Introduction
Chemotherapy usually leads to an improvement in
tumor cell death and produces a decreases in tumor mass; however,
in some patients this therapy fails due to recurrence or even death
(1). The response to chemotherapy
varies greatly from individual to individual. Some patients can
achieve a complete response with just one or two cycles, while
others require eight or more cycles (2,3).
Although progress in the treatment of breast cancer
has been made, chemoresistance still remains an obstacle to the
effective management of all breast cancer types (4). In general, two types of drug
resistance exist: i) Intrinsic, which exists before treatment; and
ii) acquired, which is generated after chemotherapy.
Chemoresistance is the major cause of breast cancer recurrence,
relapse and mortality (5). It is
caused by different mechanisms, such as drug efflux pumps,
dysregulation of apoptosis and cancer stem cells (6) and genetic mutations and/or epigenetic
changes (5). The two main
mechanisms of chemoresistance are drug efflux and DNA repair, both
of which epigenetically regulated (7).
Doxorubicin is one of the most effective
chemotherapeutic drugs used for treatment of breast cancer
(7-9).
It is a naturally occurring anthracycline antibiotic (10). One major cause of treatment failure
with doxorubicin is the development of drug resistance and even
tumor growth (11) that leads to
poor prognosis and survival (12-14).
Doxorubicin resistance is still an unresolved issue in treatment of
patients with breast cancer despite investigations into several
doxorubicin resistance mechanisms (10). Unfortunately, doxorubicin has acute
adverse effects that occur within 2 to 3 days of administration and
include nausea, vomiting, neutropenia, alopecia and/or arrhythmias
(15). Moreover, doxorubicin leads
to an increase in the risk of clinical cardiotoxicity by 5.43-fold
and the risk of heart attack by 4.94-fold when compared with other
non-anthracycline regiments (16).
The emergence of drug resistance and life-threatening cardiac
injury after doxorubicin treatment has led to limitations for
successful cancer treatment (17).
Overcoming doxorubicin resistance and minimizing its toxic cardiac
and kidney effects would represent a major improvement in the
effective management of breast cancer (18).
Quercetin is a polyphenolic compound that is found
in abundance in a number of fruits, vegetables and plants and
exerts various biological effects, such as antioxidant, antiviral,
anticancer, cell cycle modulatory and anti-angiogenesis effects
(19). It has been reported that
quercetin is effective in inhibiting cancer cell growth via the
induction of apoptosis (20,21).
In addition, quercetin has been reported to upregulate the
expression of estrogen receptors α and β.
The overall outcome on cell fate is reflected by
inhibition of cell proliferation, cell cycle arrest in the
G1 phase and reduction in cell migratory potential due
to actin cytoskeleton disorganization (22). To produce an improvement in
antitumor efficacy and a reduction in chemotherapeutic
chemoresistance, the natural flavonoid quercetin has been found to
have antitumor potential and synergistic effects when combined with
doxorubicin (23). The safety of
quercetin makes it an attractive candidate for producing a
reduction in cardiotoxicity attributed to anticancer drugs, such as
doxorubicin (24). The effects of
addition of quercetin to doxorubicin has been previously reported,
and multiple mechanisms accounting for such effects have been
suggested (24-26).
Its use in combination with doxorubicin in resistant cell lines and
the exploration of possible mechanisms of enhancing doxorubicin
toxicity toward cancer cells has yet to be explored.
The aim of the present study was to investigate the
effects of quercetin treatment in prevention of a
doxorubicin-chemoresistant phenotype in both doxorubicin-sensitive
and -resistant human MCF-7 breast cancer cell lines.
Materials and methods
Cell culture growth conditions
A wild-type MCF-7 cell line. MCF-7 breast cancer
cell line was originally obtained from the American Cell Culture
Collection (cat. no. HTB-22). The starting passage numbers was
five. Cells were grown as an attached monolayer culture in the
commercially defined RPMI 1640 medium (Euroclone SpA), supplemented
with 10-20% (v/v) heat-inactivated fetal bovine serum (FBS;
Euroclone SpA), 1% 2 mM L-glutamine (Euroclone SpA), 1% 100X
penicillin-streptomycin concentration (Euroclone SpA) and 0.2% 10
mg/ml gentamicin concentration (Euroclone SpA). The cells were
grown in 75 or 25 cm2 filter-cap culture flasks (SPL
life Sciences). The cells were then incubated at 37˚C in a 90%
humidified atmosphere of 5% CO2 and maintained in a
tissue culture incubator (BINDER GmbH).
Establishment of doxorubicin resistant
sub-line
Doxorubicin 2 mg/ml (Fresenius SE & Co.) was
used to induce resistance. A freshly prepared stock solution
consisted of 100 µM and was stored at 4˚C. The drug was further
diluted with cell culture medium to a concentration of 6.65 nM and
added to the cell media.
First, 6.65 nM of doxorubicin was added to the MCF-7
cells, and when the cells reached appropriate confluency at a
certain concentration, they were passaged and doxorubicin
concentration was increased by 0.05 nM over the previous one. This
process took ~3 months as growth of the cells usually has a lag
period to allow them to adapt to the higher concentration. To
exclude the effects associated with long term culture of MCF-7 wild
type cells, the wild-type cells were cultured under identical
conditions and maintained in culture for the same period as the
resistant cells but without doxorubicin addition.
Cell viability and proliferation
assays for the treatment of MCF-7 with doxorubicin and
quercetin
For wild-type cell lines, 5,000 MCF-7 cells from the
appropriate cell line were seeded per well in a 96-well plate.
After 24 h, six different concentrations of doxorubicin (100, 10,
1.0, 0.1, 0.01 or 0.001 µM) were added to the final volume of 200
µl per well of culture RPMI 1640 medium. For the resistant cell
lines, 7,000 MCF-7 cells from the appropriate cell line were seeded
per well in a 96-well plate. After 24 h, complete growth media was
used for doxorubicin dilution, and using a serial dilution
technique, 16 different concentrations of doxorubicin were prepared
starting at 500 and decreasing each subsequent concentration by 50%
until a concentration of 0.015 µM was reached and added to the
final volume of 200 µl per well of culture media. A stock solution
of quercetin (Sigma-Aldrich; Merck KGaA) was prepared [50 nM in
dimethylsulfoxide (DMSO); Euroclone SpA]. Complete growth media was
used for quercetin dilution and nine different concentrations of
quercetin were prepared (100, 50, 25, 12.5, 6.25, 3.125, 1.652,
0.826 and 0.413 µM). For the combination (both for wild and
resistant cell lines), the same concentrations of doxorubicin were
mixed with 20 µM of quercetin for MCF-7 cell line.
The antiproliferative effects of doxorubicin on
MCF7/WT and MCF7/D53.2 cells were analyzed using the tetrazolium
MTT dye (Sigma-Aldrich; Merck KGaA). This test is based on the
reduction of MTT, a yellow tetrazole, to a purple formazan, a
process that occurs in the mitochondria of viable cells. A total of
10 ml of MTT solution were prepared by weighing 50 mg of MTT dye
dissolved in 10 ml of phosphate-buffered saline (PBS) 1X (Euroclone
SpA).
After 72 h of incubation, media were aspirated from
the cells containing the drugs and replaced with fresh media (100
µl/well) and incubated for 30 min after which 10-13 µl of the MTT
dye solution was added to each well. The plates were returned to
the incubator at 37˚C for 4 h after which 100 µl of DMSO (Euroclone
SpA) was added to each well. Optical density (OD) at 570 nm
wavelength was recorded using a 96-well plate reader (Biotek
instruments, Inc.) after 20 min of shaking on the plate shaker
(cat. no. 130,000; Boekel Scientific). The percentage of living
cells (% living) was calculated by dividing the absorbance of
viable cells per well over the mean absorbance of the viable cells
in control wells. The mean percentage per concentration was then
calculated as mean ± standard deviation (SD). Measurements were
performed in triplicate.
Results of the MTT cell proliferation assay were
evaluated using GraphPad Prism 8 software (GraphPad Software,
Inc.). The inhibitory concentration (IC50), which is the
drug concentration at which 50% of cells are viable, was calculated
from the logarithmic trend line of the cytotoxicity graphs for each
drug alone and in combination. The degree of resistance was
estimated in terms of resistance index (R), which is calculated
according to the equation: R=IC50 resistant
cells/IC50 sensitive cells.
Phase contrast microscopy
Flasks for control wild-type MCF-7 cells and for
each concentration was reached for resistant MCF-7 cells were
observed under EVOS XL core imaging system (Thermo Fisher
Scientific, Inc.) at magnifications of 4, 10 and 20X and images
were captured.
RNA isolation
Next, 75 cm2 flasks were seeded with
7x106 MCF-7 cells. Treatment was started with 5 µM
doxorubicin or the combination of 5 µM doxorubicin with 20 µM
quercetin. One flask was allotted as control. RNA was extracted
from each individual cell pellet using RNeasy® Plus Mini
kit (Qiagen GmbH) following manufacturer's instructions.
Then RNA concentration and purity were measured
using a nanodrop 2000 spectrophotometer (Thermo Fisher Scientific,
Inc.). Furthermore, the quality of RNA was visualized under
ultraviolet light after performing gel electrophoresis.
cDNA synthesis
The first strand of cDNA was prepared using the RT2
Profiler PCR Array kit (Qiagen GmbH). According to the
manufacturer's instructions, aliquots containing 1 µg of total RNA
were used from each sample.
Gene expression profiling
Pathway-focused gene expression profiling was
performed using a 96-well human cancer drug resistance and
metabolism PCR array, RT2 Profiler PCR array (PAHS-131ZD-12), Human
Cancer Drug Resistance & Metabolism PCR Array (Qiagen, Inc.).
In this array, 84 wells contained all components required for the
PCR reaction in addition to a primer for a single gene in each
well. A total of five housekeeping genes (ACTB, B2M, GAPDH, HPRT1
and RPLP0) were used as a control for normalization. Notably,
Qiagen provide the array with their design primers and they do not
provide the sequence of such primers. These particular genes are
involved in tumor classification, signal transduction, DNA repair
and other commonly affected pathways such as the cell cycle,
angiogenesis, apoptosis adhesion and proteolysis. A diluted cDNA,
equivalent to 1 µg RNA for each plate, was mixed with the RT2
SYBR® green master mix (Qiagen, Inc.) according to the
manufacturer's instructions, and loaded into the 96-well array.
Reverse transcription-quantitative (RT-qPCR) reaction was performed
using the iCycler (Bio-Rad Laboratories, Inc.) machine by heating
the plate to 95˚C for 10 min followed by 40 cycles of 95˚C for 15
sec and 60˚C for 1 min.
The cycle threshold (Cq) values for each sample were
given by the iCycler. The threshold value was manually set on 0.01
as recommended by the PCR array user manual. The analysis was
performed automatically according to the SA Biosciences company
(Qiagen, Inc.) web portal (https://geneglobe.qiagen.com/jo/analyze), and
expression levels were expressed as a fold-increase or decrease
(27). The data were normalized
across the plates to the following housekeeping genes were used: i)
Actin β; ii) β-2-microglobulin, glyceraldehyde-3-phosphate
dehydrogenase; iii) hypoxanthine phosphoribosyltransferase 1; and
iv) ribosomal protein large, P0.
Gene functional annotation
The Gene Ontology (GO; https://david.ncifcrf.gov/tools.jsp) and pathway
database for genes whose expression levels changed by 2-fold was
investigated using the web-based application DAVID (http://david.abcc.ncifcrf.gov), which provides for
ontology and pathway mapping, annotation, and visualization of
results. Results appear in tables that describe the most affected
function and the pathway based on genes changes.
Apoptosis analysis
Apoptosis events induced by anticancer drug
treatment in MCF-7 wild-type and resistant cell lines treated with
doxorubicin alone or in combination with quercetin in addition to
no treatment were investigated. The mechanism of cell death was
determined by Annexin V/propidium iodide (PI) staining using flow
cytometry. MCF-7 wild-type and resistant cells were seeded into
six-well plates and incubated overnight at 37˚C. Wild-type MCF-7
cells were exposed to 1.3 µM doxorubicin alone or to a combination
of 1.3 µM doxorubicin combined with 20 µM of quercetin. Resistant
cells were exposed to 4 µM doxorubicin or to a combination of 4 µM
doxorubicin combined with 20 µM of quercetin. For both cell types,
control cells remained untreated. After incubation for 72 h, media
were removed, and cell cultures were washed two times with PBS.
Cells were collected into flow tubes using Accutase solution (800
µl on each well at room temperature for 10 min). After this step,
an Annexin V/PI apoptosis kit was used to stain the cell pellets
following the kit's instructions. The apoptosis assay was carried
out using the annexin v assay (Molecular Probes; Thermo Fisher
Scientific, Inc.), and cell death (either apoptosis or necrosis)
was analyzed by FACS Diva7, a fluorescein-activated sorter (FACS)
Canto II version 3 (BD Biosciences). Flow experiments was performed
using single color staining.
The results from the apoptosis assay groups were
tested for significance using a two-way analysis of variance
(ANOVA) followed by a Šidák multiple comparison test to identify
the statistical significance using GraphPad Prism. Data was
presented as mean ± standard deviation and the experiments were
repeated three times. P<0.05 was considered to indicate a
statistically significant difference. Fig. 1 presents the study flowchart.
Results
Determination of IC50 value
of doxorubicin alone and combination in wild-type and resistant
MCF-7 breast cancer cell lines
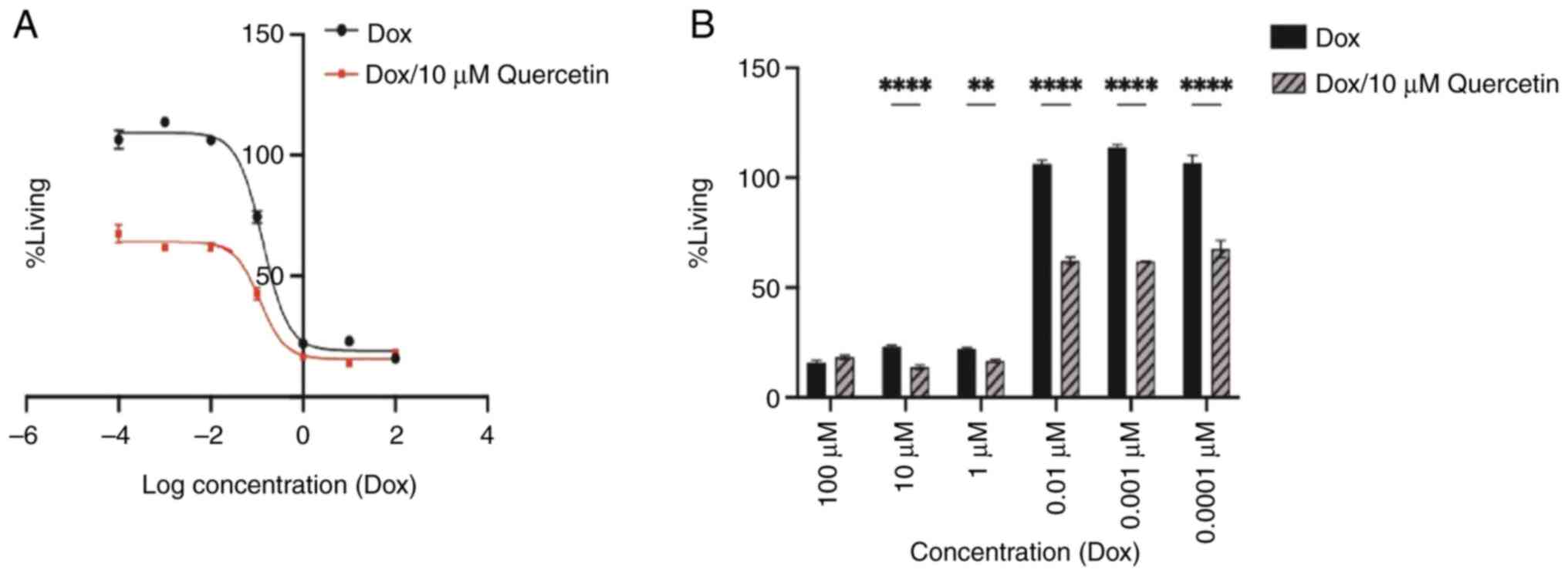
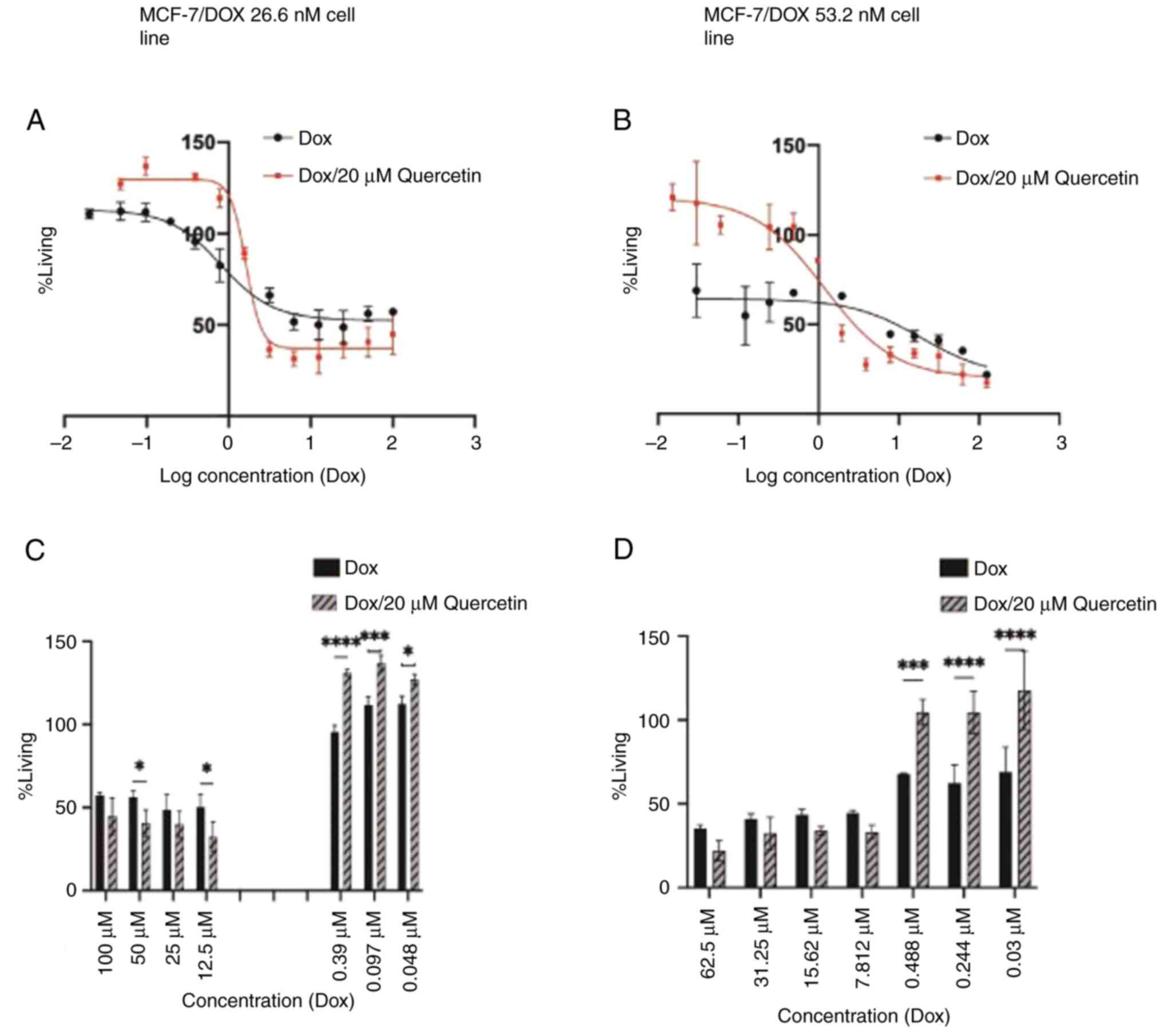
The wild MCF-7 cell viability was determined using
the MTT assay as is presented in Fig.
2. The IC50 values for doxorubicin only and
doxorubicin with 10 µM quercetin combination were 0.133 and 0.114
µM, respectively. Treatment with doxorubicin with 10 µM quercetin
at 0.01, 0.001 and 0.0001 µM only yielded significant inhibition of
the growth of MCF-7 cells when compared with the growth of cells
treated with higher doxorubicin concentrations. Overall, these
findings indicated that quercetin had a potentiation effect toward
doxorubicin against the MCF7 breast cancer cells.
Development and characterization of the MCF-7
resistant cells. The doxorubicin-resistant MCF-7 cell line was
established and examined at two different doxorubicin
concentrations: i) A final concentration of 26.6 nM; and ii) a
final concentration of 53.2 nM.
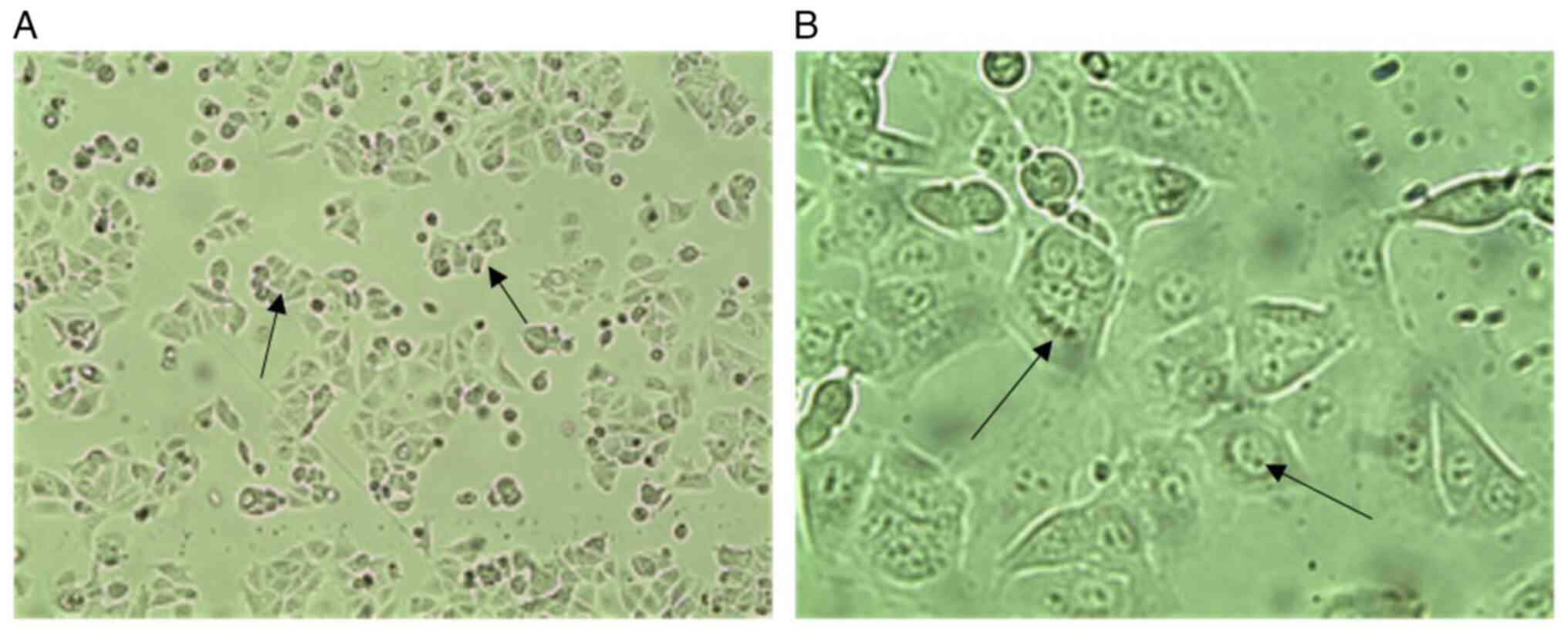
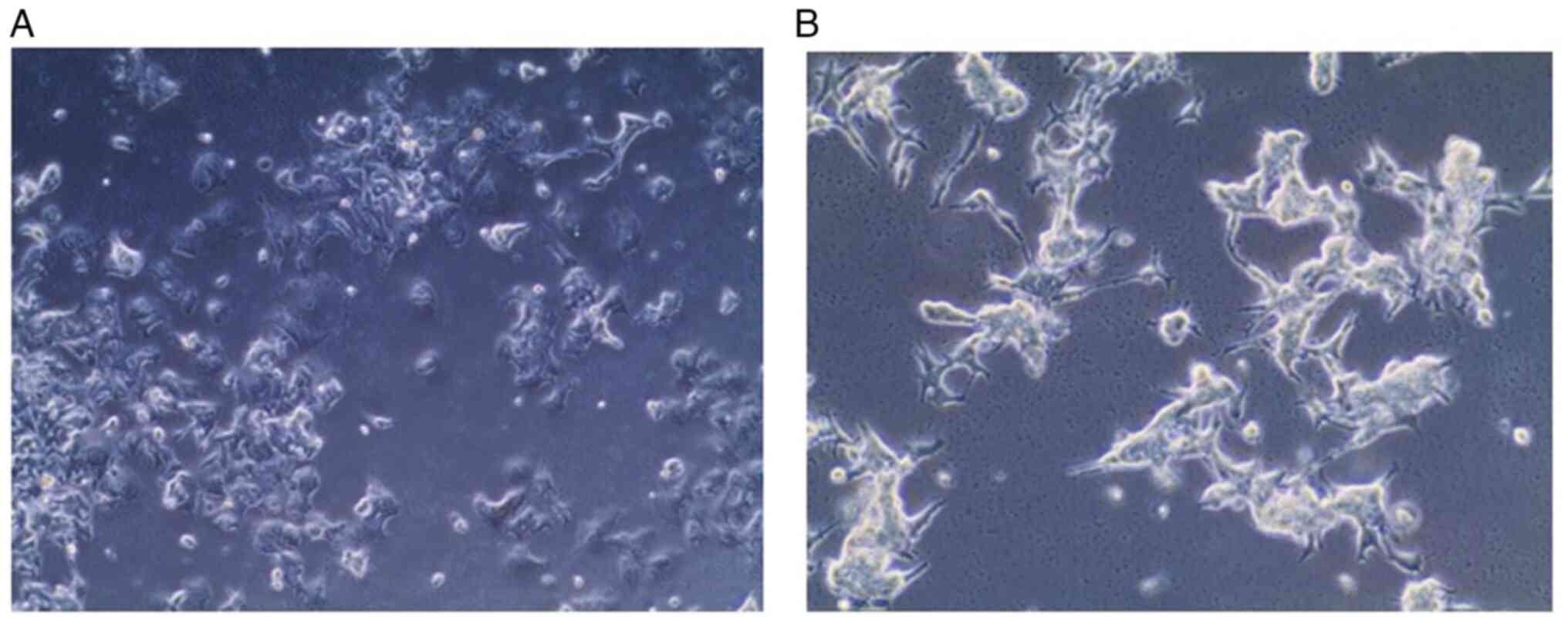
Morphological features. Fig. 3 presents phase contrast images of
wild MCF-7 cells, which present epithelial-like morphology of
small, spindle-shaped cells with a single nucleus. Cells grow in
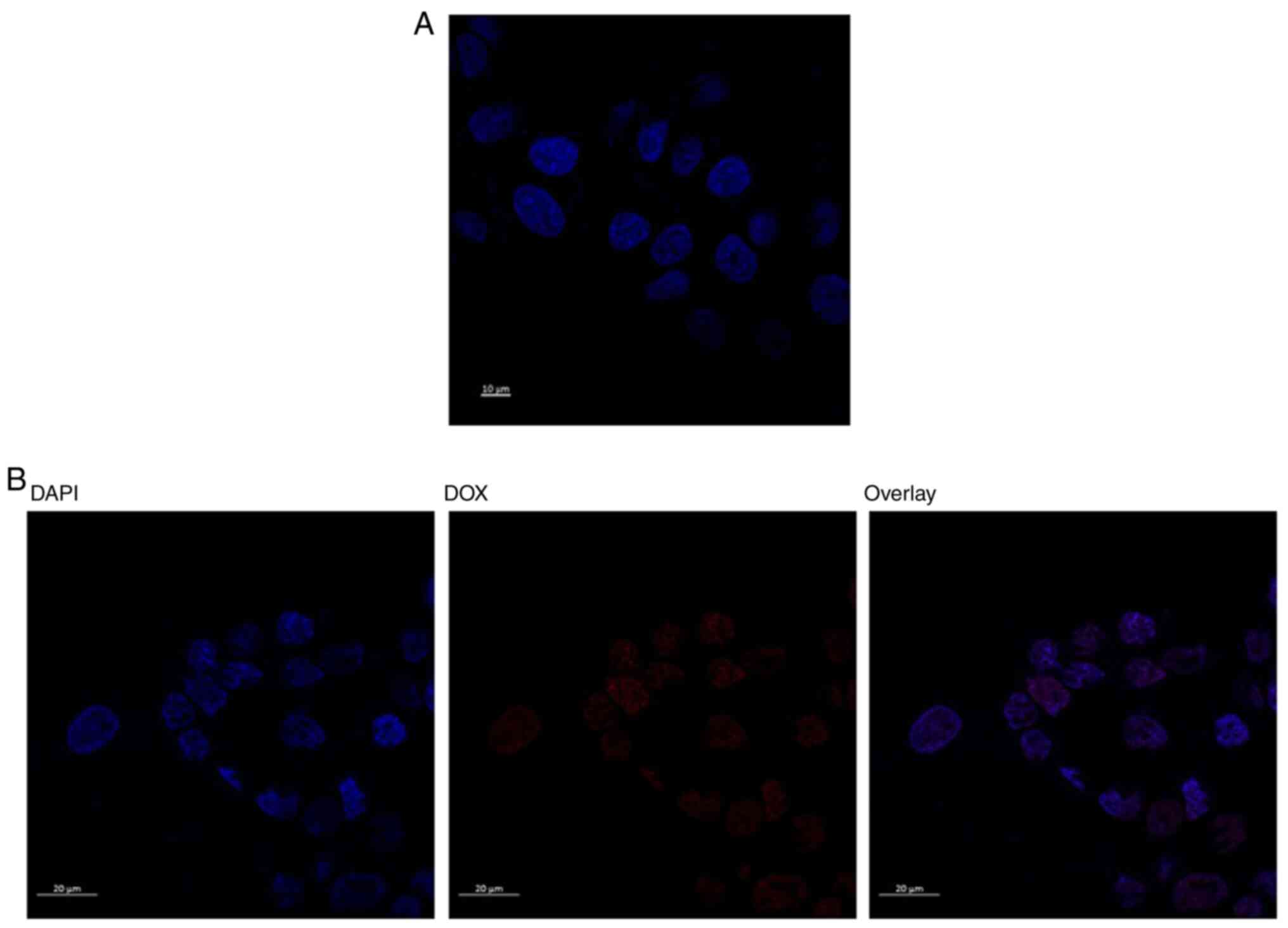
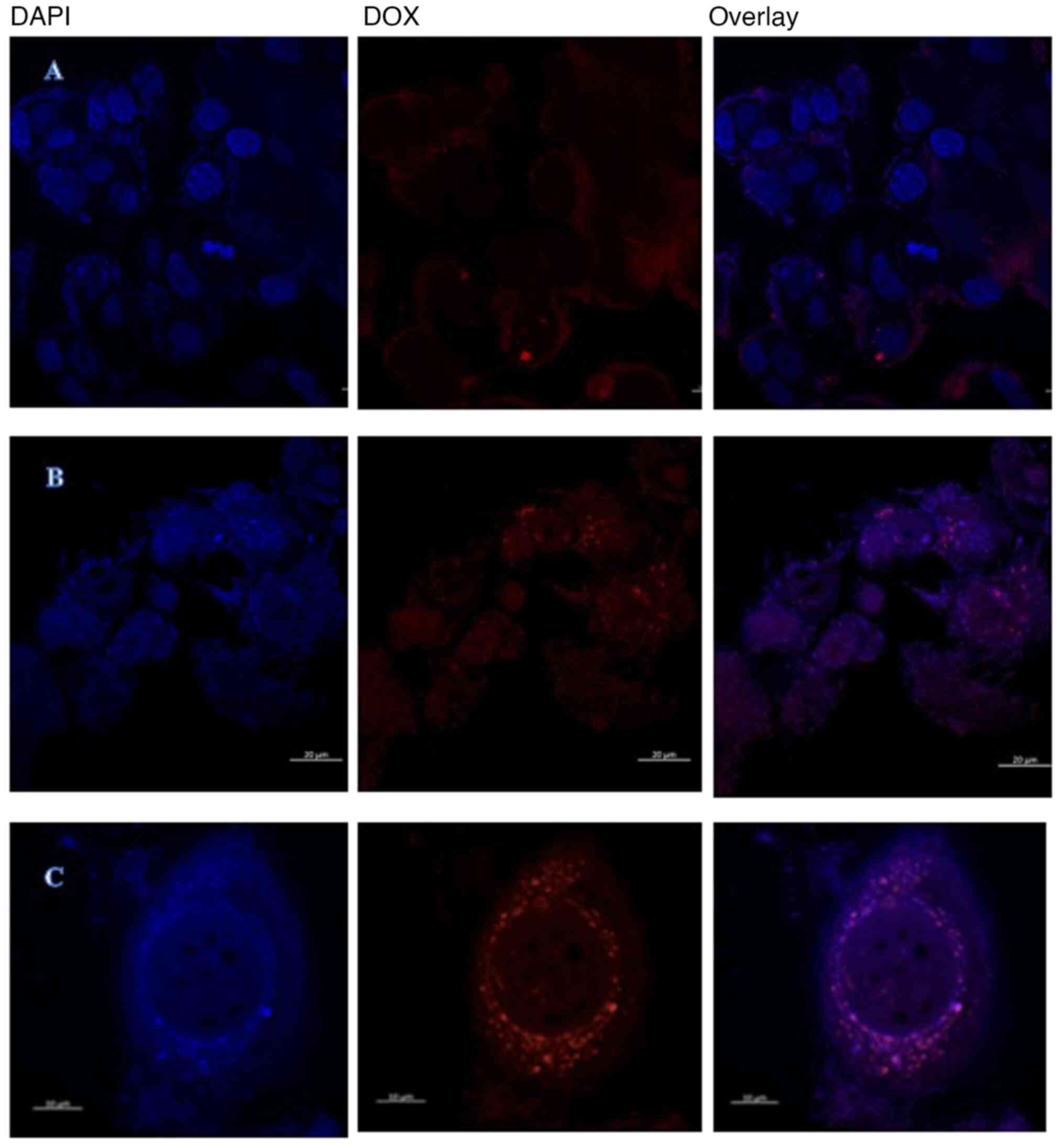
colonies, tightly packed and uniform in size. While confocal
microscopy images of DAPI-staining are presented in Fig. 4 for wild untreated and 1.3 µM
doxorubicin treated cells. It represent how the doxorubicin
accumulates within the nuclease of the cells in the treated
cells.
Sensitivity to doxorubicin
As presented in Fig.
5 the IC50 values of doxorubicin for resistant MCF-7
cell line in addition to the resistance index were calculated and
are shown in Table I.
 | Table IIC50 and R for wild and
resistant MCF-7 cell lines. |
Table I
IC50 and R for wild and
resistant MCF-7 cell lines.
| Cell line | DOX only, µM | R | DOX/20 µM
quercetin, µM |
|---|
| MCF-7 wild | 0.133 | | 0.114 |
| MCF-7/DOX 26.6
nM | 0.800 | 6.01 | 1.620 |
| MCF-7/DOX 53.2
nM | 4.000 | 30.0 | 1.290 |
Although the IC50 of the doxorubicin and
20 µM quercetin combination was higher compared with doxorubicin
alone in the MCF-7/DOX 26.6 nM group, it was observed that
quercetin enhanced anticancer activity when added to higher
doxorubicin concentrations as is presented in Fig. 5. The IC50 value of the
MCF-7/DOX 53.2 nM group was 4.0, µM, which is 30-fold higher
compared with the sensitive parent cell line. Treatment with
quercetin in the MCF-7/DOX 53.2 nM group led to a significant
decrease in the doxorubicin IC50 value and resistance
fold value from 30- to 9-fold. These findings revealed that
quercetin had significant doxorubicin re-sensitizing effects.
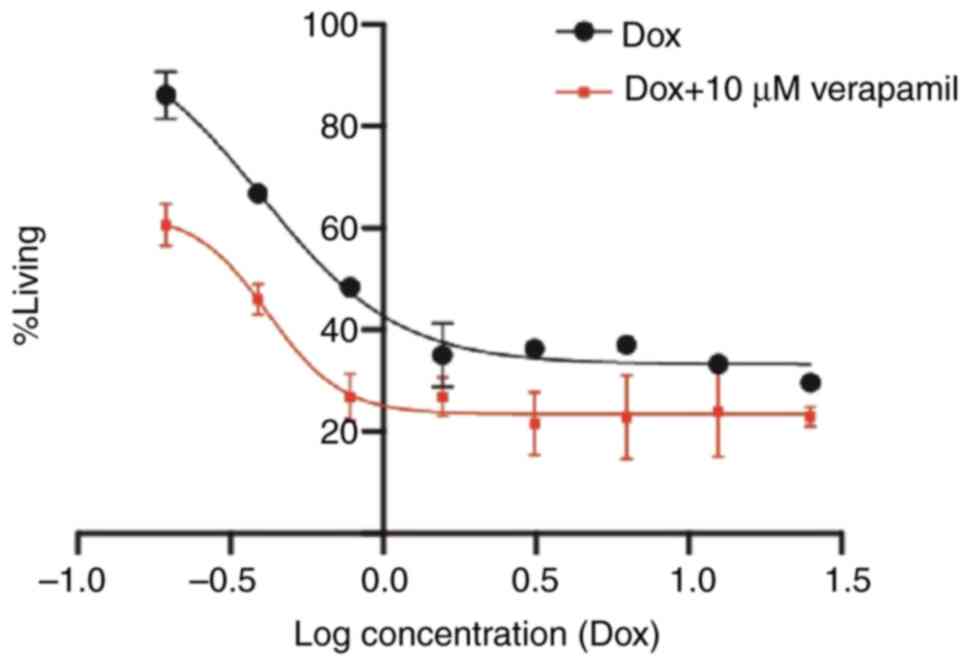
Reversal of doxorubicin resistance with verapamil
was performed to further investigate the inhibition of
P-glycoprotein (P-gp) by verapamil as a standard drug after which
proliferation of the cells was compared to the resistant MCF-7
cells with MCF-7 cells in combination (doxorubicin with 10 µM
verapamil) as presented in Fig. 6.
This figure demonstrates how the verapamil forces doxorubicin
outside the cells and therefore reduces the concentration that can
reach the nucleus.
Morphological features. In comparison with
parent wild type cells, the resistant MCF-7 cells appeared to be
larger, irregular and had a satellite-like shape with multiple
nuclei and multiple vesicles and stronger adhesion to the
underlying surface (Fig. 7).
Confocal imaging in Fig.
8 was used to investigate intracellular doxorubicin
accumulation in resistant MCF-7/DOX 53.2 nM cells. The cells were
incubated with 400 µM of doxorubicin for 4 h, it was also viewed at
0 h as a control. This demonstrated that control resistant
MCF-7/DOX 26.6 nM cells show a low level of fluorescence with a
narrow doxorubicin intracellular distribution, while in 400 µM
treated doxorubicin group the free doxorubicin surrounded the
nucleus. These figures collectively indicated that the main
resistant mechanism exemplified in effluxing doxorubicin toward the
cytoplasm and to engulf them in special type of vesicles.
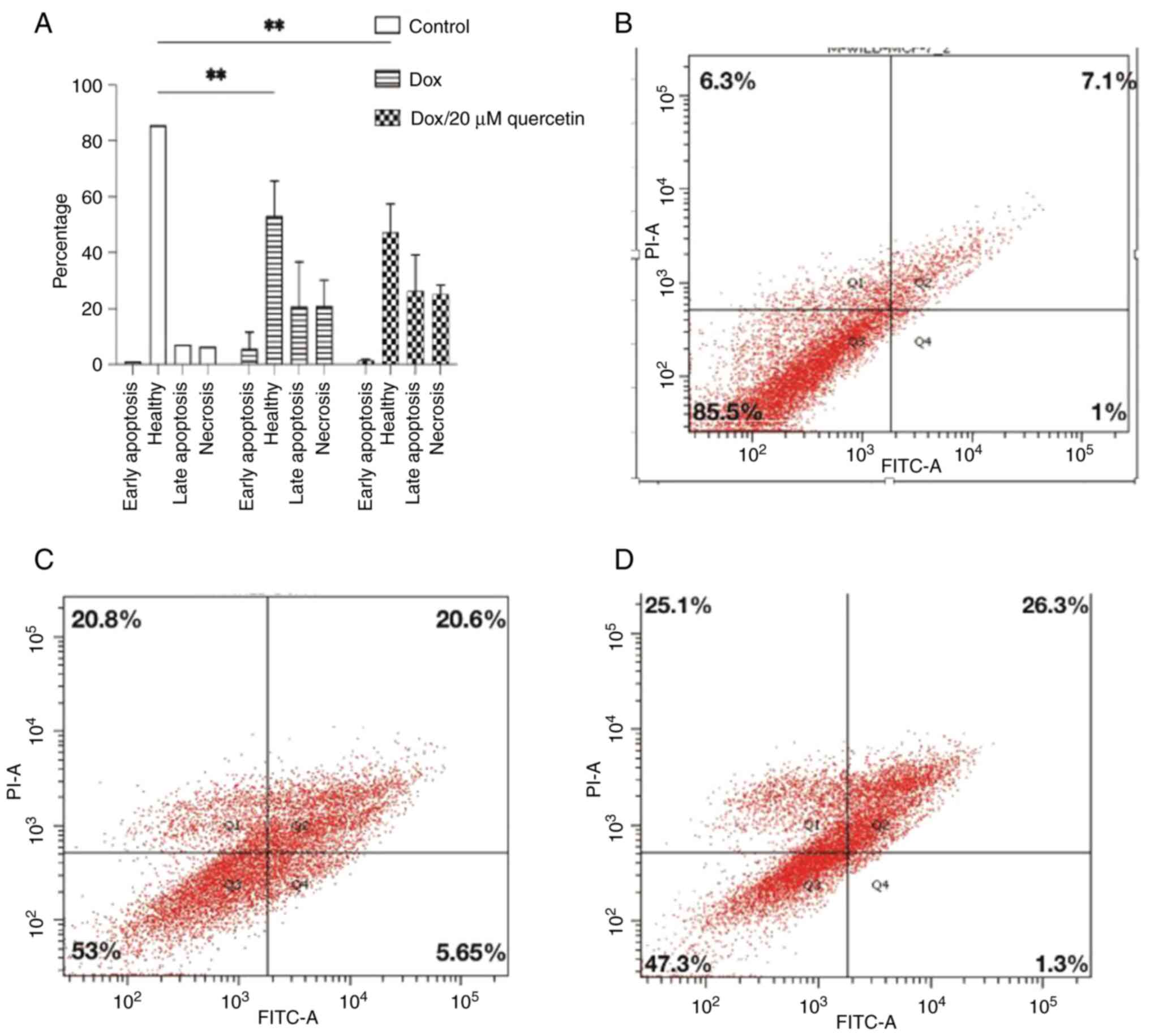
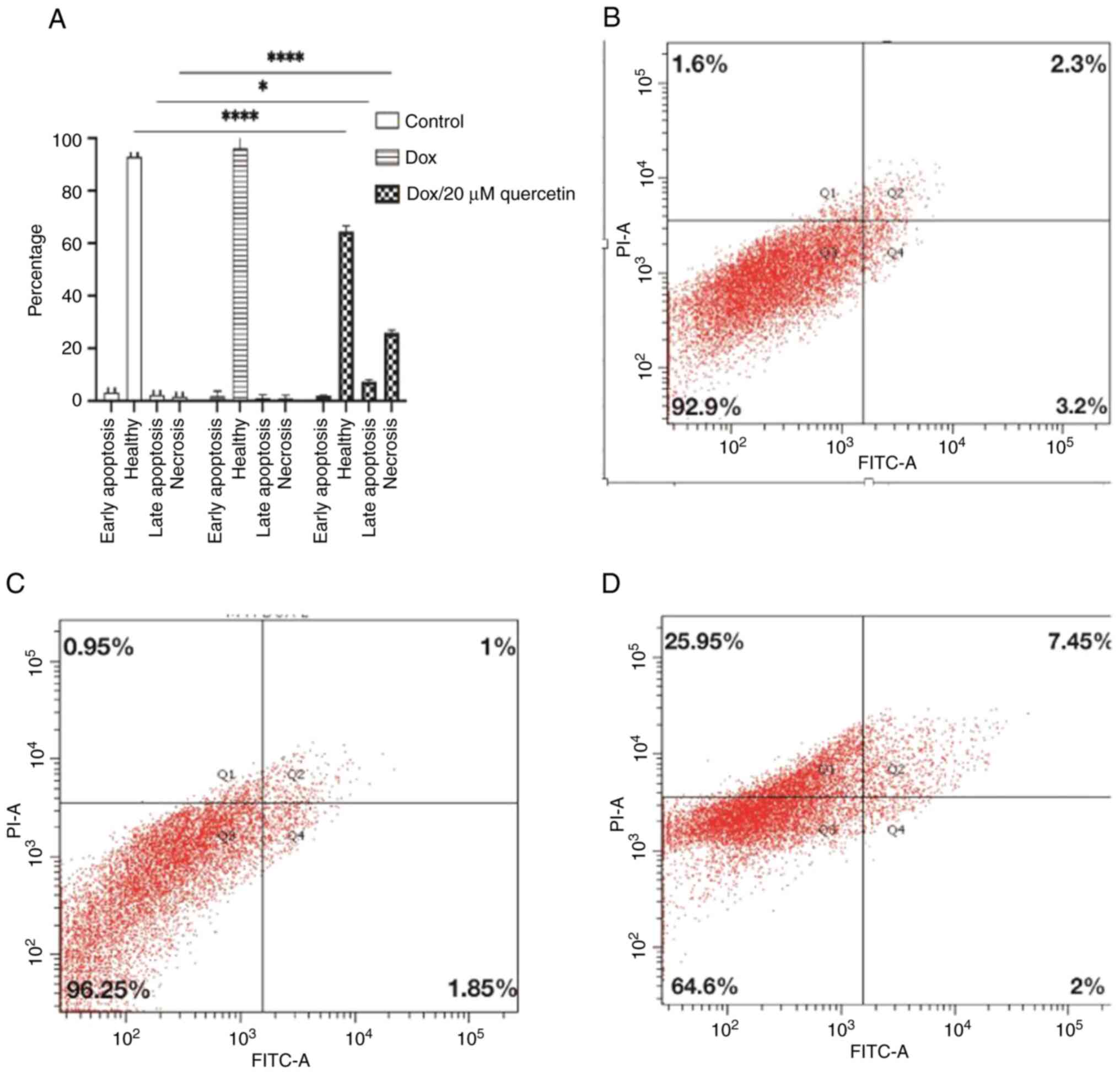
Apoptosis analysis
To determine whether the effects of quercetin on
wild-type and resistant MCF-7 cancer cell proliferation were
related to apoptosis, flow cytometry using Annexin V/PI staining
was performed. Wild-type MCF-7 cells were treated with 1.3 µM
doxorubicin only or a combination of 1.3 µM doxorubicin and 20 µM
quercetin, and resistant MCF-7 were treated with 4 µM doxorubicin
or a combination of 4 µM doxorubicin and 20 µM quercetin for 72
h.
The cells were sorted according to annexin V and PI
status into early apoptotic Q4, late apoptotic Q2, necrotic Q1 and
viable cells Q3, and results are presented in Figs. 9 and 10. In wild-type MCF-7 cells as shown in
Fig. 9, the late apoptosis rate
increased from 7.1% in control cells to 20.6 and 26.3% after
treatment corresponding to doxorubicin or the doxorubicin and
quercetin combination, respectively. Wild-type MCF-7 cells treated
with 1.3/20 µM Dox/quercetin combination had the highest apoptosis
rates. Similarly, the percentage of necrotic cells increased from
6.3% in control cells to 20.8 and 25.1% after treatment
corresponding to doxorubicin only or the doxorubicin and quercetin
combination.
In resistant MCF-7 cells (Fig. 10), the effects of quercetin on the
induction of cell apoptosis in breast cancer cells was also
measured. It was found that the late apoptosis rate changed from
2.3% in control cells to 1% or 7.25% after treatment corresponding
to doxorubicin only or to the doxorubicin and quercetin
combination, respectively. By contrast, the percentage of necrotic
cells changed from 1.6% in control cells to 0.95% or 25.95% after
treatment corresponding to doxorubicin only or the doxorubicin and
quercetin combination, respectively. Collectively, the apoptosis
assay findings indicated that the mechanism of killing for
doxorubicin differs in the presence of quercetin, especially in the
resistant MCF-7 cells.
Gene expression changes in the
resistant MCF-7 cell line
A gene expression profile analysis was performed
using the PCR array and a standard 2-fold change in expression was
used as the cut-off. The effects of 5 µM of doxorubicin on the
expression of the breast cancer genes were tested against MCF/DOX,
such treatment was called Track (1). While the combination of 5 µM dox and
20 µM quercetin were named Track (2). Analysis of track (1) revealed that 52 genes out of the 84
genes were modulated, 41 of these were upregulated, and 11 genes
were downregulated (Tables SI and
SII).
In the Track 2 analysis, it was revealed that in 46
genes out of the total 84 genes that were examined, 35 were
upregulated and 11 genes were downregulated (Tables SIII and SIV). The difference between the fold
regulation between Tracks (1) and
(2) was calculated as shown in
Tables SV and SVI using a standard of 5-fold change as
the cut-off.
Gene ontology and pathways
analysis
Gene Ontology (GO) categories for both up- and
downregulated genes for Track (1)
are shown in Tables II and
III, respectively. The data
indicated that the selection for doxorubicin resistance led to
changes in gene expression, mainly in upregulated genes, and
consisted of ‘protein binding’, ‘negative regulation of cell
proliferation’, ‘positive regulation of transcription from RNA
polymerase II promoter’, ‘negative regulation of apoptotic process’
and ‘positive regulation of cell migration’. In downregulated
genes, it was found that ‘positive regulation of transcription from
RNA polymerase II promoter’, ‘positive regulation of transcription,
DNA-templated’ and ‘positive regulation of apoptotic process’ were
affected. The most important pathways in which the up- and the
downregulated genes participated are presented in Tables IV and V, respectively. In the downregulated side,
‘pathways in cancer’, ‘microRNAs in cancer’, ‘p53 signaling
pathway’, ‘cell cycle’ and ‘signaling pathway’ are the main
pathways and ‘pathways in cancer’ is the only pathway in the
downregulated side.
 | Table IIGene Ontology groups for upregulated
genes (≥2-fold) in (MCF-7/DOX 53.2 nM). |
Table II
Gene Ontology groups for upregulated
genes (≥2-fold) in (MCF-7/DOX 53.2 nM).
| Category | Count | Genes |
|---|
| Protein
binding | 37 | CDKN1C, CDKN1A,
NOTCH1, CSF1, GSTP1, SERPINE1, PTEN, TWIST1, NR3C1, GLI1, PTGS2,
MAPK8, CCND2, PLAU, CDH1, ADAM23, SFN, SLIT2, JUN, TGFB1, CDKN2A,
MMP2, KRT5, IGF1, HIC1, ESR2, NME1, VEGFA, CCNA1, SFRP1, IL6, APC,
CCNE1, BCL2, CTNNB1, SNAI2, ABCG2 |
| Negative regulation
of cell proliferation | 14 | CDKN1A, JUN, TGFB1,
NOTCH1, CDKN2A, PTEN, PTGS2, NME1, IL6, SFRP1, APC, CDH13, CTNNB1,
RARB |
| Positive regulation
of transcription from RNA polymerase II promoter | 14 | JUN, TGFB1, NOTCH1,
CDKN2A, SERPINE1, TWIST1, IGF1, GLI1, NR3C1, VEGFA, IL6, CDH13,
CTNNB1, RARB |
| Positive regulation
of transcription, DNA- templated | 13 | CDKN1C, JUN, TGFB1,
NOTCH1, CDKN2A, IGF1, GLI1, ESR2, IL6, SFRP1, CCNE1, CDH1,
CTNNB1 |
| Negative regulation
of apoptotic process | 12 | IL6, CDKN1A, SFRP1,
MAPK8, CCND2, GSTP1, PTEN, BCL2, RARB, TWIST1, IGF1, VEGFA |
| Positive regulation
of cell proliferation | 12 | IL6, SFRP1, TGFB1,
NOTCH1, CCND2, CSF1, PTEN, BCL2, RARB, IGF1, GLI1, VEGFA |
| Positive regulation
of cell migration | 10 | TGFB1, NOTCH1, APC,
CSF1, PLAU, CDH13, SNAI2, IGF1, GLI1, VEGFA |
| Negative regulation
of transcription from RNA polymerase II promoter | 10 | CDKN1C, TGFB1,
NOTCH1, CTNNB1, RARB, TWIST1, SNAI2, HIC1, ESR2, VEGFA |
 | Table IIIGene ontology groups for
downregulated genes (≥2-fold) in (MCF-7/DOX 53.2 nM). |
Table III
Gene ontology groups for
downregulated genes (≥2-fold) in (MCF-7/DOX 53.2 nM).
| Category | Count | Genes |
|---|
| Positive regulation
of transcription from RNA polymerase II promoter | 7 | RB1, AR, XBP1, PGR,
BRCA1, ESR1, MAPK3 |
| Transcription,
DNA-templated | 7 | RB1, AR, BIRC5,
PGR, BRCA1, ESR1, MAPK3 |
| Positive regulation
of transcription, DNA-templated | 6 | RB1, AR, BRCA1,
BRCA2, ESR1, MAPK3 |
| Regulation of
apoptotic process | 3 | BIRC5, BRCA1,
ESR1 |
 | Table IVPathway analysis for upregulated
genes in (MCF-7/DOX 53.2 nM). |
Table IV
Pathway analysis for upregulated
genes in (MCF-7/DOX 53.2 nM).
| Category | Count | Genes |
|---|
| Pathways in
cancer | 19 | CDKN1A, JUN, TGFB1,
CDKN2A, GSTP1, MMP2, PTEN, IGF1, GLI1, PTGS2, VEGFA, IL6, MAPK8,
APC, CCNE1, CDH1, BCL2, CTNNB1, RARB |
| MicroRNAs in
cancer | 11 | CDKN1A, NOTCH1,
CCND2, APC, PLAU, CCNE1, CDKN2A, PTEN, BCL2, PTGS2, VEGFA |
| p53 signaling
pathway | 8 | CDKN1A, CCND2,
CCNE1, CDKN2A, SERPINE1, PTEN, SFN, IGF1 |
| Cell cycle | 8 | CDKN1C, CCNA1,
CDKN1A, TGFB1, CCND2, CCNE1, CDKN2A, SFN |
| Signaling
pathway | 7 | IL6, CDKN1A, MAPK8,
TGFB1, CCND2, PTEN, IGF1 |
 | Table VPathway analysis for downregulated
genes in (MCF-7/DOX 53.2 nM). |
Table V
Pathway analysis for downregulated
genes in (MCF-7/DOX 53.2 nM).
| Category | Count | Genes |
|---|
| Pathways in
cancer | 5 | RB1, AR, BIRC5,
BRCA2, MAPK3 |
In Track (2) the
genes that were upregulated 2-fold were functionally clustered into
main categories consisting of ‘negative regulation of cell
proliferation’, ‘negative regulation of apoptotic process’, ‘cell
cycle arrest’ and ‘regulation of cell cycle’, as shown in Table VI. In downregulated genes, it was
found that ‘positive regulation of cell migration’, ‘positive
regulation of apoptotic process’, ‘negative regulation of
transcription from RNA polymerase II promoter’ and negative
regulation of cell proliferation’ were affected, as shown in
Table VII. The main pathways in
up- and the downregulated genes are presented in Tables VIII and IX, respectively. In the upregulated side,
those pathways are ‘pathways in cancer, ‘microRNAs in cancer’, ‘p53
signaling pathway’ and ‘cell cycle’. In the downregulated side,
they are ‘microRNAs in cancer’ and ‘pathways in cancer’.
 | Table VIGene Ontology groups for upregulated
genes (≥2-fold) in (MCF-7/DOX 53.2 nM) [Track (2)]. |
Table VI
Gene Ontology groups for upregulated
genes (≥2-fold) in (MCF-7/DOX 53.2 nM) [Track (2)].
| Category | Count | Genes |
|---|
| Negative regulation
of cell proliferation | 11 | IL6, CDKN1A, SFRP1,
JUN, APC, CDKN2A, PTEN, CDH13, RARB, PTGS2, NME1 |
| Negative regulation
of apoptotic process | 10 | IL6, CDKN1A, SFRP1,
MAPK8, CCND2, GSTP1, PTEN, BCL2, RARB, IGF1 |
| Positive regulation
of apoptotic process | 9 | IL6, SFRP1, MAPK8,
APC, CDKN2A, RARB, ATM, SLIT2, PTGS2 |
| Positive regulation
of cell proliferation | 9 | IL6, SFRP1, CCND2,
CSF1, PTEN, BCL2, RARB, IGF1, GLI1 |
| Cell cycle
arrest | 5 | CDKN1C, CDKN1A,
APC, CDKN2A, ATM |
| Regulation of cell
cycle | 4 | JUN, CCNE1, PTEN,
ATM |
| DNA damage
response, signal transduction by p53 class mediator resulting in
cell cycle arrest | 3 | CDKN1A, ATM,
SFN |
 | Table VIIGene Ontology groups for
downregulated genes (≥2-fold) in (MCF-7/DOX 53.2 nM)
[Track(2)]. |
Table VII
Gene Ontology groups for
downregulated genes (≥2-fold) in (MCF-7/DOX 53.2 nM)
[Track(2)].
| Category | Count | Genes |
|---|
| Positive regulation
of cell migration | 5 | APC, CSF1, PLAU,
CDH13, SNAI2 |
| Positive regulation
of apoptotic process | 4 | APC, RARB, SLIT2,
PTGS2 |
| Negative regulation
of cell proliferation | 4 | APC, CDH13, RARB,
PTGS2 |
| Negative regulation
of transcription from RNA polymerase II promoter | 4 | RARB, SNAI2, HIC1,
ESR2 |
| Transcription,
DNA-templated | 4 | RARB, SNAI2, HIC1,
ESR2 |
 | Table VIIIPathway analysis for upregulated
genes in (MCF-7/DOX 53.2 nM) [Track(2)]. |
Table VIII
Pathway analysis for upregulated
genes in (MCF-7/DOX 53.2 nM) [Track(2)].
| Category | Count | Genes |
|---|
| Pathways in
cancer | 16 | CDKN1A, JUN,
CDKN2A, GSTP1, MMP2, PTEN, IGF1, PTGS2, GLI1, IL6, MAPK8, APC,
CCNE1, CDH1, BCL2, RARB |
| MicroRNAs in
cancer | 10 | CDKN1A, CCND2, APC,
PLAU, CCNE1, CDKN2A, PTEN, BCL2, ATM, PTGS2 |
| p53 signaling
pathway | 9 | CDKN1A, CCND2,
CCNE1, CDKN2A, SERPINE1, PTEN, ATM, SFN, IGF1 |
| Cell cycle | 8 | CDKN1C, CCNA1,
CDKN1A, CCND2, CCNE1, CDKN2A, ATM, SFN |
 | Table IXPathway analysis for downregulated
genes in (MCF-7/DOX 53.2 nM) [Track(2)]. |
Table IX
Pathway analysis for downregulated
genes in (MCF-7/DOX 53.2 nM) [Track(2)].
| Category | Count | Genes |
|---|
| MicroRNAs in
cancer | 3 | APC, PLAU,
PTGS2 |
| Pathways in
cancer | 3 | APC, RARB,
PTGS2 |
In terms of the difference between Tracks (1) and (2)
genes that were upregulated ≥5-fold, the main functional groups
included those involved in ‘negative regulation of apoptotic
process’, ‘positive regulation of cell proliferation’, ‘positive
regulation of transcription, DNA-templated’, ‘cellular response to
tumor necrosis factor’ and ‘DNA replication’ (Table X). In downregulated genes, these
groups included ‘positive regulation of cell migration’, ‘positive
regulation of apoptotic process’, ‘negative regulation of cell
proliferation’ and ‘transcription, DNA-templated’ (Table XI). The main pathways in up- and
downregulated genes are presented in Tables XII and XIII, respectively. ‘Pathways in
cancer’, ‘p53 signaling pathway’, ‘signaling pathway’ and ‘cell
cycle’ are the main pathway on the upregulated side and ‘microRNAs
in cancer’ and ‘pathways in cancer’ on the downregulated side.
 | Table XGene Ontology groups for upregulated
genes (≥5-fold) in the difference between Track (1) and Track (2). |
Table X
Gene Ontology groups for upregulated
genes (≥5-fold) in the difference between Track (1) and Track (2).
| Category | Count | Genes |
|---|
| Negative regulation
of apoptotic process | 6 | IL6, SFRP1, CCND2,
GSTP1, BIRC5, IGF1 |
| Positive regulation
of cell proliferation | 5 | IL6, SFRP1, CCND2,
BIRC5, IGF1 |
| Positive regulation
of transcription, DNA-templated | 5 | IL6, SFRP1, CDKN2A,
BRCA1, IGF1 |
| Positive regulation
of apoptotic process | 4 | IL6, SFRP1, CDKN2A,
ATM |
| Cellular response
to tumor necrosis factor | 3 | IL6, SFRP1,
BRCA1 |
| DNA
replication | 3 | ATM, BRCA1,
IGF1 |
| Replicative
senescence | 2 | CDKN2A, ATM |
 | Table XIGene Ontology groups for
downregulated genes (≥5-fold) in the difference between Track
(1) and Track (2). |
Table XI
Gene Ontology groups for
downregulated genes (≥5-fold) in the difference between Track
(1) and Track (2).
| Category | Count | Genes |
|---|
| Positive regulation
of cell migration | 5 | APC, CSF1, PLAU,
CDH13, SNAI2 |
| Positive regulation
of apoptotic process | 4 | APC, RARB, SLIT2,
PTGS2 |
| Negative regulation
of cell proliferation | 4 | APC, CDH13, RARB,
PTGS2 |
| Transcription,
DNA-templated | 4 | RARB, SNAI2, HIC1,
ESR2 |
 | Table XIIPathway analysis for upregulated
genes in the difference between Track (1) and Track (2). |
Table XII
Pathway analysis for upregulated
genes in the difference between Track (1) and Track (2).
| Category | Count | Genes |
|---|
| Pathways in
cancer | 6 | IL6, CDKN2A, GSTP1,
MMP2, BIRC5, IGF1 |
| p53 signaling
pathway | 5 | CCND2, CDKN2A,
SERPINE1, ATM, IGF1 |
| Signaling
pathway | 4 | IL6, CCND2, ATM,
IGF1 |
| Cell cycle | 3 | CCND2, CDKN2A,
ATM |
 | Table XIIIPathway analysis for downregulated
genes in the difference between Track (1) and Track (2). |
Table XIII
Pathway analysis for downregulated
genes in the difference between Track (1) and Track (2).
| Category | Count | Genes |
|---|
| MicroRNAs in
cancer | 3 | APC, PLAU,
PTGS2 |
| Pathways in
cancer | 3 | APC, RARB,
PTGS2 |
Notably, after quercetin treatment the colony
stimulating factor 1 (CSF1) gene, an apoptotic gene, decreased from
35.26- to 17.58-fold. In addition, as shown in Table II, another alteration in the
apoptotic pathway that occurred in the present study after
quercetin addition was upregulation of the ATM gene by 4.02-fold as
shown in Table VI.
In the DNA repair pathway, BRCA1 and BRCA2 gene
expression levels were downregulated in (MCF-7/DOX 53.2 nM) cells
by -9.45- and -3.94-fold compared with -2.65 and -4.04-fold in
cells treated with quercetin (Table
III).
Another valuable observations in the present study
was the upregulation of PLAU expression gene in (MCF-7/DOX 53.2 nM)
cells by 44.32-fold and downregulation by 13.7-fold after
combination treatment as shown in Table II. Moreover, SNAI2 expression
decreased by 30.06-fold to -1.17-fold after combination treatment
as shown in Table II.
Discussion
The present study was designed to investigate the
effects of quercetin in reversing the doxorubicin-chemoresistant
phenotype in a MCF-7 breast cancer cell line. First, the
doxorubicin-resistant MCF-7 cell line was established using a
series of stages of increasing doxorubicin concentrations. The
development of resistant cells was closely monitored. Sensitivity
to doxorubicin and the doxorubicin with quercetin combination was
assessed using the MTT assay. The concentration ranges of quercetin
were proven not to be cytotoxic for the cells under investigation.
Annexin V/PI staining demonstrated that the presence of quercetin
drives cells into late apoptosis and necrosis in agreement with the
present findings.
It has been suggested that the reversal property of
quercetin is due to its anti-oxidant effects and its capability to
disrupt mitochondrial membrane potential, leading to the release of
cytochrome C, which is a pro-caspase protein (16). Li et al (28) demonstrated that quercetin has little
effect on cell proliferation at concentrations <0.7 µM; however,
when combined with doxorubicin, the combination leads to
significant inhibition of cell proliferation and invasion and
suppression of the expression of HIF-1α and P-gp. In 2012, Wang
et al (29) also found that
co-treatment with quercetin (20 mM) and doxorubicin (1 mM) leads to
significant potentiation of the antitumor effects of doxorubicin in
human liver cancer cells. Shu et al (30) reported that the combination
treatment of doxorubicin and quercetin leads to a significant
promotion of doxorubicin-induced apoptosis in resistant prostate
cancer cells. Another study by Huang et al (31) showed that quercetin can inhibit oral
cancer cell proliferation through induction of apoptosis and death
in tongue squamous cell carcinoma cells.
Doxorubicin accumulation patterns or accumulation
defects have been described earlier by AbuHammad and Zihlif as
reflecting resistance to doxorubicin (11). It is assumed that doxorubicin
resistance in several P-glycoprotein-positive non-small cell lung
cancer and breast cancer multidrug resistant cell lines can be
explained by a summation of accumulation defect and alterations in
the efficacy of the drug once present in the cell (32). Furthermore, in our previous study
that describes the gene expression changes in MCF-7 cells, a
significant overexpression of p-gp gene was reported (11). The accumulation of doxorubicin in
the vesicles outside the nucleus has been reported by our group
previously (11). Resistant MCF-7
cells were treated with a combination of doxorubicin and verapamil.
Verapamil is a calcium channel inhibitor, and it has been shown to
efficiently suppress the function of P-gp, leading to the reverse
of drug resistance and eventual increase in drug accumulation and
enhanced DNA damage (33-35).
Notably, the impact of quercetin is more obvious in term of
reversing the resistance. Such impact is clear at both the necrosis
and apoptosis levels. Verapamil antiproliferation has not been
performed on wild-type MCF7 cells as it does not have an effect on
these cells by itself nor have an impact on the Doxorubicin
IC50 when there is no doxorubicin resistance (11). Verapamil impact can only be seen in
the resistant MCF7 cells that increased the P-gp expression
(36).
The gene expression investigation in the present
study revealed that quercetin produced a reversal in the
doxorubicin resistance through up- and downregulation of important
genes. Those genes are involved in a number of important cellular
processes, such as cell cycle, apoptotic pathway, DNA-repair and/or
cell migration.
CDKN2A (p16) and CDKN1A are upregulated by 5.21- and
6.11-fold, respectively. The p16 protein binds to two other
proteins, CDK4 and CDK6, which assist in cell cycle regulation.
CDK4 and CDK6 normally stimulate the cell to continue through the
cell cycle and divide; however, binding with p16 blocks cell cycle
progression (37). According to
AbuHammad and Zihlif (11), p16
regulates the G1/S cell cycle transition and has the
effect of causing cell arrest in the G1-phase. In the
present study, after treatment with quercetin, it was found that
CDKN2A (p16) was upregulated by 13.7-fold, which could be due to
the effect of quercetin in reversing doxorubicin resistance. Other
studies have showed that quercetin causes S phase arrest via
decreases in protein expression levels of CDK2 and cyclins A and B
while producing an increase in p53 and p57 protein levels (38-40).
In the present study, the CSF1 gene decreased from
35.26- to 17.58-fold after quercetin treatment, thereby leading to
a reduction in the expression of anti-apoptotic proteins and
rendering cells more sensitive to doxorubicin. In 2016, Zhang et
al (41) revealed that the CSF1
gene, which encodes M-CSF protein, induced significant doxorubicin
resistance of MCF-7 cell via inhibition of apoptosis through
activation of the PI3K/Akt/Survivin pathway.
Another alteration in the apoptotic pathway that
occurred in the present study after quercetin addition was
upregulation of the ATM gene by 4.02-fold, which further stimulated
the p53 pathway. ATM acts as a binary switch by regulating the
capability of p53 to induce cell death after chemotherapy.
AbuHammad and Zihlif (11) revealed
that ATM selectively activates p53, providing a mechanism for
controlling the cell cycle and apoptotic responses. Another study
showed that treatment with quercetin resulted in p53 upregulation,
a finding that was consistent with an earlier report (38).
Since DNA repair mechanisms play an important role
in regulating cellular sensitivity to DNA damaging agents, DNA
repair plays an important role in the development of resistance to
chemotherapy (42).
Breast cancer types 1 (BRCA) and 2 (BRCA2) genes
produce proteins that help repair damaged DNA (43). In the present study, BRCA1 and BRCA2
gene expression levels were downregulated in (MCF-7/DOX 53.2 nM)
cells by -9.45- and -3.94-fold compared with -2.65 and -4.04-fold
in cells treated with quercetin.
Variations in the studies on the role of BRCA1 and
BRCA2 proteins in doxorubicin resistance exists. One study found
that BRCA1-defective breast cancer cells are significantly less
sensitive to doxorubicin when compared with MCF-7 and
MDA-MB231(44), but another study
found that reduced BRCA1 protein expression in ovarian cancer cells
leads to an improvement in survival (45). As a result, downregulation of BRCA1
and BRCA2 gene expression levels seen in resistant MCF-7 cells may
be one of the main changes seen in the resistant phenotype. The
present results showed that quercetin led to an increase in the
expression of BRCA1 gene by 6.8-fold; this result is consistent
with those from a study by Kundur et al in 2019(46), who reported that quercetin and
curcumin lead to a dose-dependent enhancement in BRCA1
expression.
In the present study, SNAI2 expression decreased by
30.06-fold to -1.17-fold after combination treatment. This gene
encodes a member of the Snail family as described by Alves et
al (47). The encoded protein
represents a transcriptional repressor to E-cadherin transcription
in breast carcinoma. It is also involved in epithelial-mesenchymal
transitions and has antiapoptotic activity. Another study by Côme
et al (48) reported that
SNAI2 has been found to be upregulated in all endocrine resistant
cells when compared with parental cell lines. Reduction of SNAI2
expression levels lead to a disabling of cell migration and an
increase in E-cadherin levels in two fulvestrant-resistant breast
cancer cell models, proving the role of SNAI2 in controlling cell
motility and maintenance of a mesenchymal phenotype in resistant
cells (47). Another valuable
observation in the present study was the upregulation of PLAU
expression gene in (MCF-7/DOX 53.2 nM) cells by 44.32-fold and
downregulated to 13.7-fold after combination treatment.
Lin et al (49) showed that downregulation of PLAU
expression can suppresses colorectal cancer development via
inhibition of colorectal cancer cell growth, cell migration and
angiogenesis. Another study by Ai et al in 2020(50) revealed that inhibition of PLAU
expression can repress the migratory and invasive capability of
cervical cancer cells through downregulation of MMP2. This finding
may provide a clue that quercetin can cause reduction in the
metastatic phenotype of MCF-7 resistant cells as published by Hoca
et al (51) who demonstrated
that quercetin can lead to inhibition of EMT in cancer cell lines,
including oral cancer cells, breast cancer stem cells and prostate
cancer cells.
In conclusion, the results of the present study
indicated that quercetin could lead to reversal of breast cancer
cell doxorubicin resistance via downregulation of the expression of
important genes, such as SNAI2, PLAU and CSF1. Such findings may
represent a potential strategy for reversing the chemoresistance of
breast cancer. Notably, the present study needs a further protein
level confirmation for the obtained gene expression changes.
Supplementary Material
Upregulated genes in DOX resistant
cells (MCF-7/DOX53.2nM) treated with 5 μM DOX [Track (1)].
Downregulated genes in DOX resistant
cells (MCF-7/DOX53.2nM) treated with 5 μM DOX [Track (1)].
Upregulated genes in DOX resistant
cells (MCF-7/DOX53.2nM) treated with combination of 5 μM DOX with
20 μM quercetin [Track(2)].
Downregulated genes in DOX resistant
cells (MCF-7/DOX53.2nM) treated with combination of 5 μM DOX with
20 μM quercetin [Track(2)].
Upregulated genes in the difference
between Track(1) and Track (2).
Downregulated genes in the difference
between Track (1) and Track (2).
Acknowledgements
Not applicable.
Funding
Funding: This project received funding from the Deanship of
Scientific Research at the University of Jordan.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MZ and BAA confirm the authenticity of all the raw
data. MZ, RA-D and HZ conceptualized the present study. MZ, BAA and
RA-D designed the methodology. All the authors worked on the
validation. MZ, BAA and RA-D worked on the formal analysis of data.
MZ, BAA and RA-D performed the experiments. All authors worked
together on resources, writing-original draft preparation,
writing-review and editing. MZ, RA-D and HZ supervised the study.
MZ, RA-D and HZ were project administrators. All authors have read
and approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Authors' information
Ms. Hiba Zalloum (ORCID: 0000-0002-5296-2776).
References
|
1
|
Gucalp A, Gupta GP, Pilewskie ML, Sutton
EJ and Norton L: Advances in managing breast cancer: A clinical
update. F1000Prime Rep. 6(66)2014.PubMed/NCBI View
Article : Google Scholar
|
|
2
|
Wilson TR, Longley DB and Johnston PG:
Chemoresistance in solid tumours. Ann Oncol. 17 (Suppl
10):x315–x324. 2006.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Prihantono and Faruk M: Breast
cancer resistance to chemotherapy: When should we suspect it and
how can we prevent it? Ann Med Surg (Lond).
70(102793)2021.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Marquette C and Nabell L:
Chemotherapy-resistant metastatic breast cancer. Curr Treat Options
Oncol. 13:263–275. 2012.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Wang X, Zhang H and Chen X: Drug
resistance and combating drug resistance in cancer. Cancer Drug
Resist. 2:141–160. 2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Moulder S: Intrinsic resistance to
chemotherapy in breast cancer. Womens Health (Lond). 6:821–830.
2010.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Crea F, Nobili S, Paolicchi E, Perrone G,
Napoli C, Landini I, Danesi R and Mini E: Epigenetics and
chemoresistance in colorectal cancer: An opportunity for treatment
tailoring and novel therapeutic strategies. Drug Resist Updat.
14:280–296. 2011.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Barrett-Lee PJ, Dixon JM, Farrell C, Jones
A, Leonard R, Murray N, Palmieri C, Plummer CJ, Stanley A and
Verrill MW: Expert opinion on the use of anthracyclines in patients
with advanced breast cancer at cardiac risk. Ann Oncol. 20:816–827.
2009.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Shi Y, Bieerkehazhi S and Ma H:
Next-generation proteasome inhibitor oprozomib enhances sensitivity
to doxorubicin in triple-negative breast cancer cells. Int J Clin
Exp Pathol. 11:2347–2355. 2018.PubMed/NCBI
|
|
10
|
Smith L, Watson MB, O'Kane SL, Drew PJ,
Lind MJ and Cawkwell L: The analysis of doxorubicin resistance in
human breast cancer cells using antibody microarrays. Mol Cancer
Ther. 5:2115–2120. 2006.PubMed/NCBI View Article : Google Scholar
|
|
11
|
AbuHammad S and Zihlif M: Gene expression
alterations in doxorubicin resistant MCF7 breast cancer cell line.
Genomics. 101:213–220. 2013.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Li X, Lu Y, Liang K, Liu B and Fan Z:
Differential responses to doxorubicin-induced phosphorylation and
activation of Akt in human breast cancer cells. Breast Cancer Res.
7:R589–R597. 2005.PubMed/NCBI View
Article : Google Scholar
|
|
13
|
Lee ER, Kim JY, Kang YJ, Ahn JY, Kim JH,
Kim BW, Choi HY, Jeong MY and Cho SG: Interplay between PI3K/Akt
and MAPK signaling pathways in DNA-damaging drug-induced apoptosis.
Biochim Biophys Acta. 1763:958–968. 2006.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Shukla A, Hillegass JM, MacPherson MB,
Beuschel SL, Vacek PM, Pass HI, Carbone M, Testa JR and Mossman BT:
Blocking of ERK1 and ERK2 sensitizes human mesothelioma cells to
doxorubicin. Mol Cancer. 9(314)2010.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Shi Y, Moon M, Dawood S, McManus B and Liu
PP: Mechanisms and management of doxorubicin cardiotoxicity. Herz.
36:296–305. 2011.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Elmadany N, Khalil E, Vaccari L, Birarda
G, Yousef I and Abu-Dahab R: Antiproliferative activity of the
combination of doxorubicin/quercetin on MCF7 breast cancer cell
line: A combined study using colorimetric assay and synchrotron
infrared microspectroscopy. Infrared Phys Technol. 95:141–147.
2018.
|
|
17
|
Yang F, Teves SS, Kemp CJ and Henikoff S:
Doxorubicin, DNA torsion, and chromatin dynamics. Biochim Biophys
Acta. 1845:84–89. 2014.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Barbeau D, Persoons R, Marques M, Hervé C,
Laffitte-Rigaud G and Maitre A: Relevance of urinary
3-hydroxybenzo(a)pyrene and 1-hydroxypyrene to assess exposure to
carcinogenic polycyclic aromatic hydrocarbon mixtures in metallurgy
workers. Ann Occup Hyg. 58:579–590. 2014.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Huang ZP, Liu XJ, Zou BX, Wang LG and Zhou
T: The complete recanalization of PICC-related venous thrombosis in
cancer patients: A series of case reports. Exp Ther Med. 6:411–412.
2013.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Hashemzaei M, Delarami Far A, Yari A,
Heravi RE, Tabrizian K, Taghdisi SM, Sadegh SE, Tsarouhas K,
Kouretas D, Tzanakakis G, et al: Anticancer and apoptosis-inducing
effects of quercetin in vitro and in vivo. Oncol Rep.
38:819–828. 2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Slighoua M, Amrati FE, Chebaibi M, Mahdi
I, Al Kamaly O, El Ouahdani K, Drioiche A, Saleh A and Bousta D:
Quercetin and ferulic acid elicit estrogenic activities in vivo and
in silico. Molecules. 28(5112)2023.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Notas G, Nifli AP, Kampa M, Pelekanou V,
Alexaki VI, Theodoropoulos P, Vercauteren J and Castanas E:
Quercetin accumulates in nuclear structures and triggers specific
gene expression in epithelial cells. J Nutr Biochem. 23:656–666.
2012.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Liu Y, Liu C, Tang C and Yin C: Dual
stimulus-responsive chitosan-based nanoparticles co-delivering
doxorubicin and quercetin for cancer therapy. Mater Lett.
305(130826)2021.
|
|
24
|
Henidi HA, Al-Abbasi FA, El-Moselhy MA,
El-Bassossy HM and Al-Abd AM: Despite blocking doxorubicin-induced
vascular damage, quercetin ameliorates its antibreast cancer
activity. Oxid Med Cell Longev. 2020(8157640)2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Chen JY, Hu RY and Chou HC:
Quercetin-induced cardioprotection against doxorubicin
cytotoxicity. J Biomed Sci. 20(95)2013.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Li S, Yuan S, Zhao Q, Wang B, Wang X and
Li K: Quercetin enhances chemotherapeutic effect of doxorubicin
against human breast cancer cells while reducing toxic side effects
of it. Biomed Pharmacother. 100:441–447. 2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Li SZ, Li K, Zhang JH and Dong Z: The
effect of quercetin on doxorubicin cytotoxicity in human breast
cancer cells. Anticancer Agents Med Chem. 13:352–355.
2013.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Wang G, Zhang J, Liu L, Sharma S and Dong
Q: Quercetin potentiates doxorubicin mediated antitumor effects
against liver cancer through p53/Bcl-xl. PLoS One.
7(e51764)2012.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Shu Y, Xie B, Liang Z and Chen J:
Quercetin reverses the doxorubicin resistance of prostate cancer
cells by downregulating the expression of c-met. Oncol Lett.
15:2252–2258. 2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Huang CF, Liu SH, Ho TJ, Lee KI, Fang KM,
Lo WC, Liu JM, Wu CC and Su CC: Quercetin induces tongue squamous
cell carcinoma cell apoptosis via the JNK activation-regulated
ERK/GSK-3α/β-mediated mitochondria-dependent apoptotic signaling
pathway. Oncol Lett. 23(78)2022.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Schuurhuis GJ, Van Heijningen TH,
Cervantes A, Pinedo HM, de Lange JH, Keizer HG, Broxterman HJ, Baak
JP and Lankelma J: Changes in subcellular doxorubicin distribution
and cellular accumulation alone can largely account for doxorubicin
resistance in SW-1573 lung cancer and MCF-7 breast cancer multidrug
resistant tumour cells. Br J Cancer. 68:898–908. 1993.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Wang X, Li Y, Fan GF, Zhang TY, Sun B and
Fan PS: Effect of verapamil in the reversal of doxorubicin
chemotherapy resistance in advanced gastric cancer. Eur Rev Med
Pharmacol Sci. 24:7753–7763. 2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Rogan AM, Hamilton TC, Young RC, Klecker
RW Jr and Ozols RF: Reversal of adriamycin resistance by verapamil
in human ovarian cancer. Science. 224:994–996. 1984.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Bellamy WT, Dalton WS, Kailey JM, Gleason
MC, McCloskey TM, Dorr RT and Alberts DS: Verapamil reversal of
doxorubicin resistance in multidrug-resistant human myeloma cells
and association with drug accumulation and DNA damage. Cancer Res.
48:6365–6370. 1988.PubMed/NCBI
|
|
36
|
AlQudah DA, Zihlif MA and Taha MO:
Ligand-based modeling of diverse aryalkylamines yields new potent
P-glycoprotein inhibitors. Eur J Med Chem. 110:204–223.
2016.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Li J, Poi MJ and Tsai MD: Regulatory
mechanisms of tumor suppressor P16(INK4A) and their relevance to
cancer. Biochemistry. 50:5566–5582. 2011.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Srivastava S, Somasagara RR, Hegde M,
Nishana M, Tadi SK, Srivastava M, Choudhary B and Raghavan SC:
Quercetin, a natural flavonoid interacts with DNA, arrests cell
cycle and causes tumor regression by activating mitochondrial
pathway of apoptosis. Sci Rep. 6(24049)2016.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Suh DK, Lee EJ, Kim HC and Kim JH:
Induction of G(1)/S phase arrest and apoptosis by quercetin in
human osteosarcoma cells. Arch Pharm Res. 33:781–785.
2010.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Choi JA, Kim JY, Lee JY, Kang CM, Kwon HJ,
Yoo YD, Kim TW, Lee YS and Lee SJ: Induction of cell cycle arrest
and apoptosis in human breast cancer cells by quercetin. Int J
Oncol. 19:837–844. 2001.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Zhang M, Zhang H, Tang F, Wang Y, Mo Z,
Lei X and Tang S: Doxorubicin resistance mediated by cytoplasmic
macrophage colony-stimulating factor is associated with switch from
apoptosis to autophagic cell death in MCF-7 breast cancer cells.
Exp Biol Med (Maywood). 241:2086–2093. 2016.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Torgovnick A and Schumacher B: DNA repair
mechanisms in cancer development and therapy. Front Genet.
6(157)2015.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Roy R, Chun J and Powell SN: BRCA1 and
BRCA2: Different roles in a common pathway of genome protection.
Nat Rev Cancer. 12:68–78. 2011.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Tassone P, Tagliaferri P, Perricelli A,
Blotta S, Quaresima B, Martelli ML, Goel A, Barbieri V, Costanzo F,
Boland CR and Venuta S: BRCA1 expression modulates chemosensitivity
of BRCA1-defective HCC1937 human breast cancer cells. Br J Cancer.
88:1285–1291. 2003.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Quinn JE, James CR, Stewart GE, Mulligan
JM, White P, Chang GK, Mullan PB, Johnston PG, Wilson RH and Harkin
DP: BRCA1 mRNA expression levels predict for overall survival in
ovarian cancer after chemotherapy. Clin Cancer Res. 13:7413–7420.
2007.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Kundur S, Prayag A, Selvakumar P, Nguyen
H, McKee L, Cruz C, Srinivasan A, Shoyele S and Lakshmikuttyamma A:
Synergistic anticancer action of quercetin and curcumin against
triple-negative breast cancer cell lines. J Cell Physiol.
234:11103–11118. 2019.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Alves CL, Elias D, Lyng MB, Bak M and
Ditzel HJ: SNAI2 upregulation is associated with an aggressive
phenotype in fulvestrant-resistant breast cancer cells and is an
indicator of poor response to endocrine therapy in estrogen
receptor-positive metastatic breast cancer. Breast Cancer Res.
20(60)2018.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Côme C, Magnino F, Bibeau F, De Santa
Barbara P, Becker KF, Theillet C and Savagner P: Snail and slug
play distinct roles during breast carcinoma progression. Clin
Cancer Res. 12:5395–5402. 2006.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Lin M, Zhang Z, Gao M, Yu H, Sheng H and
Huang J: MicroRNA-193a-3p suppresses the colorectal cancer cell
proliferation and progression through downregulating the PLAU
expression. Cancer Manag Res. 11:5353–5363. 2019.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Ai C, Zhang J, Lian S, Ma J, Győrffy B,
Qian Z, Han Y and Feng Q: FOXM1 functions collaboratively with PLAU
to promote gastric cancer progression. J Cancer. 11:788–794.
2020.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Hoca M, Becer E, Kabadayı H, Yücecan S and
Vatansever HS: The effect of resveratrol and quercetin on
epithelial-mesenchymal transition in pancreatic cancer stem cell.
Nutr Cancer. 72:1231–1242. 2020.PubMed/NCBI View Article : Google Scholar
|