Introduction
Traditional homemade remedies, such as garlic, have
been used for the treatment of pain, inflammation and
cardiovascular disease. Scientific research exploring the medicinal
properties of garlic, focuses on allicin, diallyl sulfate and other
diallyls (1). The Allium
species, including garlic, contain sulfoxides with unique medicinal
properties, including antioxidant (2), anti-cancer (3), anti-viral, anti-microbial (4) and anti-fungal properties (5), and have been used in the treatment of
diabetes (6,7) and periodontal disease (8,9) and
for potentially preventing cardiovascular (10-12)
and neurodegenerative diseases (13,14).
This great variety in therapeutic properties is the factor that has
motivated such extensive research into garlic.
Allicin (diallyl thiosulfinate) is a prominent study
molecule linked to various beneficial properties; for instance, it
plays a protective role in cardiovascular diseases (15-17).
Its structure was described in 1948(18). Produced upon tissue damage by
garlic, allicin is a molecule that contributes to the defense of
the plant with a wide range of biological actions. Allicin is
almost exclusively responsible for the antimicrobial action of
freshly ground garlic (19) and it
also presents antifungal activity (20).
Despite all the interest in allicin, not all enzymes
involved in its biosynthetic pathway have been identified. In the
final steps of the biosynthetic pathway of allicin, γ-glutamyl
transpeptidases catalyze the removal of glutamyl from
γ-glutamyl-S-allyl-L-cysteine to produce S-allyl-L-cysteine (SAC)
(21), which in turn undergoes an
S-oxygenation, catalyzed by the flavin-dependent S-monooxygenase
(FMO) enzyme, resulting in the production of alliin
(S-allylcysteine sulfoxide) (22).
This non-proteinogenic amino acid is converted to allicin in a
reaction catalyzed by the enzyme alliinase. As a major precursor of
allicin, alliin is also crucial to scientific research in order to
further explore the biosynthetic pathway of allicin (23).
Thus far, three genes [Allium sativum
γ-glutamyl-transpeptidase (AsGGT)1, AsGGT2 and AsGGT3] encoding
γ-glutamyl transpeptidases, have been identified in garlic
(24). Recombinant peptides of
AsGGT1, AsGGT2 and AsGGT3 have exhibited notable deglutamylation
activity towards alliin's intermediate,
γ-glutamyl-S-allyl-L-cysteine. These proteins can function as
hydrolases without a suitable substrate; however, their activity
increases with glycylglycine. The three peptides, AsGGT1, AsGGT2
and AsGGT3, differ in their affinity for the
γ-glutamyl-S-allyl-L-cysteine substrate. AsGGT1 and AsGGT2 have a
high affinity for γ-glutamyl-S-allyl-L-cysteine and contribute to
alliin biosynthesis in leaves during bulb formation and maturation
(25). AsGGT3 may contribute to
alliin biosynthesis in bulbs upon dormancy termination (25). Additionally, AsGGT2 localizes to the
vacuole, while AsGGT1 and AsGGT3 lack a signal peptide for
intracellular organelles (24).
These γ-glutamyl transferases may contribute differently to alliin
biosynthesis in garlic and may act synergistically (25).
The size of the garlic nuclear genome is ~16.9 Gbp,
organized into eight chromosomes, and the number of predicted genes
thus far is 57,561(25). The garlic
genome owes this increased quantity primarily to polyploidy caused
by whole genome duplication events and transposable element
proliferation. The main aim of the present study was to map known
genes which code γ-glutamyl transpeptidases on the garlic genome
and to search for unidentified ones, as it was proposed that
differences in the expression of enzymes involved in the
biosynthesis of allicin, may be related to variant allicin
production levels between garlic cultivars.
Materials and methods
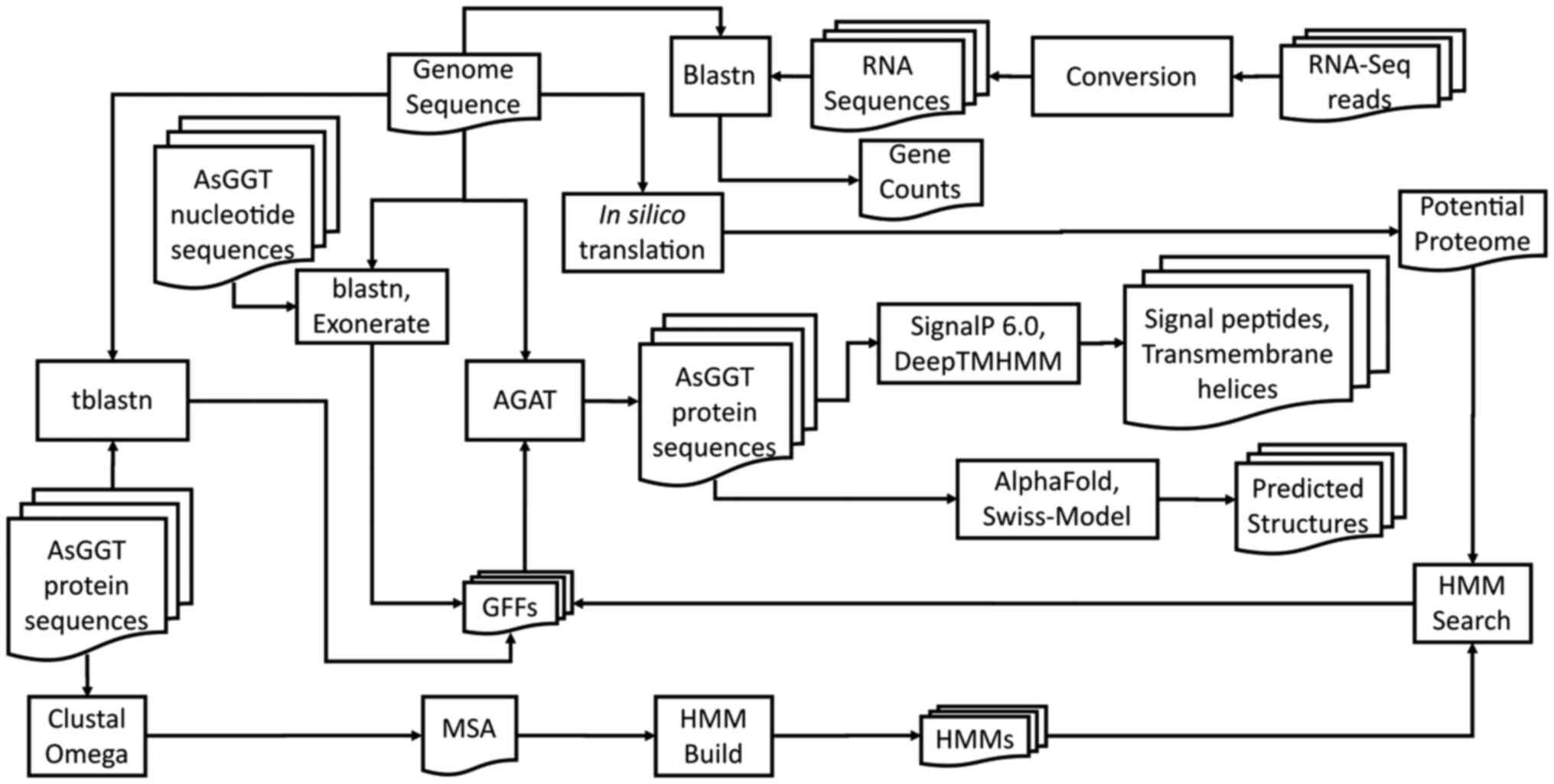
For the comprehensive analysis of all AsGGT genes
and gene products, a bioinformatics pipeline was followed (Fig. 1).
Genome mapping of characterized AsGGT
genes
To search for the nucleotide sequences of garlic
γ-glutamyl transpeptidase transcripts in the National Center for
Biotechnology Information (NCBI) GenBank (26), the following query was used:
‘gamma-glutamyl transpeptidase’ AND ‘Allium
sativum’[porgn:__txid4682]. The three resulting nucleotide
sequences were stored in FASTA format. To identify and download
their corresponding peptide sequences in FASTA format, the link of
each NCBI GenBank entry was followed to its corresponding NCBI
Protein (https://www.ncbi.nlm.nih.gov/protein/) entry.
The garlic genome sequence in FASTA format, as well
as its corresponding annotation in general feature format (GFF)
format (27), were downloaded from
NCBI Genome (https://www.ncbi.nlm.nih.gov/genome/). The GenBank
accession of the genome assembly used in the analysis in the
present study was GCA_014155895.2(25). Due to the size of the Allium
sativum genome chromosomes, it was deemed necessary to fragment
them, as basic local alignment search tool (BLAST)+ (28) that was to be used to search for
sequences of interest in the garlic genome cannot handle sequences
longer than 1 Gbp. For this reason, a script in PHP programming
language was created to split the chromosome sequences into
sequences with maximum size 1 Gbp. The preliminary identification
of the genomic regions of the three known genes was based on the
pairwise alignments between their nucleotide and protein sequences,
and those of the genome, as obtained from BLAST+ searches: The
boundaries of the AsGGT exons were identified using BLASTN search
for the nucleotide sequences. Similarly, TBLASTN searches for the
peptide sequences revealed the genomic coordinates of the coding
sequence (CDS) of each exon for every AsGGT gene. Exonerate
(29) was then used for the precise
mapping of the nucleotide and protein sequences on the genome.
Exonerate determined the exact boundaries for each gene, exon, CDS
and 5' and 3' untranslated regions (UTRs), and produced a GFF file
for each gene. Finally, a manual inspection of these boundaries was
performed using Integrative Genomics Viewer (IGV, Version: 2.16.1)
(30). Exonerate and manual
inspection ensured that the splice sites of the introns belong to
the GU-AG group (31).
To compare the manual annotation for the three genes
with the annotations produced automatically (25), all relevant genomic features from
the automatic GFF file were extracted to create the GFF
specifically for these genes. The automatically and manually
created exon-intron boundaries were visually inspected using IGV
and the protein sequences were extracted from the GFF files and the
garlic genome with another Gtf/Gff analysis toolkit (AGAT)
(32). Global pairwise alignments
(33) between the automatically and
manually produced protein sequences were performed.
Identification of AsGGT-coding exons
on chromosomes and scaffolds using hidden Markov models (HMMs)
A multiple sequence alignment of the three protein
sequences was performed using Clustal Omega (34). It was noted that apart from the
first exon CDSs which did not present a high degree of conservation
among the three peptides, the boundaries of the CDSs of all other
exons matched perfectly. Based on this finding, HMM profiles for
the multiple sequence alignments of the six conserved CDSs were
built using HMMER (35). All
characterized Allium sativum proteins were collected from
NCBI Protein. This search yielded 804 peptide sequences which were
stored as a single FASTA file. A search for the HMM of each exon
against all characterized Allium sativum proteins was
performed. This search did not yield any new peptide sequence
beyond the three already known proteins; therefore, it was assumed
that no other characterized protein was homologous to the known
AsGGTs.
With the aid of a PHP script, the translation of the
whole Allium sativum genome into the six open reading frames
(ORFs) was performed as previously described (36). With this procedure, all possible
peptides, as well as their corresponding GFFs were generated. To
search for homologous sequences potentially encoding AsGGT enzymes
in addition to those already characterized ones, a PHP script
searched the HMM of each exon against all potentially genome-coded
peptides. This produced a GFF file per chromosome or scaffold which
contained all genomic regions that could code for AsGGT CDSs. To
identify chromosomes or scaffolds coding AsGGT genes, a manual
check of their corresponding GFF files was performed. The GFF files
for chromosome 8 (CM031537.1), chromosome 5 (CM031533.1) and
chromosome 4 (CM031532.1) contained the CDSs of the already
characterized AsGGT1, AsGGT2 and AsGGT3 genes, respectively. The
GFF file for chromosome 6 (CM031534.1) contained consecutive CDSs
which corresponded to exons 2-7, suggesting the existence of a near
complete gene structure. Mapping the three known protein sequences
on chromosome 6 with Exonerate and consequently performing manual
inspection with IGV, a GFF file containing genomic features (exons
2-7) of this candidate gene was created.
Discovering the first exon of the
AsGGT-like gene on chromosome 6
The protein sequence of the newly discovered
AsGGT-like (AsGGT4) gene was extracted from its GFF and the
Allium sativum genome sequence, using AGAT. A BLASTP search
limited to Liliopsida (monocots) was performed in UniProt (37) to examine whether the peptide in
question had already been identified and to identify homologous
peptide sequences in related species of Allium sativum. The
peptide had not been previously identified and the UniProt BLASTP
search revealed that its amino terminal end was occasionally
aligned with the signal peptide of homologous proteins, suggesting
that the potential enzyme may contain a signal peptide at its amino
terminal end. Thus, SignalP 6.0(38) and DeepTMHMM (39) were used to check for the existence
of a eukaryotic signal peptide in the known AsGGT enzymes, as well
as the potential one.
The confirmation of the existence of a signal
peptide in the new enzyme allowed for the prediction of the length
of the CDS region of its missing first exon: As signal peptides
usually have a length of 22-25 amino acids and 19 amino acids are
already found in the CDS region of exon 2 in this enzyme, it was
expected that the CDS region of the first exon would encode ~4
amino acids, beginning with a methionine (AUG codon). In addition,
the boundaries of the intron between exon 1 and exon 2 of the gene
might be of GU-AG type, as explained above. Visually scanning the
genomic area upstream exon 2 in IGV, a sequence matching the above
criteria was discovered, thus completing the potential gene
sequence. GFF features of the genomic region where AsGGT4 gene is
located, were extracted from the automatically produced GFF file
(25). A comparison between the
automatically and manually produced annotations as they were
depicted in their respective GFFs, was performed by visual
inspection in IGV.
Mapping of RNA-seq reads on AsGGT
transcript sequences with BLAST+
To ensure that AsGGT4 is actually transcribed, an
alignment of all available Allium sativum short RNA-seq
reads was performed on its potential transcript, using a PHP
script. As a positive control, the same alignment was performed on
the AsGGT3 transcript. AsGGT3 was selected as it was found that it
was the most similar known AsGGT to AsGGT4. SRA (40) was searched for all Illumina RNA-Seq
runs of Allium sativum samples. SRR IDs of these entries
were used for the download of their corresponding fastq.gz files
from the European Nucleotide Archive (ENA) (41). These files were then unzipped and
converted into FASTA files using EMBOSS (42). Each FASTA file was converted into a
BLAST database and the two transcript sequences were
BLASTN-searched against the RNA-Seq read database. The outputs of
each search were then manually examined to identify reads which
could correspond to the AsGGT3 or AsGGT4 transcripts.
Prediction of the tertiary structure
of the AsGGT peptides
As no solved 3D structures for AsGGT proteins are
available, AlphaFold (43) was used
for structure predictions from primary peptide sequences. The
predicted structures of the known AsGGTs were downloaded from the
AlphaFold Protein Structure Database (44). A prediction of the tertiary
structure of the AsGGT4 was performed within the ColabFold website
(45), which uses a simplified
version of AlphaFold v2.3.2. For AsGGT4 structure prediction,
homology modelling was also performed using the SWISS-MODEL
Workspace (46).
Construction of a phylogenetic tree
for the four genes
Since SWISS-MODEL used the solved structure of
Bacillus licheniformis γ-glutamyl-transpeptidase [Protein Data Bank
(PDB) ID: 4Y23] to predict the structure of the AsGGT4 peptide by
homologous modelling, the bacterial peptide sequence was used as a
distant evolutionary relative (outgroup) of Allium sativum
AsGGT peptides. This sequence was downloaded from PDB (47) and was then searched using BLASTP in
UniProt. Its corresponding sequence in UniProt ID was Q65KZ6_BACLD.
MUSCLE (48) was used to generate a
multiple sequence alignment and a phylogenetic tree of the peptide
sequences of the four AsGGTs and their distant relative,
Bacillus licheniformis γ-glutamyl transpeptidase.
Visualization of the multiple sequence alignment was performed with
JalView (49). The phylogenetic
tree of the five peptides in Newick format (50) was visualized with Dendroscope
(51).
Results
Mapping of the three already
characterized AsGGT genes
The constructed GFF files for the AsGGT1, AsGGT2 and
AsGGT3 genes were visualized (Fig.
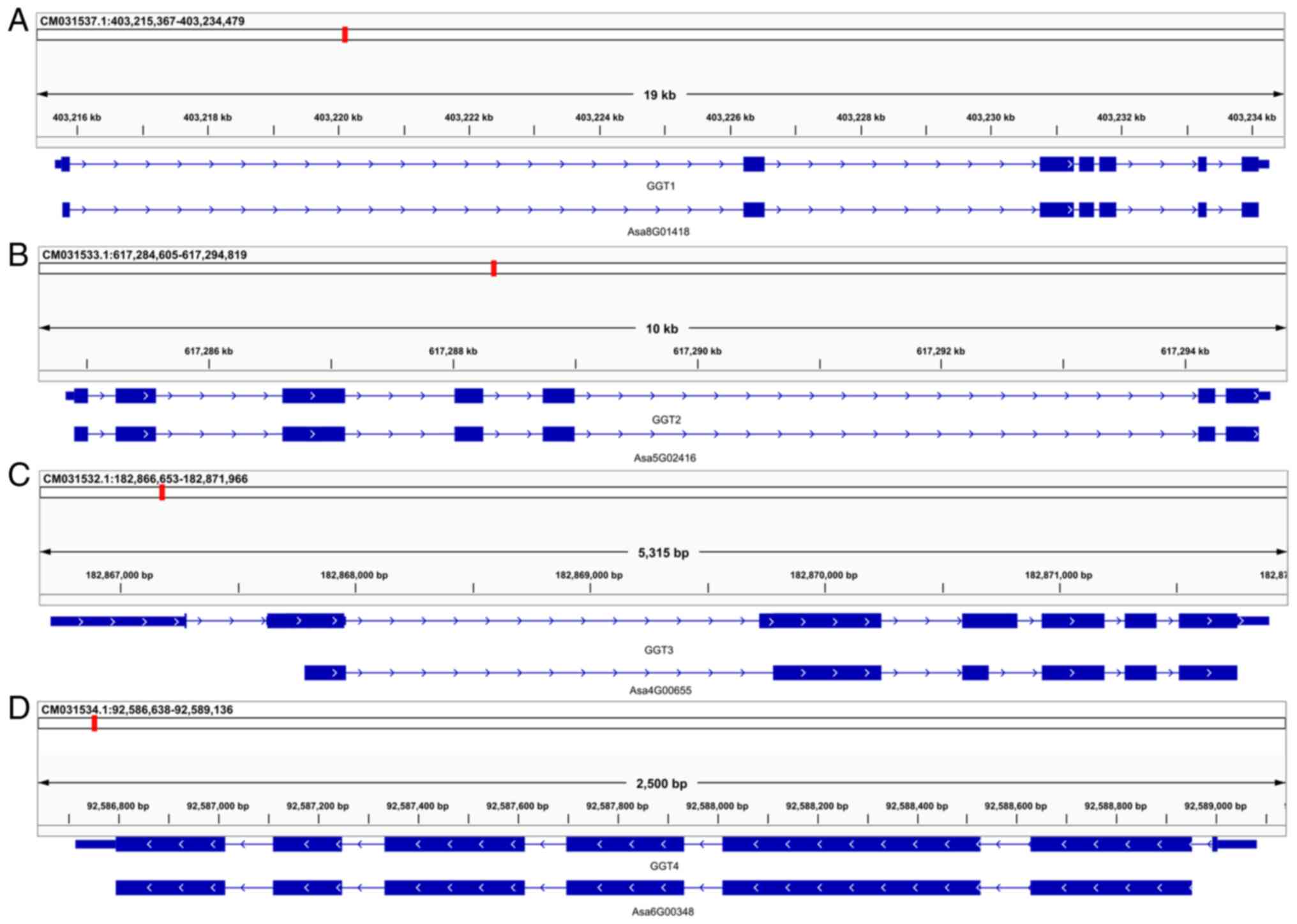
2A-C). The analysis revealed that AsGGT1 is located on
chromosome 8: 403,215,661-403,234,269, AsGGT2 on chromosome 5:
617,284,828-617,294,697 and AsGGT3 on chromosome 4:
182,866,703-182,871,895. All three genes consist of seven exons of
comparable size. Although the size of each corresponding exon is
comparable among the three genes, the size of the corresponding
introns varies, resulting in a considerable difference in gene
size: The AsGGT1 gene size is 18,609 bp, that of AsGGT2 is 9,870 bp
and that of AsGGT3 is 5,193 bp.
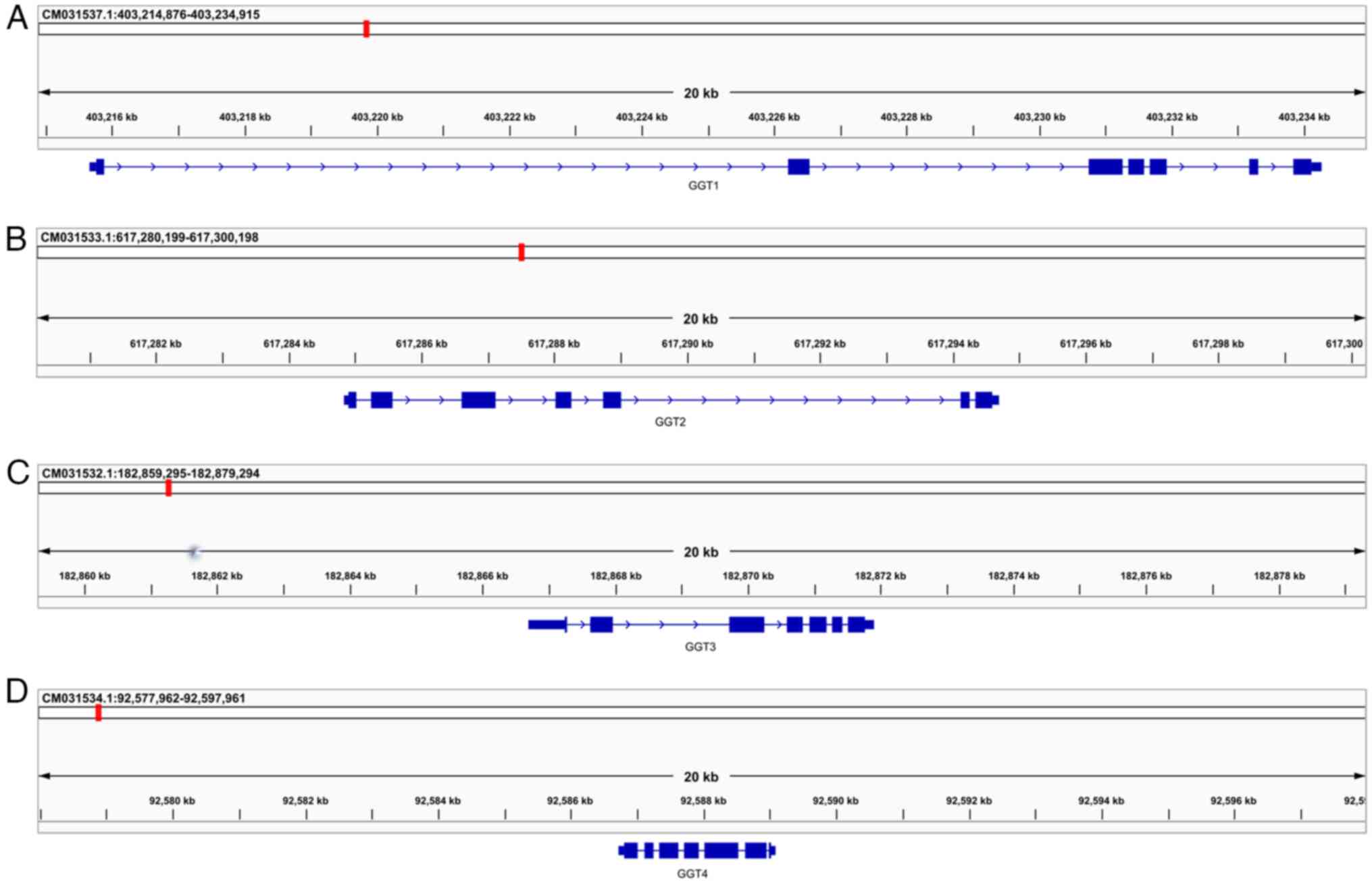 | Figure 2Genome mapping of AsGGT genes. (A)
AsGGT1 is located on chromosome 8: 403,215,661-403,234,269, (B)
AsGGT2 on chromosome 5: 617,284,828-617,294,697 and (C) AsGGT3 on
chromosome 4: 182,866,703-182,871,895. (D) AsGGT4 is located on
chromosome 6: 92,586,715-92,589,081. All four genes consist of
seven exons of comparable size, though the size of the
corresponding introns varies, resulting in a considerable
difference in gene size: AsGGT1 is 18,609 bp, AsGGT2 is 9,870 bp
and AsGGT3 is 5,193 bp. AsGGT, Allium sativum
γ-glutamyl-transpeptidase. |
Through a manual inspection of the genomic features
of AsGGT3 on chromosome 4, it became apparent that there are four
loci in the genomic sequence where single base deletions or
insertions (indel events) occurred (Fig. 3). An addition of an adenine appears
at position 182,867,809 in the second exon, an addition of a
guanine appears at position 182,869,779 in the third exon, a
deletion of an adenine appears at position 182,870,778 in the
fourth exon and an addition of a guanine appears at position
182,870,929 in the seventh exon.
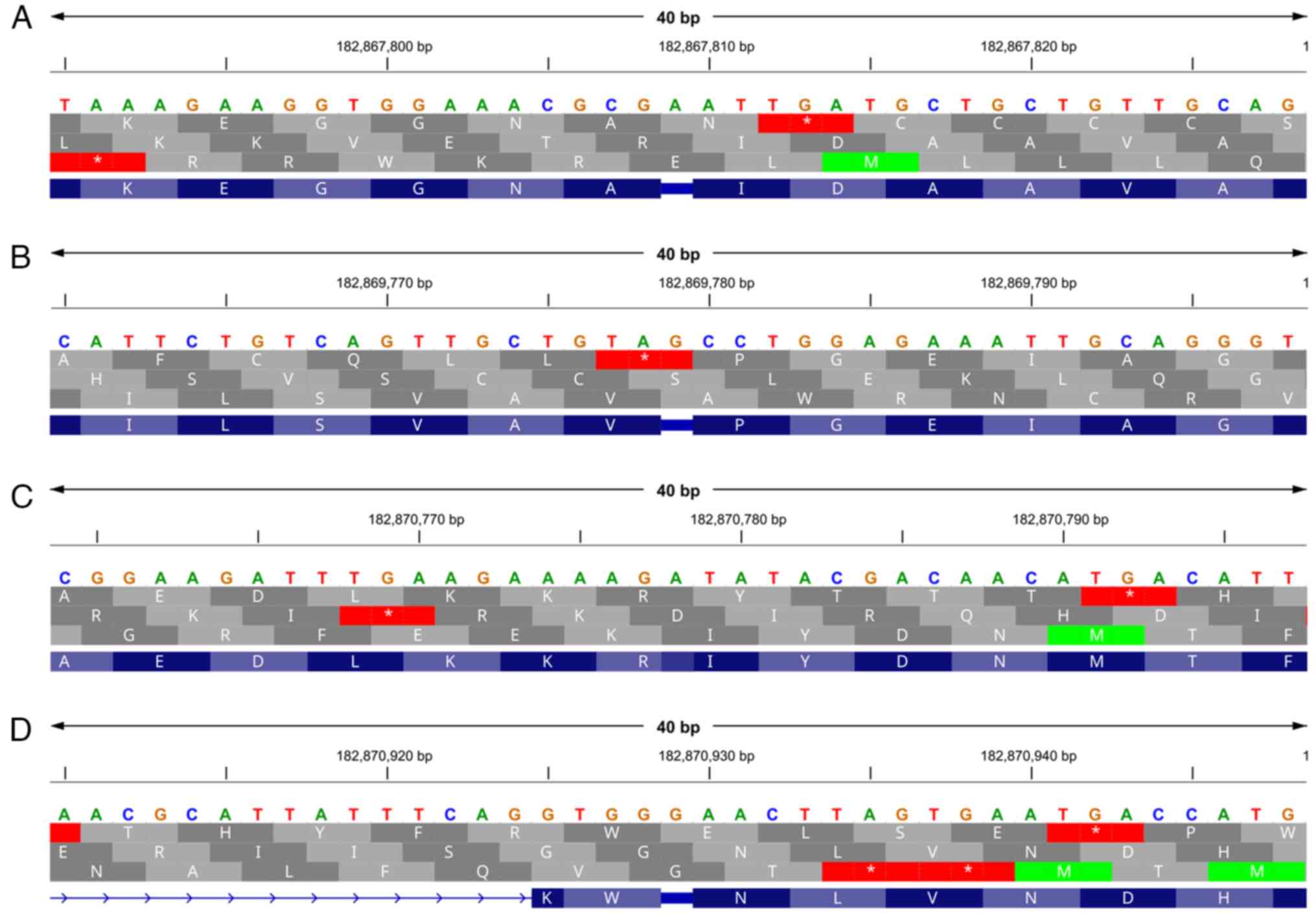 | Figure 3Discovery of deletions and insertions
in the coding region of the AsGGT3 gene. (A) An addition of an
adenine appears at position 182,867,809 in the second exon, (B) an
addition of a guanine appears at position 182,869,779 in the third
exon, (C) a deletion of an adenine appears at position 182,870,778
in the fourth exon and (D) an addition of a guanine appears at
position 182,870,929 in the seventh exon. The top line indicates
the genomic sequence, the lines in grey indicate the translated
sequence in the +1, +2 and +3 open reading frames, and the line in
purple indicates the reading frames that correctly correspond to
AsGGT3 mRNA. AsGGT, Allium sativum
γ-glutamyl-transpeptidase. |
Mapping of the potential γ-glutamyl
transpeptidase gene (AsGGT4)
Search for the HMMs of the six coding sequences
which correspond to already characterized AsGGT exons in the
potential proteome, which resulted from the in silico
translation of the genome into the six ORFs, in addition to
identifying the coding regions of known genes, allowed for the
discovery of novel genomic regions that could encode parts of
γ-glutamyl transpeptidase enzymes.
By filtering the search results and keeping only
those containing adjacent regions corresponding to all exons that
comprise a potential AsGGT gene, a crude GFF was generated that
described the coding regions of a potential fourth γ-glutamyl
transpeptidase gene (AsGGT4). The complete GFF file for AsGGT4 was
constructed by optimizing the primary GFF. AsGGT4 is located on
chromosome 6: 92,586,715-92,589,081 and has a size of 2,336 bp.
Similar to the three already characterized genes (AsGGT1, AsGGT2
and AsGGT3), the AsGGT4 (Figs. 2D
and 4) gene consists of seven
exons. Unlike the already characterized genes, AsGGT4 is
transcribed in the reverse direction.
Signal peptide in AsGGTs
A BLASTP search in UniProt for homologous monocot
protein sequences with the potential γ-glutamyl-transpeptidase
sequence revealed a number of sequences belonging to different
species (Fig. 5). The visual
observation of the produced pairwise alignments revealed that a
number of homologous peptides possessed a signal peptide. It was
therefore examined whether the three known enzymes and the
potential one, contain a signal peptide in their amino terminal
sequence using SignalP and DeepTMHMM. SignalP did not predict a
signal peptide for AsGGT1 or AsGGT2 peptides. DeepTMHMM predicted a
single transmembrane helix in 32-47 and 30-46 amino acid regions of
AsGGT1 and AsGGT2, respectively. It also predicted that their
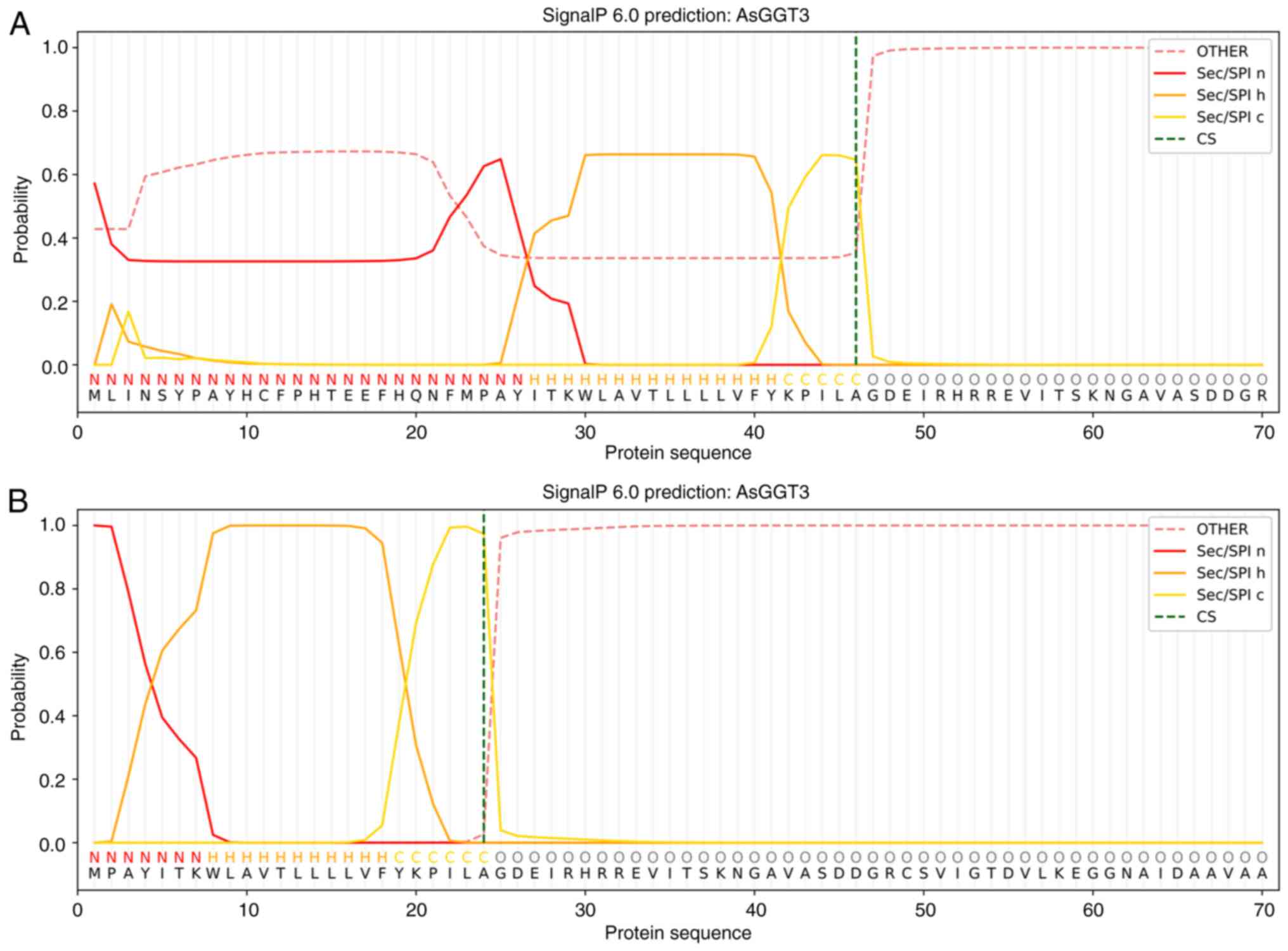
C-terminus is extracellular. SignalP revealed a potential signal
peptide profile, mainly in the 23-46 amino acid region of AsGGT3
(Fig. 6A), beginning with a
methionine at position 23. When the first 22 amino acids were
removed from the sequence, SignalP revealed the presence of a
signal peptide, where amino acids 1-7 form its amino terminal
region, amino acids 8-18 are hydrophobic and finally amino acids
19-24 form its carboxy terminal end (Fig. 6B). DeepTMHMM also predicted a signal
peptide for both the untruncated and truncated AsGGT3 peptide.
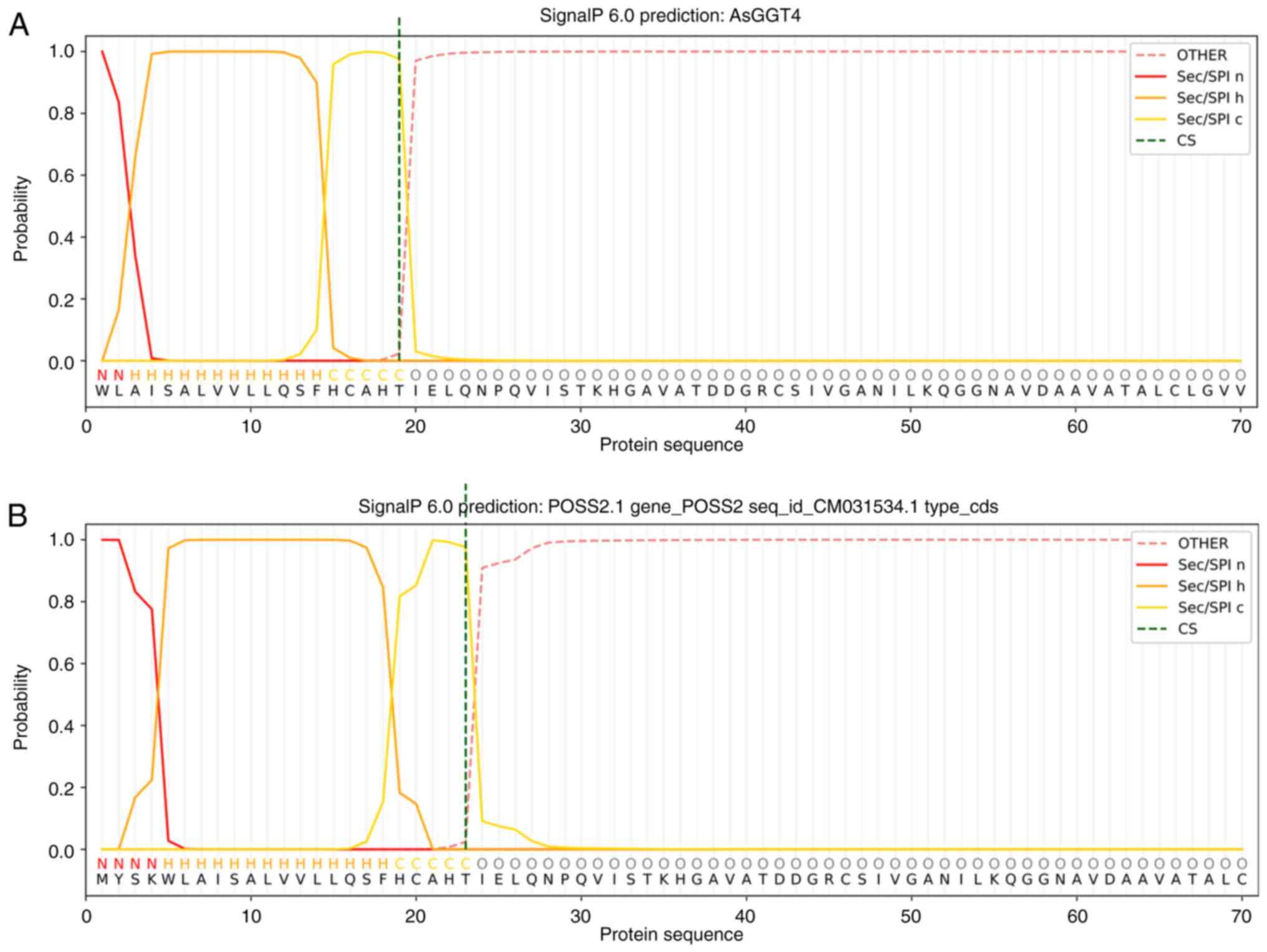
SignalP predicted a 19 amino acid long signal
peptide in the amino terminal coding region of the second exon
(Fig. 7A), implying that the coding
sequence of the first exon of AsGGT4 would code for approximately
another four amino acids. By manually inspecting the genomic region
upstream of the second exon, a first exon which could code for four
amino acids was proposed. SignalP predicted a signal peptide of 23
amino acids in the full length AsGGT4 peptide, where the first four
amino acids form its amino terminal region, amino acids 5-18 are
hydrophobic, while amino acids 19-23 form its carboxy terminus
(Fig. 7B). DeepTMHMM also predicted
a signal peptide.
Comparison of automatic and manual
genome annotation
The coding regions of AsGGT1 and AsGGT2 are
identical to the coding regions obtained by automated genomic
annotation; i.e., from the use of the GFF file that was provided by
the group that performed the genome sequencing (Fig. 8A and B); by contrast, automated genomic
annotation failed to correctly assign the exon boundaries and
coding regions of AsGGT3 (Fig. 8C).
The extracted protein sequence also differed from the one
previously reported (24).
Pairwise alignments between the protein sequences
extracted from manual and automatic annotation revealed that the
peptide sequences of the AsGGT1 and AsGGT2 genes from the automatic
annotation matched almost perfectly with the sequences obtained
experimentally and with the annotation performed in the present
study-small differences may be due to point mutations. By contrast,
marked differences were observed between the automatic annotation
of the peptides of the AsGGT3 and AsGGT4 genes and the annotation
performed the present study. In AsGGT3, differences appear at the
four points where indel events occur in the genomic region, as
expected, but the exon boundaries are generally correctly described
beyond these points. By contrast, the automatic annotation of
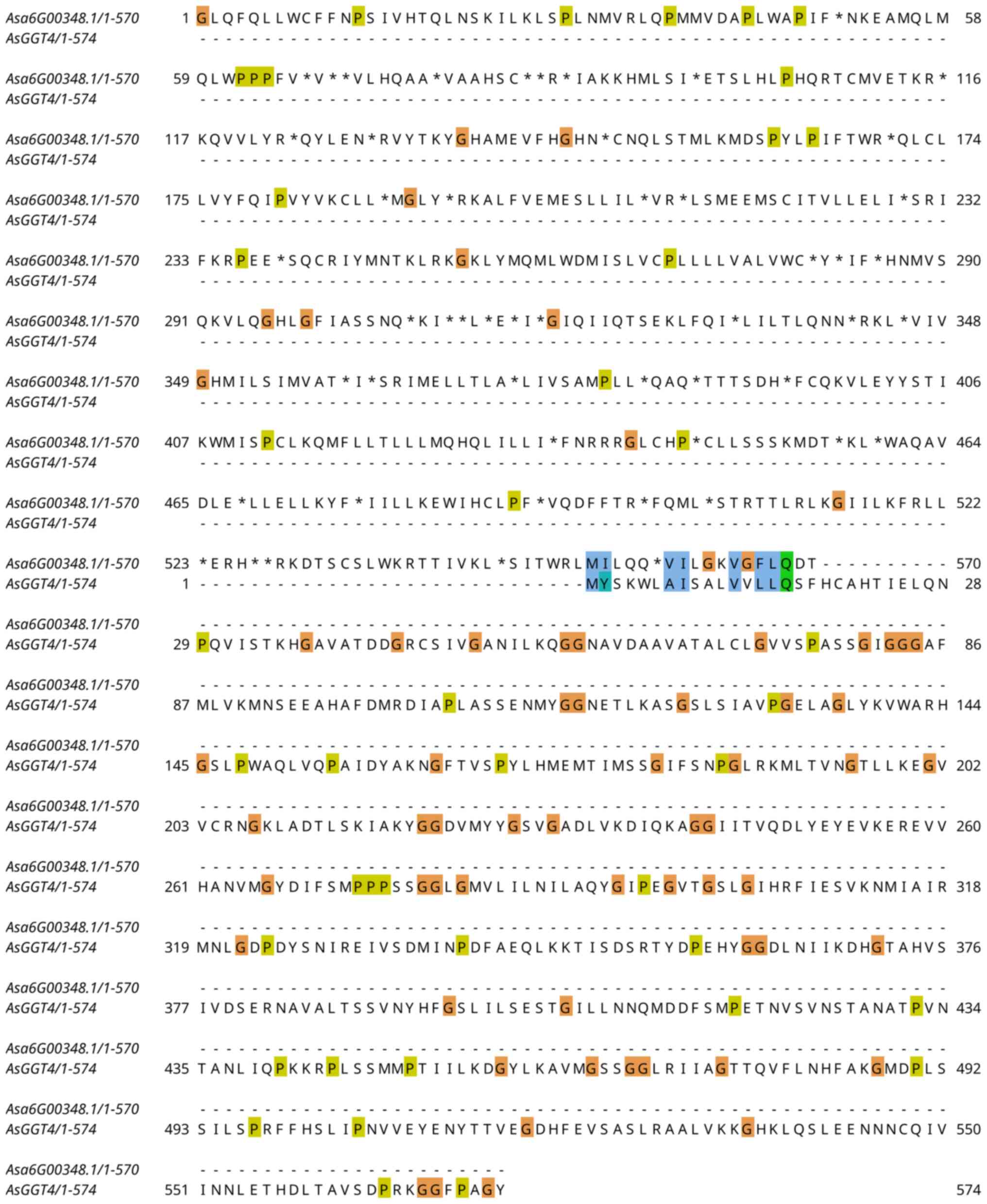
AsGGT4 appears to have failed almost completely. Although the exon
boundaries are correctly predicted (Fig. 8D), the ORF is incorrect in all
exons, and the first exon is completely missing-the start of the
code region is incorrectly located in the second exon. Thus,
pairwise alignment does not detect any similarity-apart from a
minute region that is probably accidental, between the two
extracted peptide sequences (Fig.
9). By changing the phase from 2 to 1 in the automatically
produced GFF, the resulting peptide sequence is the correct
one.
Phylogenetic analysis
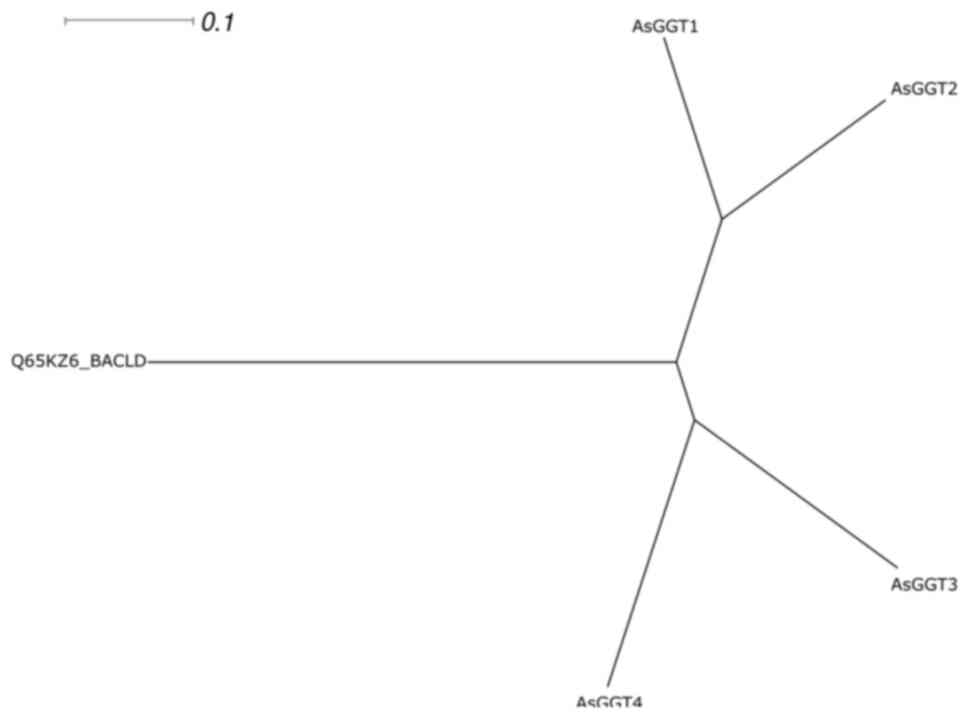
A multiple sequence alignment (Fig. 10) of the four garlic AsGGT peptide
sequences and the Bacillus licheniformis γ-glutamyl transpeptidase
sequence was performed and the radial phylogram of the five
peptides was constructed (Fig.
11). In the radial phylogram, the root of the AsGGTs subtree is
the point at which the outgroup (the Bacillus licheniformis
peptide) connects to the subtree of the AsGGT peptides. The root,
which represents the common ancestor of the four AsGGTs, appears to
have given rise to two ancestral peptides, which subsequently gave
rise to the peptides AsGGT1 and AsGGT2, and the peptides AsGGT3 and
AsGGT4.
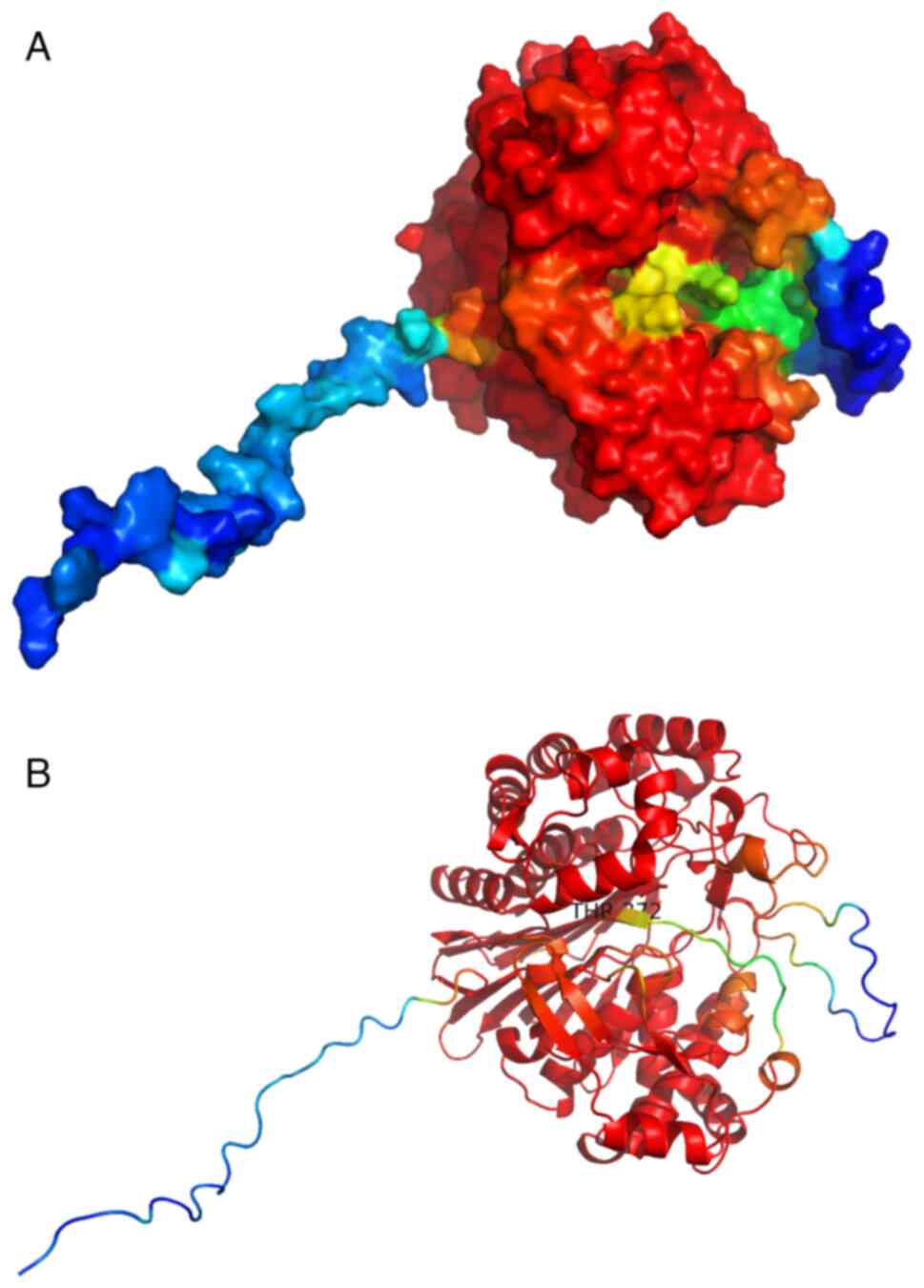
Prediction of the tertiary structure
of the AsGGTs
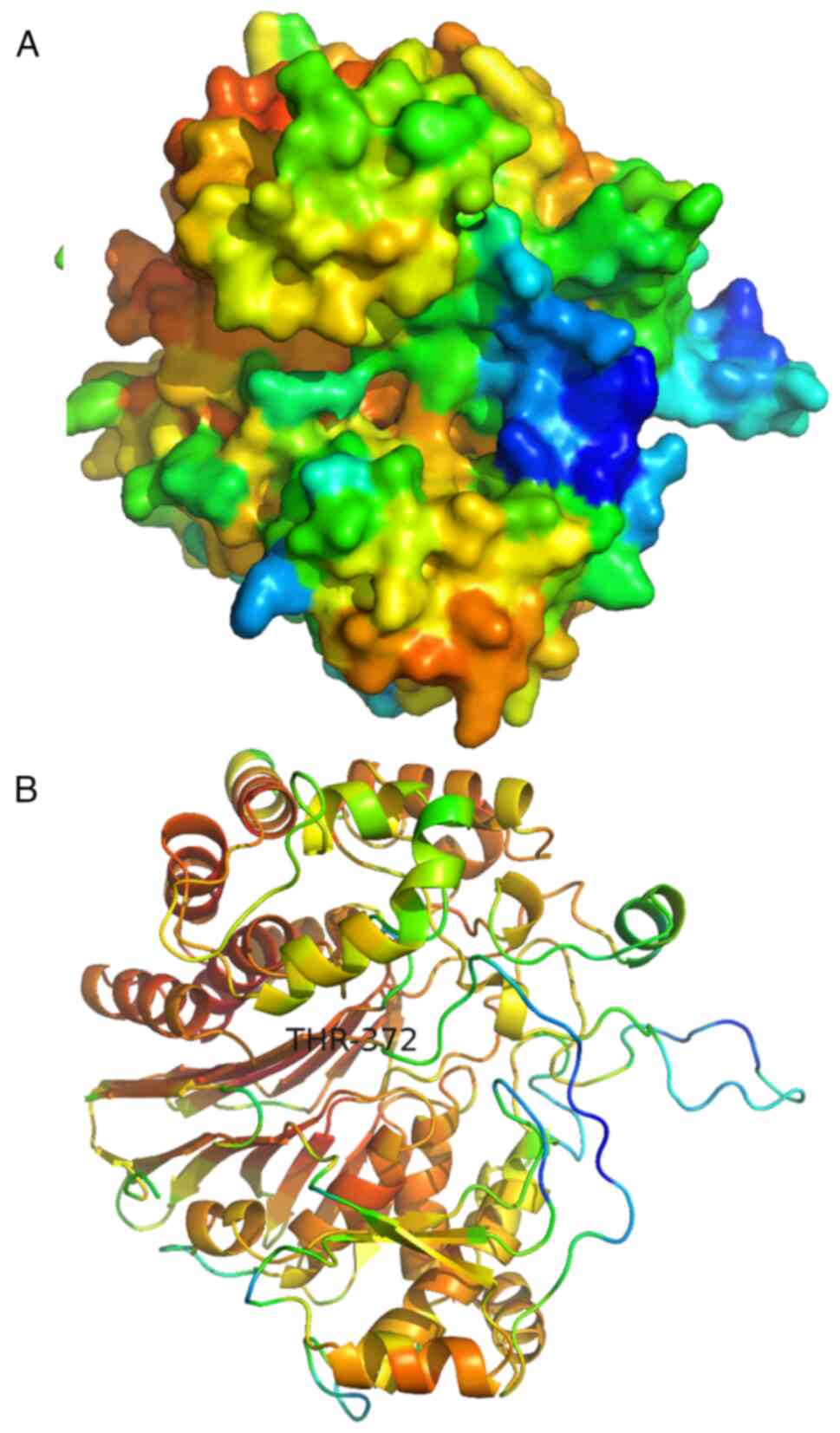
The predicted structures of all AsGGTs are
relatively similar and consist of one β-sandwich which is
surrounded by clusters of interacting α-helices. The two
cartoon-like visualizations of AsGGT4 produced by SWISS-MODEL
(Fig. 12 and AlphaFold (Fig. 13) (the color spectrum in both
visualizations reflects the confidence of prediction) were
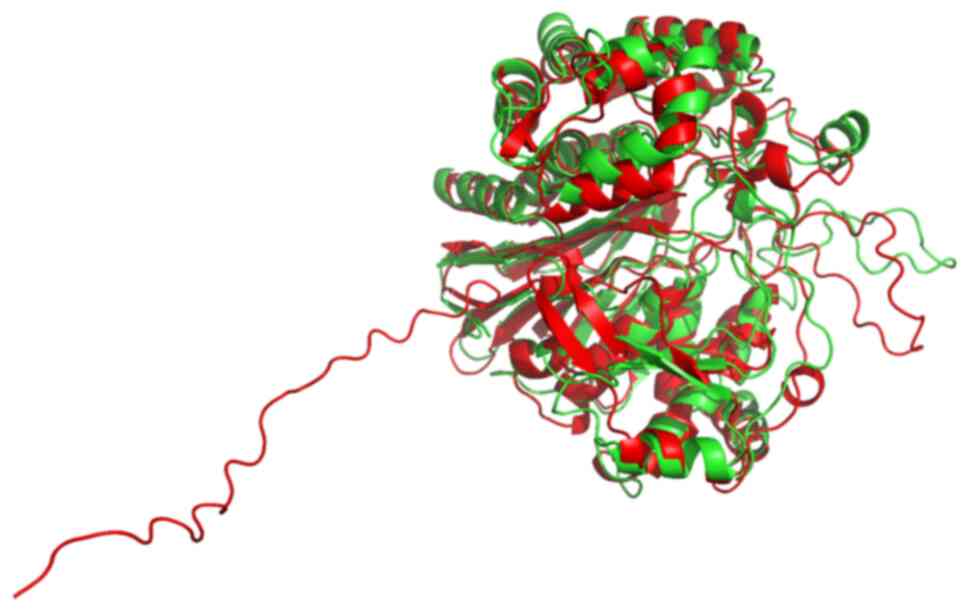
superimposed (Fig. 14) with PyMOL
software (52). This revealed that
predicted structures by both algorithms are very similar. Most
importantly, the catalytic T372 remains at the same position in the
active site in both predictions. The position of a flexible loop
relative to the active site appears displaced between the two
structures, with low prediction confidence in both structures. The
main difference between the two predictions is that an unstructured
sequence which corresponds to the predicted signal peptide, only
appears in the AlphaFold structure, with low prediction confidence.
As with AsGGT4, AlphaFold also fails to predict any secondary
structure for the predicted signal peptide of AsGGT3. Notably,
AlphaFold predicts a helical structure for the DeepTMHHM-predicted
transmembrane helix of AsGGT1, but not for the same feature in
AsGGT2.
Detection of AsGGT4 expression
A total of 366 paired-end and five single-end
Illumina RNA-Seq runs were identified (until March 24, 2023) in
garlic. In the vast majority of these, reads corresponding to
AsGGT3 transcripts were identified. On the other hand, a small
proportion of runs contained AsGGT4 reads. The majority of these
were identified on the SRR13219906 run, which corresponded to a
leaf tissue replicate of garlic grown under normal soil moisture
conditions. Other runs containing reads which corresponded to
AsGGT4 transcripts included those of the PRJNA566287 and
PRJNA489986 BioProjects. Their runs derive from long-vernalization,
short-vernalization and non-vernalized stored clove and leaf and
clove samples, respectively.
Discussion
To map known genes and discover new ones, to
characterize fully their genomic structure, to predict the tertiary
structure of the proteins they encode and to discover their
evolutionary history, an integrated workflow based on a number of
cutting-edge bioinformatics tools was developed.
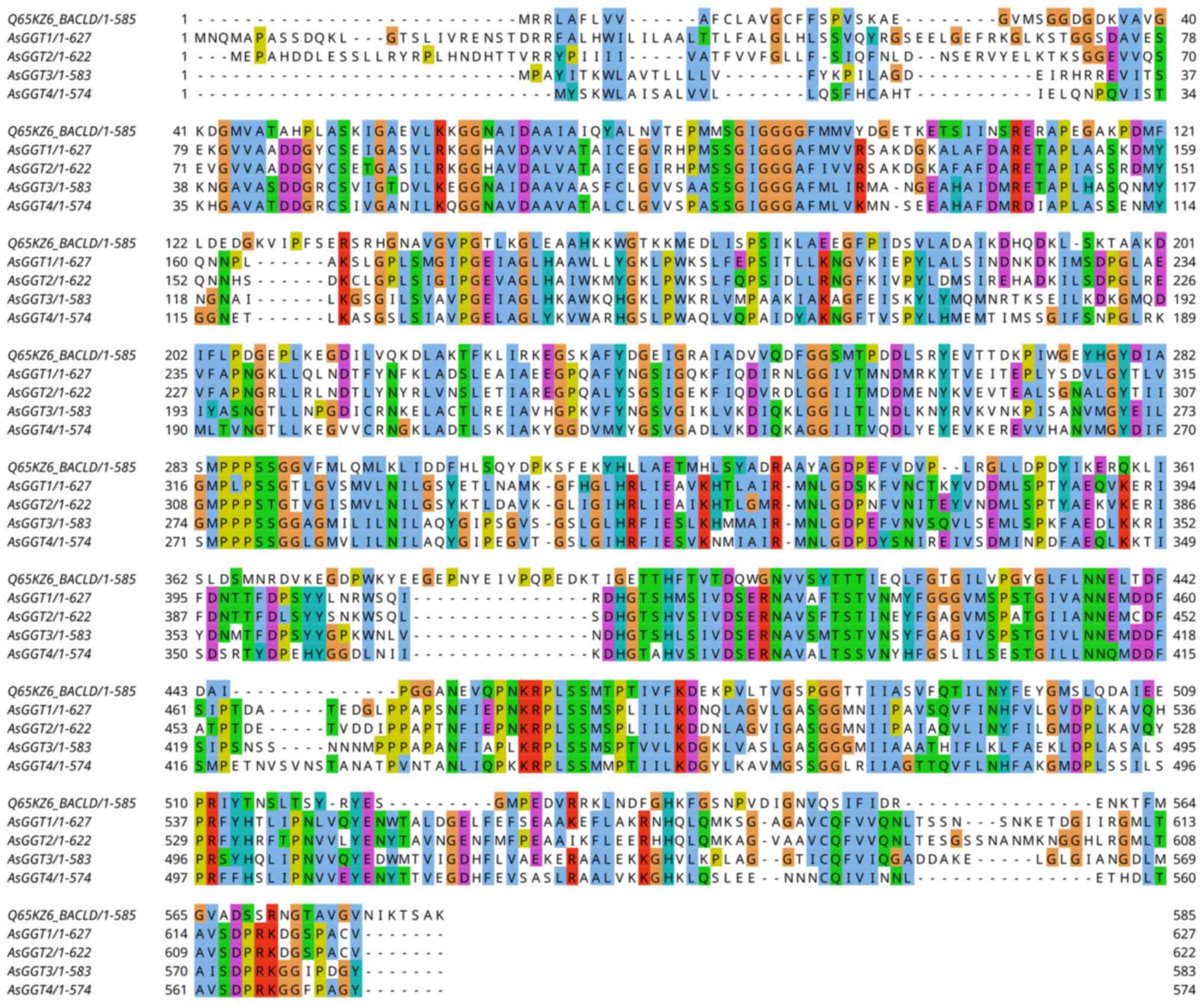
All four AsGGT genes consist of seven exons, and
there appears to be a conservation in the overall structure of
these exons. All introns of all four genes belong to the GT-AG
splice site group. There is, however, a large discrepancy in length
between the four genes, due to the large difference in size of
their introns.
Frameshift mutations are base insertions or
deletions (indels) within the coding region of a gene that disrupt
the reading frame in such a manner that the entire set of triplets
following the mutation site is altered. Often a termination codon
forms within the coding sequence, leading to premature termination
of mRNA translation and hence to a shorter non-functional peptide
(53). In the case of the AsGGT3
gene, four indel events were observed during the alignment of the
transcript and its protein sequence onto the genome. A possible
explanation, at least for three of the four incomplete alignments,
is that sequencers may be mistaken in their estimation of the
repeat length of the same nucleotide (54). Therefore, in the first insertion,
the sequencer estimated that there were two adenines when there may
have been one. Similarly, in the last insertion, it considered that
there were three guanines, whereas there were probably two.
Finally, in the deletion, it predicted the presence of one adenine,
whereas there were probably two. These potential errors in the
sequencing and assembly of the genomic sequence disoriented
automatic annotation and made manual mapping of this gene
particularly challenging.
If the observed single-nucleotide indel events are
not errors during sequencing, then they lead to successive
frameshifts of the ORF. The presence of only one of the four
frameshift mutations occurring in the AsGGT3 coding region is
sufficient to render the gene inactive, let alone the accumulation
of all four. Therefore, if these mutations are indeed present,
AsGGT3 gene is in fact a pseudogene, at least in the garlic
cultivar Ershuizao used in the genome sequencing (25). By contrast, in the garlic cultivar
Fukuchi-howaito, which was used to discover and characterize the
three AsGGT genes (24), AsGGT3
still appears functional. The existence of GGT pseudogenes in
garlic cannot be excluded, as a GGT-homolog in humans, GGT2, does
not encode for a functional enzyme (55).
This accumulation of mutations in the AsGGT3 gene
may be due to garlic domestication. During the domestication of an
organism, the selection is based on specific traits. In garlic,
this process has led, among others, to the loss of native
reproduction, so that garlic is now exclusively clonal (56). If a strain with inactivated AsGGT3
had been selected, all offspring would have inherited it. Once the
first mutation that leads to the inactivation of the gene has
occurred, an accumulation of new mutations can occur, as there is
no longer any natural selection pressure to prevent mutations. This
inactivation of AsGGT3 may possibly partially explain the observed
differences in the amount of allicin produced by different garlic
cultivars (57). When constructing
the GFF file for AsGGT3, it was difficult to represent the ORF
shift in a manner that would be recognized by visualization
software. In the GFF3 format, the gap representation is represented
by the ‘Gap’ attribute. The ‘Gap’ feature representation consists
of a series of pairs (mode and length) separated by a space, for
example ‘M8 D3 M6’, where each mode is represented by a code
(27). IGV does not support mapping
of frameshifts, as its developers consider that there is no
critical mass demanding this feature (determined by personal
communication).
The majority of proteins intended for transport to
subcellular compartments have a signal peptide at their amino
terminal end. The newly synthesized precursor proteins are
localized and transported across the membrane through a multitude
of possible pathways (58-60).
The signal peptide is responsible for transporting the remaining
polypeptide across the membrane. After proteins have crossed the
membrane or during transport, their signal peptides must be removed
to activate the mature proteins when they reach their destination
(60).
Signal peptides consist of three distinct regions:
The amino-terminal end, the hydrophobic core and the carboxy
terminal end. The hydrophobic core constitutes the largest part of
the signal peptide and contains 10-15 amino acid residues. The part
of the hydrophobic residues appears to adopt an α-helical
configuration across the plasma membrane (61).
In Escherichia coli (E. coli), the
sequence of the γ-glutamyl-transpeptidase gene contains a single
ORF, encoding a signal peptide at the amino-terminal end and some
large and small functional regions. The E. coli
γ-glutamyl-transpeptidase signal peptide, consisting of 25 amino
acids, is cleaved and the mature GGT localizes to the periplasmic
space without anchoring to the membrane (62). By contrast, in mammalian cells, GGT
is located on the outside of the plasma membrane with the amino
terminal end of the large functional domain anchored to the cell
membrane (63). Of note, of the
four AsGGT isoenzymes, only AsGGT3 and AsGGT4 have a signal
peptide, suggesting that they probably do not perform exactly the
same role as AsGGT1 and AsGGT2.
SWISS-MODEL failed to predict any structure of the
AsGGT4 signal peptide, whereas AlphaFold did not predict any
secondary structure for the signal peptide. The signal peptides
contained in the enzymes encoding the AsGGT3 and AsGGT4 genes are
expected to perform a role similar to that of the signal peptides
of E. coli γ-glutamyl transpeptidase or mammalian cells.
They may signal the need to transport the newly synthesized protein
across membranes. The prediction of a single transmembrane helix
close to the N-terminus of AsGGT1 and AsGGT2 suggests that their
location is transmembrane and that their C-termini are
extracellular. Hints for the subcellular location of a protein can
be provided if the subcellular compartment of its homologs in other
species is characterized: According to the Human Protein Atlas
(64), human GGT1 may be localized
to the vesicles, GGT5 to nucleoli fibrillar center and GGT7 to the
vesicles and the nucleoplasm. All three human homologs are
predicted by DeepTMHMM to contain a single transmembrane helix,
like the one predicted for garlic GGT1 and GGT2.
During automated genome annotation in eukaryotes, it
is assumed, marginally arbitrarily, that the first methionine codon
of a transcript is the start codon of its coding region (CDS). This
can lead to an incorrect annotation, since translation may be
initiated by a subsequent methionine codon. Presumably such an
error occurred in the case of the automatic genomic annotation of
AsGGT3, which resulted in the presence of a signal peptide being
ignored (24). A similar case to
that of AsGGT3 was observed in glutathione hydrolase of Oryza
meyeriana var. granulata (UniProt: A0A6G1BRJ1_9ORYZ). As in the
case of AsGGT3, the first amino acids of the rice glutathione
hydrolase are probably not part of the protein and the peptide
begins from a later methionine. Thus, the AsGGT3 sequence was
modified, deleting the first 28 amino acids.
The fact that the pHMMs constructed from the garlic
GGT peptides identify even bacterial γ-glutamyl transpeptidase
sequences and that AsGGT4 homology modelling was based on bacterial
solved GGT structure means that γ-glutamyl-transpeptidases have
appeared in evolutionary history before the prokaryote-eukaryote
split over three and a half billion years ago and they remain
conserved ever since. Such conserved genes confer properties
necessary for the survival and adaptation of organisms (65). Generally, in bacteria, yeasts,
plants and mammals, GGT is a heteromeric protein, consisting of
large and small subunits, both produced from a common inactive
polypeptide precursor through a process of autocatalysis (66-68).
Some plants, such as tomato, onion and radish, are considered to
possess GGT enzymes composed of a single polypeptide, although
their sequences remain unknown (69,70).
The amino acid sequences of AsGGT1, AsGGT2 and AsGGT3 possess a
conserved threonine residue required for autocatalysis and amino
acid residues essential for GGT activity, which have been
identified by biochemical analyses in human and E. coli
(71,72). Plant GGTs are classified in the same
evolutionary clade, which is further divided into two distinct
subclades. AsGGT1 and AsGGT2 belong to the subclade containing
Arabidopsis thaliana AtGGT4, which is involved in the degradation
of S-glutathione in the vacuole (73-75),
whereas AsGGT3 belongs to the subclade containing Arabidopsis
thaliana AtGGT1 and AtGGT2, which are involved in the
degradation of extracellular glutathione (74-76)
along with onion AcGGT (77). In
the present study, the phylogenetic analysis revealed that AsGGT1
(BAQ21911.1) and AsGGT2 (BAQ21912.1) peptides have a common
ancestor which probably lacked a signal peptide and that AsGGT3
(BAQ21913.1) shares a common ancestor with AsGGT4 which probably
contained a signal peptide.
The transcriptomic analysis displayed that while
AsGGT3 is ubiquitously expressed, AsGGT4 expression is probably
tissue-, condition- and/or developmental stage-specific. Thus, it
comes as no surprise that the first attempt for the identification
of garlic GGT genes (24) prior to
the knowledge of the genomic sequence, was able to identify AsGGT1,
AsGGT2 and AsGGT3, but failed to identify AsGGT4. Therefore, it is
possible that AsGGT4 plays a different role than that of the other
AsGGTs, which has yet to be discovered.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
IM, CP and DAS conceived the study. The study
methodology was proposed by IM. Data were retrieved, curated and
analyzed by EB and IM. Software was developed by IM. Visualization
was performed by EB. IM and CP supervised the study. The manuscript
was written by EB and IM. EB and IM confirm the authenticity of all
the raw data. All authors contributed to the revision of the work,
and have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
DAS is the Editor-in-Chief for the journal, but had
no personal involvement in the reviewing process, or any influence
in terms of adjudicating on the final decision, for this article.
The other authors declare that they have no competing
interests.
References
|
1
|
Tesfaye A: Revealing the therapeutic uses
of garlic (Allium sativum) and its potential for drug
discovery. Sci World J. 2021:1–7. 2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Tsuneyoshi T: BACH1 mediates the
antioxidant properties of aged garlic extract. Exp Ther Med.
19:1500–1503. 2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Kanamori Y, Via LD, Macone A, Canettieri
G, Greco A, Toninello A and Agostinelli E: Aged garlic extract and
its constituent, S-allyl-L-cysteine, induce the apoptosis of
neuroblastoma cancer cells due to mitochondrial membrane
depolarization. Exp Ther Med. 19:1511–1521. 2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Nakamoto M, Kunimura K, Suzuki JI and
Kodera Y: Antimicrobial properties of hydrophobic compounds in
garlic: Allicin, vinyldithiin, ajoene and diallyl polysulfides. Exp
Ther Med. 19:1550–1553. 2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Khounganian RM, Alwakeel A, Albadah A,
Nakshabandi A, Alharbi S and Almslam AS: The antifungal efficacy of
pure garlic, onion, and lemon extracts against Candida
albicans. Cureus. 15(e38637)2023.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Hutchins E, Shaikh K, Kinninger A,
Cherukuri L, Birudaraju D, Mao SS, Nakanishi R, Almeida S,
Jayawardena E, Shekar C, et al: Aged garlic extract reduces left
ventricular myocardial mass in patients with diabetes: A
prospective randomized controlled double-blind study. Exp Ther Med.
19:1468–1471. 2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Shaikh K, Kinninger A, Cherukuri L,
Birudaraju D, Nakanishi R, Almeida S, Jayawardena E, Shekar C,
Flores F, Hamal S, et al: Aged garlic extract reduces low
attenuation plaque in coronary arteries of patients with diabetes:
A randomized, double-blind, placebo-controlled study. Exp Ther Med.
19:1457–1461. 2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Ohtani M and Nishimura T: The preventive
and therapeutic application of garlic and other plant ingredients
in the treatment of periodontal diseases. Exp Ther Med.
19:1507–1510. 2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Mann J, Bernstein Y and Findler M:
Periodontal disease and its prevention, by traditional and new
avenues. Exp Ther Med. 19:1504–1506. 2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Gruenwald J, Bongartz U, Bothe G and
Uebelhack R: Effects of aged garlic extract on arterial elasticity
in a placebo-controlled clinical trial using EndoPAT™
technology. Exp Ther Med. 19:1490–1499. 2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Matsutomo T: Potential benefits of garlic
and other dietary supplements for the management of hypertension.
Exp Ther Med. 19:1479–1484. 2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Ried K: Garlic lowers blood pressure in
hypertensive subjects, improves arterial stiffness and gut
microbiota: A review and meta-analysis. Exp Ther Med. 19:1472–1478.
2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Kosuge Y: Neuroprotective mechanisms of
S-allyl-L-cysteine in neurological disease. Exp Ther Med.
19:1565–1569. 2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Sripanidkulchai B: Benefits of aged garlic
extract on Alzheimer's disease: Possible mechanisms of action. Exp
Ther Med. 19:1560–1564. 2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Rahman K and Lowe GM: Garlic and
cardiovascular disease: A critical review. J Nutr. 136 (Suppl
3):736S–740S. 2006.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Gruhlke MCH, Nicco C, Batteux F and
Slusarenko AJ: The effects of allicin, a reactive sulfur species
from garlic, on a selection of mammalian cell lines. Antioxidants
(Basel). 6(1)2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Kita T, Kume N, Minami M, Hayashida K,
Murayama T, Sano H, Moriwaki H, Kataoka H, Nishi E, Horiuchi H, et
al: Role of oxidized LDL in atherosclerosis. Ann N Y Acad Sci.
947:199–206. 2001.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Stoll A and Seebeck E: About alliin, the
genuine mother substance of garlic oil. Helv Chim Acta. 31:189–210.
1948.PubMed/NCBI View Article : Google Scholar : (In German).
|
|
19
|
Cavallito CJ and Bailey JH: Allicin, the
antibacterial principle of Allium sativum I. isolation,
physical properties and antibacterial action. J Am Chem Soc.
66:1950–1951. 1944.
|
|
20
|
Kim YS, Kim KS, Han I, Kim MH, Jung MH and
Park HK: Quantitative and qualitative analysis of the antifungal
activity of allicin alone and in combination with antifungal drugs.
PLoS One. 7(e38242)2012.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Yoshimoto N and Saito K:
S-Alk(en)ylcysteine sulfoxides in the genus Allium: Proposed
biosynthesis, chemical conversion, and bioactivities. J Exp Bot.
70:4123–4137. 2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Valentino H, Campbell AC, Schuermann JP,
Sultana N, Nam HG, LeBlanc S, Tanner JJ and Sobrado P: Structure
and function of a flavin-dependent S-monooxygenase from garlic
(Allium sativum). J Biol Chem. 295:11042–11055.
2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Borlinghaus J, Albrecht F, Gruhlke MC,
Nwachukwu ID and Slusarenko AJ: Allicin: Chemistry and biological
properties. Molecules. 19:12591–12618. 2014.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Yoshimoto N, Yabe A, Sugino Y, Murakami S,
Sai-Ngam N, Sumi S, Tsuneyoshi T and Saito K: Garlic γ-glutamyl
transpeptidases that catalyze deglutamylation of biosynthetic
intermediate of alliin. Front Plant Sci. 5(758)2014.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Sun X, Zhu S, Li N, Cheng Y, Zhao J, Qiao
X, Lu L, Liu S, Wang Y, Liu C, et al: A chromosome-level genome
assembly of garlic (Allium sativum) provides insights into
genome evolution and allicin biosynthesis. Mol Plant. 13:1328–1339.
2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Sayers EW, Cavanaugh M, Clark K, Pruitt
KD, Sherry ST, Yankie L and Karsch-Mizrachi I: GenBank 2023 update.
Nucleic Acids Res. 51 (D1):D141–D144. 2023.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Stein L: Generic feature format version 3
(GFF3). GitHub, 2020.
|
|
28
|
Camacho C, Coulouris G, Avagyan V, Ma N,
Papadopoulos J, Bealer K and Madden TL: BLAST+: architecture and
applications. BMC Bioinformatics. 10(421)2009.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Slater GS and Birney E: Automated
generation of heuristics for biological sequence comparison. BMC
Bioinformatics. 6(31)2005.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Thorvaldsdóttir H, Robinson JT and Mesirov
JP: Integrative genomics viewer (IGV): High-performance genomics
data visualization and exploration. Brief Bioinform. 14:178–192.
2013.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Brown TA: Chapter 10 synthesis and
processing of RNA. In: Genomes. 2nd edition Oxford: Wiley-Liss,
2002.
|
|
32
|
Dainat J: AGAT: Another Gff Analysis
Toolkit to handle annotations in any GTF/GFF format (version
v0.4.0). Zenodo, 2020.
|
|
33
|
Needleman SB and Wunsch CD: A general
method applicable to the search for similarities in the amino acid
sequence of two proteins. J Mol Biol. 48:443–453. 1970.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Sievers F, Wilm A, Dineen D, Gibson TJ,
Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al:
Fast, scalable generation of high-quality protein multiple sequence
alignments using Clustal Omega. Mol Syst Biol.
7(539)2011.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Eddy SR: HMMER development team: HMMER
user's guide. Biological sequence analysis using profile hidden
Markov models, version 3.3.2. http://hmmer.org/. Accessed Nov 2020, 2020.
|
|
36
|
Kanost MR, Arrese EL, Cao X, Chen YR,
Chellapilla S, Goldsmith MR, Grosse-Wilde E, Heckel DG, Herndon N,
Jiang H, et al: Multifaceted biological insights from a draft
genome sequence of the tobacco hornworm moth, Manduca sexta. Insect
Biochem Mol Biol. 76:118–147. 2016.PubMed/NCBI View Article : Google Scholar
|
|
37
|
UniProt Consortium: UniProt: The universal
protein knowledgebase in 2023. Nucleic Acids Res. 51
(D1):D523–D531. 2023.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Teufel F, Almagro Armenteros JJ, Johansen
AR, Gíslason MH, Pihl SI, Tsirigos KD, Winther O, Brunak S, von
Heijne G and Nielsen H: SignalP 6.0 predicts all five types of
signal peptides using protein language models. Nat Biotechnol.
40:1023–1025. 2022.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Hallgren J, Tsirigos KD, Damgaard Pedersen
M, Almagro Armenteros JJ, Marcatili P, Nielsen H, Krogh A and
Winther O: DeepTMHMM predicts alpha and beta transmembrane proteins
using deep neural networks. bioRxiv: doi: https://doi.org/10.1101/2022.04.08.487609.
|
|
40
|
Katz K, Shutov O, Lapoint R, Kimelman M,
Brister JR and O'Sullivan C: The sequence read archive: A decade
more of explosive growth. Nucleic Acids Res. 50 (D1):D387–D390.
2022.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Burgin J, Ahamed A, Cummins C, Devraj R,
Gueye K, Gupta D, Gupta V, Haseeb M, Ihsan M, Ivanov E, et al: The
European nucleotide archive in 2022. Nucleic Acids Res. 51
(D1):D121–D125. 2023.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Rice P, Longden I and Bleasby A: EMBOSS:
The European molecular biology open software suite. Trends Genet.
16:276–277. 2000.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Jumper J, Evans R, Pritzel A, Green T,
Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A,
Potapenko A, et al: Highly accurate protein structure prediction
with AlphaFold. Nature. 596:583–589. 2021.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Varadi M, Anyango S, Deshpande M, Nair S,
Natassia C, Yordanova G, Yuan D, Stroe O, Wood G, Laydon A, et al:
AlphaFold protein structure database: Massively expanding the
structural coverage of protein-sequence space with high-accuracy
models. Nucleic Acids Res. 50 (D1):D439–D444. 2022.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Mirdita M, Schütze K, Moriwaki Y, Heo L,
Ovchinnikov S and Steinegger M: ColabFold: Making protein folding
accessible to all. Nat Methods. 19:679–682. 2022.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Waterhouse A, Bertoni M, Bienert S, Studer
G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C,
Bordoli L, et al: SWISS-MODEL: Homology modelling of protein
structures and complexes. Nucleic Acids Res. 46 (W1):W296–W303.
2018.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Berman HM, Westbrook J, Feng Z, Gilliland
G, Bhat TN, Weissig H, Shindyalov IN and Bourne PE: The protein
data bank. Nucleic Acids Res. 28:235–242. 2000.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Edgar RC: MUSCLE: Multiple sequence
alignment with high accuracy and high throughput. Nucleic Acids
Res. 32:1792–1797. 2004.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Waterhouse AM, Procter JB, Martin DMA,
Clamp M and Barton GJ: Jalview version 2-a multiple sequence
alignment editor and analysis workbench. Bioinformatics.
25:1189–1191. 2009.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Archie J, Day WH, Maddison W, Meacham C,
Rohlf FJ, Swofford D and Felsenstein J: The Newick tree format.
http://evolution.genetics.washington.edu/phylip/newicktree.html.
|
|
51
|
Huson DH and Scornavacca C: Dendroscope 3:
An interactive tool for rooted phylogenetic trees and networks.
Syst Biol. 61:1061–1067. 2012.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Schrödinger LLC: The PyMOL molecular
graphics system. PyMOL, 2023.
|
|
53
|
Strauss BS: Frameshift mutation,
microsatellites and mismatch repair. Mutat Res. 437:195–203.
1999.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Treangen TJ and Salzberg SL: Repetitive
DNA and next-generation sequencing: Computational challenges and
solutions. Nat Rev Genet. 13:36–46. 2011.PubMed/NCBI View Article : Google Scholar
|
|
55
|
West MB, Wickham S, Parks EE, Sherry DM
and Hanigan MH: Human GGT2 does not autocleave into a functional
enzyme: A cautionary tale for interpretation of microarray data on
redox signaling. Antioxid Redox Signal. 19:1877–1888.
2013.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Bradley K, Rieger MA and Collins GG:
Classification of Australian garlic cultivars by DNA
fingerprinting. Aust J Exp Agric. 36:613–618. 1996.
|
|
57
|
González RE, Soto VC, Sance MM, Camargo AB
and Galmarini CR: Variability of solids, organosulfur compounds,
pungency and health-enhancing traits in garlic (Allium
sativum L.) cultivars belonging to different ecophysiological
groups. J Agric Food Chem. 57:10282–10288. 2009.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Dalbey RE and Robinson C: Protein
translocation into and across the bacterial plasma membrane and the
plant thylakoid membrane. Trends Biochem Sci. 24:17–22.
1999.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Driessen AJ, Manting EH and van der Does
C: The structural basis of protein targeting and translocation in
bacteria. Nat Struct Biol. 8:492–498. 2001.PubMed/NCBI View
Article : Google Scholar
|
|
60
|
Tjalsma H, Bolhuis A, Jongbloed JD, Bron S
and van Dijl JM: Signal peptide-dependent protein transport in
Bacillus subtilis: A genome-based survey of the secretome.
Microbiol Mol Biol Rev. 64:515–547. 2000.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Briggs MS, Cornell DG, Dluhy RA and
Gierasch LM: Conformations of signal peptides induced by lipids
suggest initial steps in protein export. Science. 233:206–208.
1986.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Suzuki H, Kumagai H and Tochikura T:
gamma-Glutamyltranspeptidase from Escherichia coli K-12: Formation
and localization. J Bacteriol. 168:1332–1335. 1986.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Tate SS and Meister A: gamma-Glutamyl
transpeptidase: Catalytic, structural and functional aspects. Mol
Cell Biochem. 39:357–368. 1981.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Thul PJ, Åkesson L, Wiking M, Mahdessian
D, Geladaki A, Ait Blal H, Alm T, Asplund A, Björk L, Breckels LM,
et al: A subcellular map of the human proteome. Science.
356(eaal3321)2017.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Koonin E and Galperin M: Chapter 2
evolutionary concept in genetics and genomics. In:
Sequence-evolution-function: Computational approaches in
comparative genomics. Kluwer Academic, Boston, 2003.
|
|
66
|
Jones MG, Hughes J, Tregova A, Milne J,
Tomsett AB and Collin HA: Biosynthesis of the flavour precursors of
onion and garlic. J Exp Bot. 55:1903–1918. 2004.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Penninckx MJ and Jaspers CJ: Molecular and
kinetic properties of purified γ-glutamyl transpeptidase from yeast
(Saccharomyces cerevisiae). Phytochemistry. 24:1913–1918.
1985.
|
|
68
|
Storozhenko S, Belles-Boix E, Babiychuk E,
Hérouart D, Davey MW, Slooten L, Van Montagu M, Inzé D and Kushnir
S: Gamma-glutamyl transpeptidase in transgenic tobacco plants.
Cellular localization, processing, and biochemical properties.
Plant Physiol. 128:1109–1119. 2002.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Lancaster JE and Shaw ML: Characterization
of purified γ-glutamyl transpeptidase in onions: Evidence for in
vivo role as a peptidase. Phytochemistry. 36:1351–1358. 1994.
|
|
70
|
Nakano Y, Okawa S, Yamauchi T, Koizumi Y
and Sekiya J: Purification and properties of soluble and bound
gamma-glutamyltransferases from radish cotyledon. Biosci Biotechnol
Biochem. 70:369–376. 2006.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Ikeda Y, Fujii J, Taniguchi N and Meister
A: Human gamma-glutamyl transpeptidase mutants involving conserved
aspartate residues and the unique cysteine residue of the light
subunit. J Biol Chem. 270:12471–12475. 1995.PubMed/NCBI
|
|
72
|
Okada T, Suzuki H, Wada K, Kumagai H and
Fukuyama K: Crystal structures of gamma-glutamyltranspeptidase from
Escherichia coli, a key enzyme in glutathione metabolism, and its
reaction intermediate. Proc Natl Acad Sci USA. 103:6471–6476.
2006.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Grzam A, Martin MN, Hell R and Meyer AJ:
gamma-Glutamyl transpeptidase GGT4 initiates vacuolar degradation
of glutathione S-conjugates in Arabidopsis. FEBS Lett.
581:3131–3138. 2007.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Ohkama-Ohtsu N, Radwan S, Peterson A, Zhao
P, Badr AF, Xiang C and Oliver DJ: Characterization of the
extracellular gamma-glutamyl transpeptidases, GGT1 and GGT2, in
Arabidopsis. Plant J. 49:865–877. 2007.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Ohkama-Ohtsu N, Zhao P, Xiang C and Oliver
DJ: Glutathione conjugates in the vacuole are degraded by
gamma-glutamyl transpeptidase GGT3 in Arabidopsis. Plant J.
49:878–888. 2007.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Martin MN, Saladores PH, Lambert E, Hudson
AO and Leustek T: Localization of members of the gamma-glutamyl
transpeptidase family identifies sites of glutathione and
glutathione S-conjugate hydrolysis. Plant Physiol. 144:1715–1732.
2007.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Shaw ML, Pither-Joyce MD and McCallum JA:
Purification and cloning of a gamma-glutamyl transpeptidase from
onion (Allium cepa). Phytochemistry. 66:515–522.
2005.PubMed/NCBI View Article : Google Scholar
|