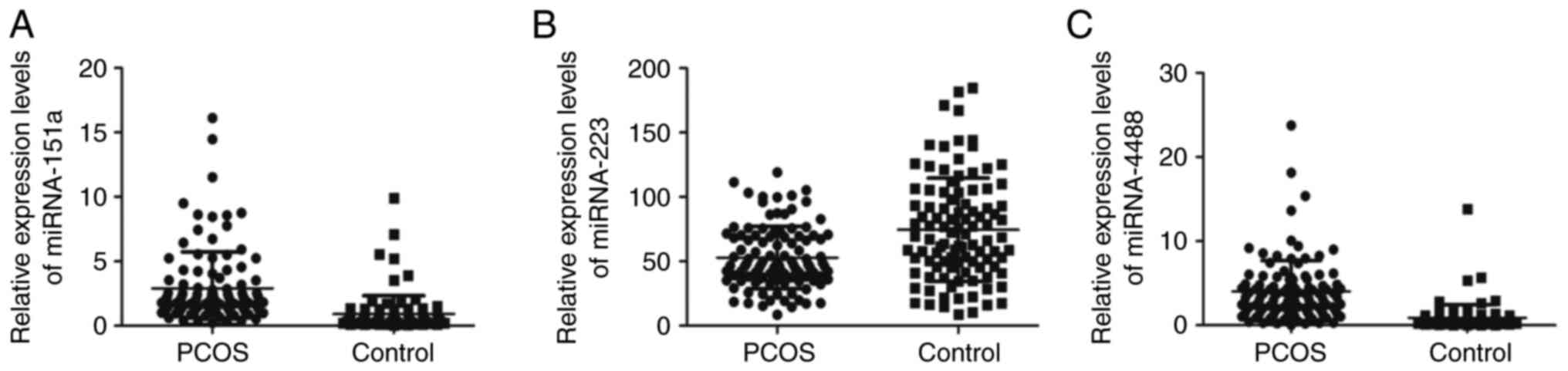
quantified using RT-qPCR. The miR-151a-5p levels in the PCOS group
were 4x that in the control subjects. Compared to control subjects,
the expression levels of miR-4488 in the PCOS group were increased
by ~570% (P<0.001; Fig. 1A and
C). The relative expression of
miR-223-3p in the PCOS group was reduced ~31% vs. control subjects
(P<0.001; Fig. 1B).
To demonstrate the specificity of exosomal miR-4488,
miR-151a-5p, and miR-223-3p to PCOS, the expression of these miRNAs
in patients with MPS and AUB were also detected. The results
provided in Table II revealed that
the expression levels of the three miRNAs differed significantly in
patients with MPS and AUB compared with those in the PCOS and
control groups (P<0.01). Moreover, the levels of these miRNAs
were several times higher in the PCOS group than in the MPS and AUB
patients. This supports the relative specificity of these three
miRNAs in PCOS vs. other gynecological diseases.
 | Table IIRelative expression levels of
miR-151a-5p, miR-223-3p, and miR-4488 in PCOS patients were
compared with the CON group, MPS group and AUB group. |
Table II
Relative expression levels of
miR-151a-5p, miR-223-3p, and miR-4488 in PCOS patients were
compared with the CON group, MPS group and AUB group.
| Variable | PCOS,
n=107b | Control,
n=101b | MPS,
n=10b | AUB,
n=10b | P-value |
|---|
| miR-151a-5p | 1.92
(1.36-3.20) | 0.48
(0.33-0.91) | 0.02
(0.01-0.05) | 0.01
(0.01-0.04) |
<0.01a |
| miR-4488 | 3.42
(1.73-4.89) | 0.51
(0.32-0.95) | 2.03
(1.04-4.81) | 2.36
(1.67-3.09) |
<0.01a |
| miR-223-3p | 47.01
(36.25-70.03) | 68.83
(45.78-100.9) | 0.20
(0.01-0.32) | 0.35
(0.18-0.51) |
<0.01a |
Correlations between miRNA levels and
other clinical parameters in PCOS patients
To evaluate the possible association between miRNA
levels and glycometabolic parameters, samples from the PCOS and
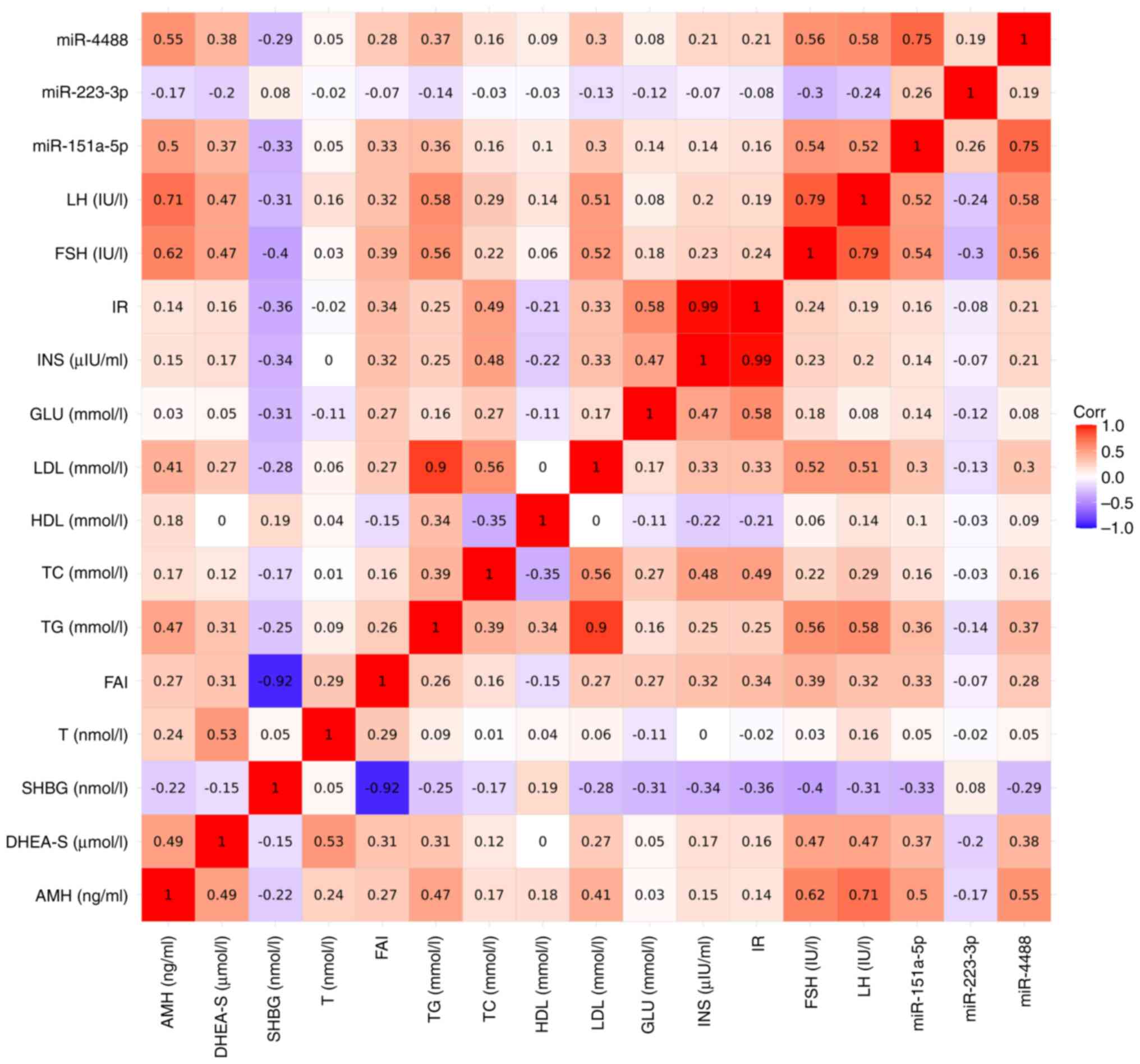
control groups were studied to determine any correlations (Fig. 2). The results revealed that the
miRNAs and clinical parameters were weak-moderately correlated with
various parameters. miRNA-151a-5p and miRNA-4488 were primarily
positively correlated with AMH, FSH, and LH, and miRNA-223-3p was
negatively correlated with FSH, LH, and DHEA-S. In addition, the
parameters measured showed notable correlations with each other
including FSH, LH, AMH, triglyceride, low-density lipoproteins,
glucose, and insulin resistance (Fig.
2). The results of the Spearman's correlation analyses are
shown in Table SI.
ROC curve analyses
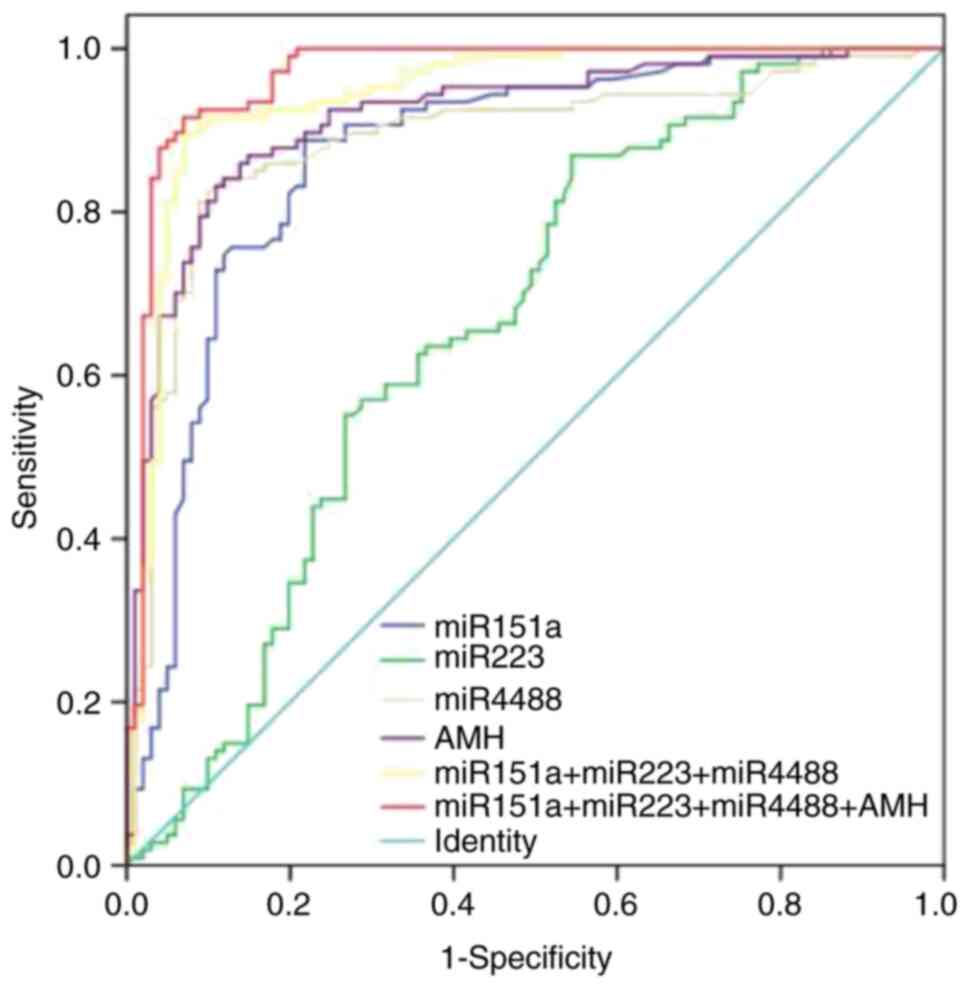
ROC analyses were performed to evaluate the value of
differentially expressed miRNAs in discriminating PCOS patients
from control subjects. The highest AUC value was observed for
miR-4488 (AUC=0.889; P<0.001), followed by miR-151a-5p
(AUC=0.871; P<0.001). The lowest value was observed for
miR-223-3p (AUC=0.664; P<0.001) (Fig. 3). Accordingly, miR-4488 had a
sensitivity and specificity of 81.3 and 91.1%, respectively.
miR-151a-5p showed 88.8% sensitivity and 78.2% specificity, whereas
miR-223-3p showed 86.9% sensitivity and 45.5% specificity (Fig. 3).
Among miR-151a-5p and miR-223-3p, miR-4488 had an
AUC of 0.889 which increased the sensitivity to 89.7% and slightly
increased the specificity to 93.1%. Evaluation of AMH yielded an
AUC value of 0.926 which increased further when combined with the
three miRNAs (AUC=0.967; P<0.001), with a sensitivity and
specificity of 91.6 and 93.1%, respectively, indicating this may be
a more potent diagnostic tool. The results of the ROC curve
analysis for miRNA and clinical measurement discrimination are
shown in Table SII.
Functional analysis of predicted
targets of miR-151a-5p, miR-223-3p, and miR-4488
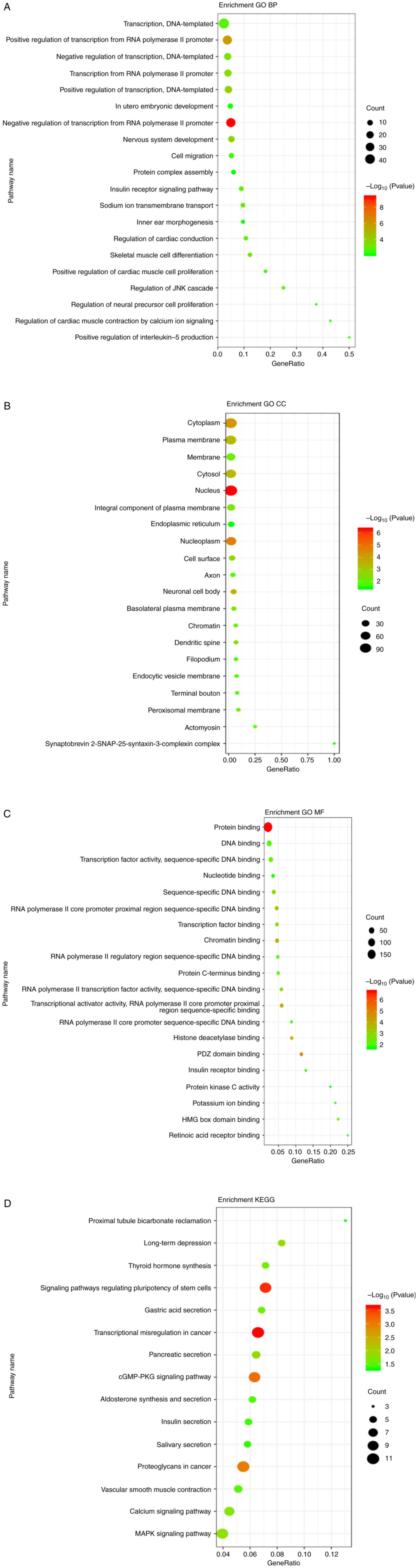
GO and KEGG pathway analyses were performed using
TarBase to identify the predicted conserved targets and biological
functions potentially influenced by the three miRNAs. In the GO
analysis, the number of target genes corresponding to GO entries
was determined, and the enrichment score was regarded as the-log
(P-value); the target genes of the three analyzed miRNAs are
summarized in Fig. 4A-C. For the
biological process enrichment, the most significant term was
organelle organization, which was significantly enriched in the
independent targets of both miR-223-3p and miR-4488. In the
cellular component, the term with the most genes was ‘protein
complex’, and the most significantly enriched term was ‘cytosol’.
For molecular functions, the most significantly enriched term was
‘binding’. Thus, the three mRNAs were involved in metabolic
processes, growth, and development.
The KEGG pathway analysis identified the top 15
significantly enriched pathways (Fig.
4D). The three miRNAs were involved in multiple signaling
pathways such as those regulating the pluripotency of stem cells,
transcriptional miscegenation in cancer, the cGMP-protein kinase A
signaling pathway, proteoglycans in cancer, and the MAPK signaling
pathway, which is related to cancer, hormone secretion, and
pluripotency of stem cells.
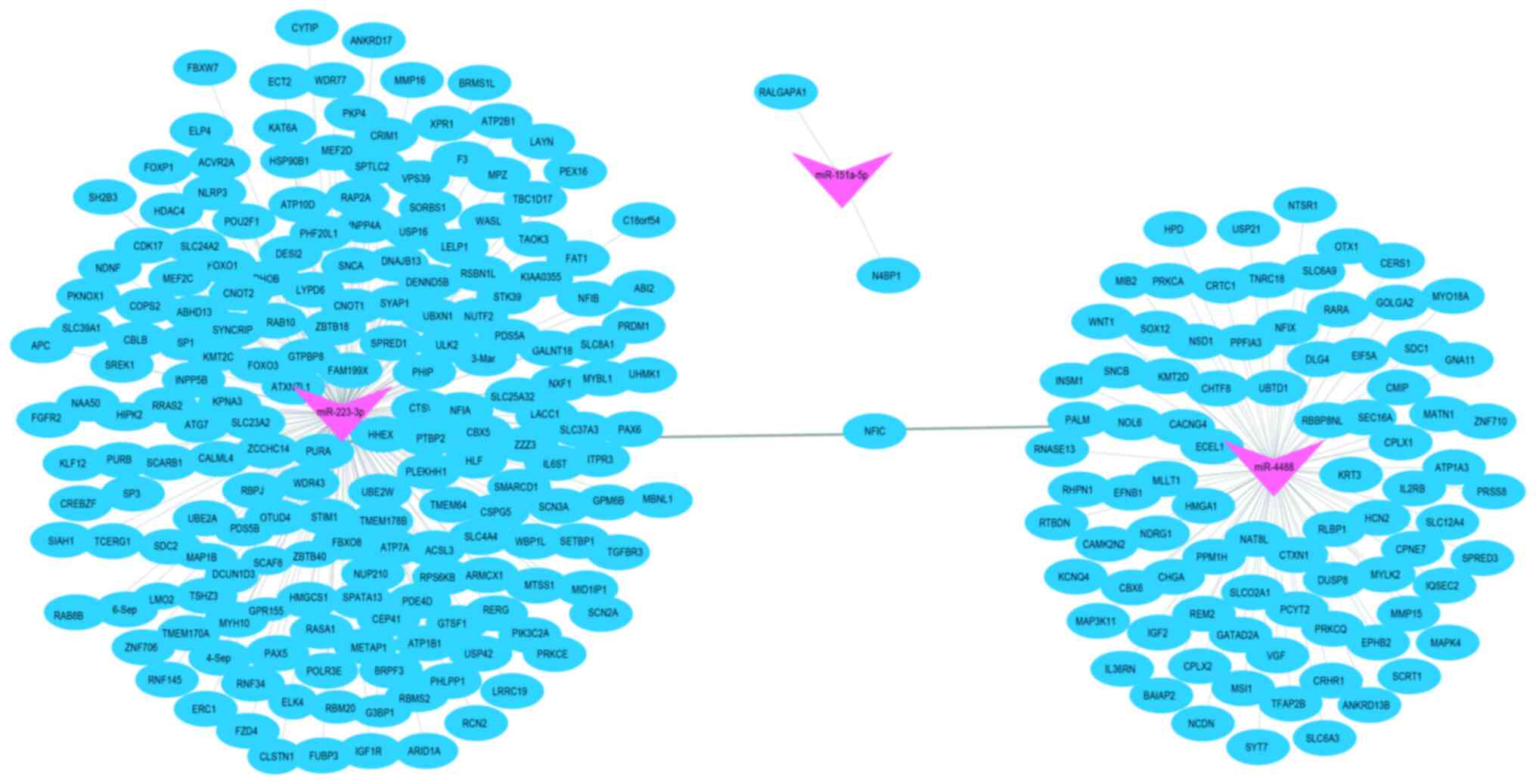
The mapping analysis using Cytoscape software
indicated that miR-151a-5p, miR-223-3p, and miR-4488 may influence
the expression of certain genes (such as N4BP1, PURA, and ECEL1)
that were primarily enriched in several pathways associated with
mRNA binding activity, single-stranded DNA-binding proteins,
cellular responses to ultraviolet radiation, and negative
regulation of viral genome replication. These genes were predicted
to be located in the cytosol and nucleolus, and to be regulated by
neuropeptides and peptide hormones. Interestingly, there were no
common genes among the three miRNAs (Fig. 5), suggesting that these three miRNAs
may play independent roles in the regulation of biological
processes through different pathways.
Discussion
Here, for the first time, it was shown that exosomal
miRNAs with increased/decreased expression, namely miR-151a-5p,
miR-4488, and miR-223-3p, were associated with the risk of PCOS. To
evaluate the diagnostic value of the miRNAs for PCOS, ROC curves
and AUCs were used, revealing that a combination of the three
miRNAs and AMH may offer a potent diagnostic tool to distinguish
between patients with PCOS and controls, with a sensitivity and
specificity of 91.6 and 93.1%, respectively. This suggests that
exosomal miRNA expression patterns in PCOS samples differ from
those in controls.
The experimental results demonstrated that serum
miRNAs are differentially expressed in PCOS. Compared with other
gynecological diseases such as MPS and AUB, the expression of
miR-151a-5p, miR-4488, and miR-223-3p was relatively specific to
PCOS. The increased fasting blood glucose and insulin levels found
in the PCOS samples could partly explain the changes in the miRNAs;
miRNA-151a-5p, miRNA-4488, and miRNA-223-3p were notably correlated
with other parameters, particularly AMH, FSH, and LH, which were
significantly associated with the upregulated miRNAs. In addition,
bioinformatics analyses showed that miR-151a-5p, miR-4488, and
miR-223-3p are related to insulin metabolism processes. Further
analysis showed that glycometabolic and hormone profiles had a
weak-moderate correlation with miRNAs. Of note, the changes in the
number of individuals for subsequent miRNA validation and
correlation analysis are not expected to have an impact on the
present findings and conclusions.
Previous studies have shown the involvement of
miRNAs in the pathophysiological mechanism of PCOS. For example,
one study showed that miR-222, miR-164a, and miR-30c were highly
expressed in PCOS patients, and miR-222 was strongly positively
correlated with serum insulin, whereas miR-164a was negatively
correlated with serum testosterone (28). In addition, researchers observed
decreased serum expression levels of miR-320 and showed
downregulated expression when treated with TGF-1, and increased
insulin resistance in PCOS subjects, while inconsistent expression
was observed in follicular fluid in PCOS and increased expression
of granulosa cells (9,29,30).
Interestingly, Mohammad, Naji et al (31) found that there was no significant
change in the expression of miRNAs in the serum samples from PCOS
individuals. In contrast to the significant decrease in follicular
fluid, the levels of miR-93 and miR-21 were significantly increased
in granuloma cells compared with those in normal androgenic
patients. Furthermore, serum miR-21, miR-27b, and miR-103 levels
were associated with PCOS, metabolic disorders, and low-level
inflammation. However, it has been suggested that the expression
profile of serum miRNAs does not necessarily reflect local changes
in the ovaries (9). As such,
interpreting miRNA expression in PCOS requires careful
consideration of various confounders, and further exploration of
the miRNA profiles is needed to understand the causality and
changes in correlation between miRNAs and glycolipid metabolism,
and to clarify how miRNAs change over time as the disease
progresses.
Numerous miRNAs are expressed in the ovaries and
regulate granulosa cell proliferation and apoptosis, follicular
growth, atresia, ovulation, luteinization and spermatogenesis, and
play an important role in ovarian disorders such as PCOS (32,33).
Manuela et al (18) compared
the differences in miRNA expression between follicular fluid
exosomes and plasma in women of reproductive age and found that the
expression of 37 miRNAs was upregulated in follicular fluid, 32 of
which existed in exosomes and may be involved in several key
signaling pathways of follicular development and egg cell
maturation, such as the WNT, MAPK, ErbB, and TGF-ß1 signaling
pathways.
Using a bioinformatics approach, it was found that
miR-4488 and miR-223-3p targeted numerous genes. The target genes
of miR-4488 and miR-223-3p were involved in various biological
processes, among which the most important were metabolic, growth,
and developmental processes, which likely affect the occurrence and
development of PCOS. In addition, these genes were associated with
multiple pathways such as insulin secretion, and the cGMP-PKG and
MAPK signaling pathways; the latter is one of the two
best-characterized insulin signaling pathways.
A recent study reported that plasma exosomal miRNAs
are involved in the proliferation and differentiation of insulin
target cells causing insulin resistance in women with PCOS
(34). Reproductive cellular
processes were involved and the p38 MAPK protein was found to be
expressed in oocytes and granular cells, which is central to
regulating oocyte maturation and fertilization (35-37).
Through the p38 MAPK and protein kinase A signaling pathways, FSH
and cAMP were interrelated and promoted the expression of AMH
(38). Thus, the differentially
expressed miRNAs enriched in this pathway may directly target the
ovaries, leading to substantial changes in patients with PCOS.
Both environmental and genetic factors contribute to
the etiology of PCOS (39), which
exhibits a range of symptoms such as oligomenorrhea, amenorrhea,
infertility, obesity, hirsutism, alopecia, acne vulgaris, and
insulin resistance (1-3).
Recent studies have reported that PCOS and depression share certain
similar clinical symptoms that can further affect the quality of
life (40,41).
In conclusion, as research on PCOS is still in its
relative infancy, an urgent direction for future studies is to
increase sample sizes and perform more functional analyses to
confirm the etiological, diagnostic, prognostic, and therapeutic
significance of miRNAs in PCOS. Collectively, the present study
should encourage further research and improve our understanding of
this topic.
Supplementary Material
Spearman’s correlation matrix.
Results of receiver operating curve
analysis for clinical measurement discrimination between the
control and polycystic ovarian syndrome groups.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Science and
Technology Commission of Hangzhou (grant no. 20150633B22) and the
Medical Health Science and Technology Project of Zhejiang
Provincial Health Commission (grant no. 2020KY763).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XS designed the study and analyzed and interpreted
the data. YL performed the experiments, analyzed the data and
drafted and revised the manuscript. BX and ZW performed the
experiments. YC and MD acquired and interpreted the data. All
authors have read and approved the final version of the manuscript.
XS and YL confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
All participants were recruited from the Hangzhou
Women's Hospital. Written informed consent was obtained from the
patients or their legal guardians. All experimental protocols were
approved by the Medical Ethics Committee of the Hangzhou Women's
Hospital (Hangzhou, China) and performed in accordance with
relevant guidelines and regulations [(2015) Scientific research
medical Review approval no. (002)-02].
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Dunaif A: Insulin resistance and the
polycystic ovary syndrome: Mechanism and implications for
pathogenesis. Endocr Rev. 18:774–800. 1997.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Yildiz BO, Bozdag G, Yapici Z, Esinler I
and Yarali H: Prevalence, phenotype and cardiometabolic risk of
polycystic ovary syndrome under different diagnostic criteria. Hum
Reprod. 27:3067–3073. 2012.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Vitek W, Hoeger K and Legro RS: Treatment
strategies for infertile women with polycystic ovary syndrome.
Minerva Ginecol. 68:450–457. 2016.PubMed/NCBI
|
|
4
|
Skog J, Würdinger T, van Rijn S, Meijer
DH, Gainche L, Sena-Esteves M, Curry WT Jr, Carter BS, Krichevsky
AM and Breakefield XO: Glioblastoma microvesicles transport RNA and
protein that promote tumor growth and provide diagnostic
biomarkers. Nat Cell Biol. 10:1470–1476. 2008.PubMed/NCBI View
Article : Google Scholar
|
|
5
|
Ohshima K, Inoue K, Fujiwara A, Hatakeyama
K, Kanto K, Watanabe Y, Muramatsu K, Fukuda Y, Ogura S, Yamaguchi K
and Mochizuki T: Let-7 microRNA family is selectively secreted into
the extracellular environment via exosomes in a metastatic gastric
cancer cell line. PLoS One. 5(e13247)2010.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Sun Z, Shi K, Yang S, Liu J, Zhou Q, Wang
G, Song J, Li Z, Zhang Z and Yuan W: Effect of exosomal miRNA on
cancer biology and clinical applications. Mol Cancer.
17(147)2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Hossain MM, Cao M, Wang Q, Kim JY,
Schellander K, Tesfaye D and Tsang BK: Altered expression of miRNAs
in a dihydrotestosterone-induced rat PCOS model. J Ovarian Res.
6(36)2013.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Sirotkin AV, Lauková M, Ovcharenko D,
Brenaut P and Mlyncek M: Identification of microRNAs controlling
human ovarian cell proliferation and apoptosis. J Cell Physiol.
223:49–56. 2010.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Sørensen AE, Wissing ML, Salö S, Englund
AL and Dalgaard LT: MicroRNAs related to polycystic ovary syndrome
(PCOS). Genes (Basel). 5:684–708. 2014.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Ambros V: MicroRNAs: Tiny regulators with
great potential. Cell. 107:823–826. 2001.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Bartel D: MicroRNAs: Genomics, biogenesis,
mechanism, and function. Cell. 116:281–297. 2004.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Gallo A, Tandon M, Alevizos I and Illei
GG: The majority of microRNAs detectable in serum and saliva is
concentrated in exosomes. PLoS One. 7(e30679)2012.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Hunter MP, Ismail N, Zhang X, Aguda BD,
Lee EJ, Yu L, Xiao T, Schafer J, Lee ML, Schmittgen TD, et al:
Detection of microRNA expression in human peripheral blood
microvesicles. PLoS One. 3(e3694)2008.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Diez-Fraile A, Lammens T, Tilleman K,
Witkowski W, Verhasselt B, De Sutter P, Benoit Y, Espeel M and
D'Herde K: Age-associated differential microRNA levels in human
follicular fluid reveal pathways potentially determining fertility
and success of in vitro fertilization. Hum Fertil (Camb). 17:90–98.
2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Da Silveira JC, Veeramachaneni DN, Winger
QA, Carnevale EM and Bouma GJ: Cell-secreted vesicles in equine
ovarian follicular fluid contain miRNAs and proteins: A possible
new form of cell communication within the ovarian follicle. Bio
Reprod. 86(71)2012.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Sang Q, Yao Z, Wang H, Feng R, Wang H,
Zhao X, Xing Q, Jin L, He L, Wu L and Wang L: Identification of
microRNAs in human follicular fluid: Characterization of microRNAs
that govern steroidogenesis in vitro and are associated with
polycystic ovary syndrome in vivo. J Clin Endocrinol Metab.
98:3068–3079. 2013.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Simons M and Rapose G: Exosmes-vesicular
carriers for intercellular communication. Curr Opin Cell Biol.
21:575–581. 2009.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Santonocito M, Vento M, Guglielmino MR,
Battaglia R, Wahlgren J, Ragusa M, Barbagallo D, Borzì P, Rizzari
S, Maugeri M, et al: Molecular characterization of exosomes and
their microRNA cargo in human follicular fluid: Bioinformatic
analysis reveals that exosomal microRNAs control pathways involved
in follicular maturation. Fertil Steril. 102:1751–1761.e1.
2014.PubMed/NCBI View Article : Google Scholar
|
|
19
|
World Medical Association. World Medical
Association Declaration of Helsinki: Ethical principles for medical
research involving human subjects. JAMA. 310:2191–2194.
2013.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Xiong W, Lin Y, Xu L, Tamadon A, Zou S,
Tian F, Shao R, Li X and Feng Y: Circulatory microRNA 23a and
microRNA 23b and polycystic ovary syndrome (PCOS): The effects of
body mass index and sex hormones in an Eastern Han Chinese
population. J Ovarian Res. 10(10)2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Xiao CC, Liu YF, Wang TF, Xu MB, Du HL,
Han YH, et al: International clinical practice guideline of chinese
medicine climacteric syndrome. World J Tradit Chin Med. 7:276–279.
2021.
|
|
22
|
Gynecologic Endocrinology Subgroup,
Chinese Society of Obstetrics and Gynecology, Chinese Medical
Association. Guideline on diagnosis and treatment of abnormal
uterine bleeding: 2022 revisions. Zhonghua Fu Chan Ke Za Zhi.
57:481–490. 2022.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|
|
23
|
Vermeulen A, Verdonck L and Kaufman JM: A
critical evaluation of simple methods for the estimation of free
testosterone in serum. J Clin Endocrinol Metab. 84:3666–3672.
1999.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Albareda M, Rodríguez-Espinosa J, Murugo
M, de Leiva A and Corcoy R: Assessment of insulin sensitivity and
beta-cell function from measurements in the fasting state and
during an oral glucose tolerance test. Diabetologia. 43:1507–1511.
2000.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Rana S, Yue S, Stadel D and Zöller M:
Toward tailored exosomes: the exosomal tetraspanin web contributes
to target cell selection. Int J Biochem Cell Biol. 44:1574–1584.
2012.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Blanchard N, Lankar D, Faure F, Regnault
A, Dumont C, Raposo G and Hivroz C: TCR activation of human T cells
induces theproduction of exosomes bearing the TCR/CD3/zeta complex.
J Immunol. 168:3235–3241. 2002.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Long W, Zhao C, Ji C, Ding H, Cui Y, Guo
X, Shen R and Liu J: Characterization of serum microRNAs profile of
PCOS and identification of novel non-invasive biomarkers. Cell
Physiol Biochem. 33:1304–1315. 2014.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Yin M, Wang X, Yao G, Lü M, Liang M, Sun Y
and Sun F: Transactivation of miR-320 by miR-383 regulates
granulosa cell functions by targeting E2F1 and SF-1 proteins. J
Biol Chem. 289:18239–18257. 2014.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Ling HY, Ou HS, Feng SD, Zhang XY, Tuo QH,
Chen LX, Zhu BY, Gao ZP, Tang CK, Yin WD, et al: Change in microRNA
(miR) profile and effects of miR-320 in insulin-resistant 3T3-L1
adipocytes. Clin Exp Pharmacol Physiol. 36:e32–e39. 2009.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Naji M, Aleyasin A, Nekoonam S, Arefian E,
Mahdian R and Amidi F: Differential Expression of miR-93a and
miR-21 in granulosa cells and follicular fluid of polycystic ovary
syndrome associating with diferent phenotypes. Sci Rep.
7(14671)2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Li Y, Fang Y, Liu Y and Yang X: MicroRNAs
in ovarian function and disorders. J Ovarian Res.
8(51)2015.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Deswal R and Dang AS: Dissecting the role
of micro-RNAs as a diagnostic marker for polycystic ovary syndrome:
A systematic review and meta-analysis. Fertil Steril.
113:661–669.e2. 2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Jiang X, Li J, Zhang B, Hu J, Ma J, Cui L
and Chen ZJ: Differential expression profile of plasma exosomal
microRNAs in women with polycystic ovary syndrome. Fertil Steril.
115:782–792. 2021.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Bost F, Aouadi M, Caron L and Binétruy B:
The role of MAPKs in adipocyte differentiation and obesity.
Biochimie. 87:51–56. 2005.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Wu W, Zhang J, Zhao C, Sun Y, Pang W and
Yang G: CTRP6 regulates porcine adipocyte proliferation and
differentiation by the AdipoR1/MAPK signaling pathway. J Agric Food
Chem. 65:5512–5522. 2017.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Fan HY and Sun QY: Involvement of
mitogen-activated protein kinase cascade during oocyte maturation
and fertilization in mammals. Biol Reprod. 70:535–547.
2004.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Taieb J, Grynberg M, Pierre A, Arouche N,
Massart P, Belville C, Hesters L, Frydman R, Catteau-Jonard S,
Fanchin R, et al: FSH and its second messenger cAMP stimulate the
transcription of human anti-Mullerian hormone in cultured granulosa
cells. Mol Endocrinol. 25:645–655. 2011.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Ajmal N, Khan SZ and Shaikh R: Polycystic
ovary syndrome (PCOS) and genetic predisposition: A review article.
Eur J Obstet Gynecol Reprod Biol X. 3(100060)2019.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Cooney LG, Lee I, Sammel MD and Dokras A:
High prevalence of moderate and severe depressive and anxiety
symptoms in polycystic ovary syndrome: A systematic review and
meta-analysis. Hum Reprod. 32:1075–1091. 2017.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Kolhe JV, Chhipa AS, Butani S, Chavda V
and Patel SS: PCOS and Depression: Common links and potential
targets. Reprod Sci. 29:3106–3123. 2022.PubMed/NCBI View Article : Google Scholar
|