Introduction
Prenatal testing is an integral part of daily
obstetric practice in most developed countries (1). The current methods include
non-invasive prenatal screening, recommended in all pregnancies and
prenatal diagnosis, which can include invasive analysis of fetal
material with minimal risk of miscarriage (2). Based on this evidence, novel
approaches and non-invasive methods of obtaining fetal material are
systematically sought, which can result in the identification of
predictive or diagnostic indicators for the detection or prediction
of chromosomal abnormalities, genetic diseases, or pathological
conditions of pregnancy that lead to adverse events of the
fetus/newborn or the pregnant mother (3). In recent years, the application of
non-invasive prenatal testing (NIPT) is based on the isolation of
cell-free fetal DNA (cffDNA) from maternal blood samples (4). Potential sources of cffDNA include the
fetal nucleated red blood cells that undergo apoptosis in maternal
circulation, but the most likely source of origin is the placenta
(4,5). cffDNA is used as a diagnostic or
predictive biomarker and its application has attracted considerable
research interest (5-10).
Several studies that examined the application of
NIPT indicated that the number of embryonic cells and the cffDNA
concentration changed not only with gestational age, but also in
the presence of various pregnancy complications, such as preterm
birth, idiopathic hydramnios, placenta previa, intrauterine growth
retardation (IUGR), vaginal bleeding, threatened miscarriage and
recurrent pregnancy loss (11-19).
Moreover, a well-documented association has been observed between
altered concentration of cffDNA and preeclampsia (8,20,21)
haemolysis, elevated liver enzyme activity levels and low platelet
count syndrome (HELLP), or eclampsia (22,23).
Previous studies that examined the application of cffDNA in
pregnancy complications are controversial with regard to the
successful use of this marker in predicting adverse pregnancy
outcomes (24).
The present prospective study aimed to investigate
the association between the levels of cffDNA in the serum of
pregnant women and the occurrence of pregnancy complications. In
addition, the potential application of the predictive value of
cffDNA was explored. Pregnant women were monitored at the
Department of Obstetrics and Gynecology University Hospital of
Heraklion, Crete, particularly at the Human Reproduction Unit, the
Embryo Medicine Unit and the Outpatient unit of the Obstetrics
Clinic.
Patients and methods
Peripheral blood samples were obtained at 11-13
weeks of pregnancy from all the participants at the Department of
Obstetrics and Gynecology, University Hospital of Heraklion, Crete,
Greece. During the first trimester, a routine screening test was
used to assess aneuploidy by the use of biochemical markers
[β-human chorionic gonadotropin (β-hCG) and pregnancy associated
plasma protein-A (PAPP-A)] and sonographic markers, such as nuchal
translucency (NT) thickness, nasal bone presence, ductus venosus
blood flow and examination for tricuspid regurgitation.
The study group comprised pregnant women aged 19 to
41 years old with an age range of 22 years who presented with
pregnancy complications, such as preterm birth, idiopathic
hydramnios, placenta previa, IUGR, vaginal bleeding, threatened
miscarriage, preeclampsia/eclampsia and HELLP syndrome eclampsia.
The control group comprised women who did not present with any of
the aforementioned complications and had a normal pregnancy course
and an optimal perinatal outcome. Recruitment and sample collection
started on November, 2014 and was completed October, 2016.
The samples were collected in tubes with EDTA and
stored at -80˚C. In addition, data were collected regarding the
somatometric and demographic characteristics of pregnant women, the
individual and medical history, as well as the previous obstetric
history. In pregnancies, the following parameters were recorded: i)
The parameters of fetal development as well as other elements of
the course of pregnancy, including the presentation of any
complications (aforementioned) and the perinatal outcome; ii) the
parameters of childbirth, such as birth weight at birth, gestation
week at delivery, mode of birth and condition of the newborn and
iii) the clinical, sonographic and laboratory parameters used to
assess the pregnancy course and those that are indicative or
diagnostic for the pathological function of the placenta (Doppler
of uterine and fetal vessels), the cardiotocograph, the biophysical
activities of the fetus, the amount of amniotic fluid and specific
biochemical indicators.
Sequencing and analysis of the samples was performed
by Natera, Inc. Cell-free DNA was amplified, sequenced, and
analyzed using a custom Natera collection kit. The test contains an
SNP algorithm to determine fetal fraction and ploidy status. Fetal
fraction was measured using an SNP-based cfDNA prenatal test
(PanoramaTM; Natera, Inc.) as previously described
(25-27).
In brief, 13,926 SNPs in maternal plasma cfDNA were amplified and
sequenced. Custom QC metrics based on the sequencing data were used
and 50-bp single-end reads were used. A maximum likelihood method
which differentiates maternal from fetal alleles was used to
determine the presence or absence of fetal aneuploidy, and
simultaneously return a fetal fraction measurement. NextSeq™
500/550 High Output Kit v2.5 (75 cycles; cat. no. 20024911;
Illumina, Inc.) was used as sequencing kit. A custom data pipeline
and algorithms were used to analyze the data (PanoramaTM; Natera,
Inc.) (25-27)
In the present study, a total of 2x10 ml (a single
draw) of blood was used from each subject for analysis. An average
of 1.22x107 reads were mapped for each sample when
sequenced at normal depth-of-read. Samples not generating
sufficient information were resequenced at a higher depth of read
(average, 2.45x107 mapped reads per sample). The fetal
fraction was reported as a percentage of the identified fetal
(placental) DNA to the total cell-free DNA present in maternal
blood.
The research protocol was approved (approval no.
882, 20/29-10-1014) by the Ethics and Bioethics Scientific
Committee of University Hospital of Heraklion-PAGNI (Heraklion,
Greece).
Statistical analysis
Statistical analysis was performed with Statistical
Package for the Social Sciences (SPSS) v.26 (IBM Corp.) using
non-parametric methods. The parameters were normalized by the
multiples of median (MoM) method (28,29),
which is the standard for reporting serum maternity test results
(5). Shapiro-Wilk normality test
was performed and the data indicated that the demographic
parameters did not follow a normal distribution. Non-parametric
tests, such as χ2, Mann-Whitney U and Kruskal-Wallis
were used. The correlations between the demographic factors and the
measurements of the parameters examined were investigated using
Spearman correlation analysis. The correlation coefficient was
considered to be weak (<0.4), moderate (0.5-0.7) and strong
(>0.7). Statistical significance was defined as a value of
P<0.01.
Results
Statistical analysis was performed in 494 out of the
500 samples collected from pregnant women. The demographics for all
available data are presented in Table
I. A total of 248 out of 494 (50.2%) neonates were male with an
average weight of 3,308 g, while 242 were females (49%) with an
average weight of 3,135 g. The average difference in weight was
significantly different, with male infants being heavier by 173 g
than females (P<0.001).
 | Table IDemographics for available data of
enrolled pregnancies. |
Table I
Demographics for available data of
enrolled pregnancies.
| Parameters | | Ν | N% | χ2 |
|---|
| Delivery type | Normal | 461 | 93.3 | P<0.001 |
| | C-section | 31 | 6.3 | |
| Delivery week | 33 | 2 | 0.4 | P<0.001 |
| | 36 | 114 | 23.1 | |
| | 37 | 124 | 25.1 | |
| | 38 | 122 | 24.7 | |
| | 39 | 113 | 22.9 | |
| | 40 | 16 | 3.2 | |
| Induced | Yes | 28 | 5.7 | P<0.001 |
| | No | 424 | 85.8 | |
| Sex | Male | 248 | 50.2 | P=0.786 |
| | Female | 242 | 49.0 | |
| ICU | No | 486 | 98.4 | P<0.001 |
| | Yes | 8 | 1.6 | |
| Conception | Automatic
conception | 471 | 95.3 | P<0.001 |
| | IVF | 21 | 4.3 | |
A total of 461 newborns were delivered naturally and
31 by cesarean section, while induction of delivery was performed
in 28 neonates (Table I). Only 8
infants were placed in the intensive care unit (ICU) (1.6%),
whereas conception was performed via in vitro fertilization
(IVF) for 21 cases (4.3%). The fetal development parameters
(biometrics) and the data on the course of pregnancy (amniotic
fluid volume and fetal biophysical activities) were monitored. A
total of 459 out of 494 infants (92.9%) did not present with a
pathological condition (Table II).
The demographic information of the relevant pathological
pregnancies is listed in Table
II.
 | Table IIPathological pregnancy conditions and
demographics. |
Table II
Pathological pregnancy conditions and
demographics.
| |
Pathological conditions |
|---|
| | Normal | Preeclampsia | Gestational
hypertension | Bleeding-placental
abruption | IUGR | Gestational
diabetes with with diet | Gestational
diabetes with insulin | Preterm
delivery |
Oligohydramnios | Polyhydramnios |
|---|
| Parameters | | N | % | N | % | N | % | N | % | N | % | N | % | N | % | N | % | N | % | N | % |
|---|
| Delivery | Νormal | 437 | 95.2 | 11 | 78.6 | 2 | 50 | 3 | 75 | 0 | 0 | 3 | 100 | 0 | 0 | 1 | 100 | 2 | 40 | 2 | 100 |
| | C-section | 21 | 4.6 | 2 | 14.3 | 2 | 50 | 1 | 25 | 1 | 100 | 0 | 0 | 1 | 100 | 0 | 0 | 3 | 60 | 0 | 0 |
| Delivery | 33 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 100 | 0 | 0 | 0 | 0 | 1 | 100 | 0 | 0 | 0 | 0 |
| week | 36 | 112 | 24.4 | 2 | 14.3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| | 37 | 122 | 26.6 | 2 | 14.3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| | 38 | 112 | 24.4 | 2 | 14.3 | 2 | 50 | 1 | 25 | 0 | 0 | 1 | 33.3 | 0 | 0 | 0 | 0 | 2 | 40 | 2 | 100 |
| | 39 | 98 | 21.4 | 8 | 57.1 | 2 | 50 | 0 | 0 | 0 | 0 | 1 | 33.3 | 1 | 100 | 0 | 0 | 3 | 60 | 0 | 0 |
| | 40 | 12 | 2.6 | 0 | 0 | 0 | 0 | 3 | 75 | 0 | 0 | 1 | 33.3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Induced | Pre-term | 19 | 4.1 | 3 | 21.4 | 1 | 25 | 3 | 75 | 0 | 0 | 1 | 33.3 | 0 | 0 | 0 | 0 | 1 | 20 | 0 | 0 |
| | Normal | 408 | 88.9 | 9 | 64.3 | 1 | 25 | 0 | 0 | 0 | 0 | 2 | 66.7 | 0 | 0 | 1 | 100 | 1 | 20 | 2 | 100 |
| Sex | Male | 232 | 50.5 | 8 | 57.1 | 2 | 50 | 1 | 25 | 1 | 100 | 1 | 33.3 | 0 | 0 | 1 | 100 | 2 | 40 | 0 | 0 |
| | Female | 223 | 48.6 | 6 | 42.9 | 2 | 50 | 3 | 75 | 0 | 0 | 2 | 66.7 | 1 | 100 | 0 | 0 | 3 | 60 | 2 | 100 |
| ICU | No | 458 | 99.8 | 11 | 78.6 | 4 | 100 | 3 | 75 | 0 | 0 | 3 | 100 | 1 | 100 | 0 | 0 | 4 | 80 | 2 | 100 |
| | Yes | 1 | 0.2 | 3 | 21.4 | 0 | 0 | 1 | 25 | 1 | 100 | 0 | 0 | 0 | 0 | 1 | 100 | 1 | 20 | 0 | 0 |
| Conception | Natural
conception | 443 | 96.5 | 12 | 85.7 | 4 | 100 | 3 | 75 | 0 | 0 | 3 | 100 | 0 | 0 | 1 | 100 | 3 | 60 | 2 | 100 |
| | IVF | 15 | 3.3 | 2 | 14.3 | 0 | 0 | 1 | 25 | 1 | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 40 | 0 | 0 |
The pathological complications were recorded in 14
out of 494 (2.8%) of the fetuses, where preeclampsia was noted in
their corresponding mothers. In addition, a total of 4 pregnant
women suffered from pre-hypertension and placental
abruption/bleeding, whereas oligohydramnios was present in 5 and
polyhydramnios in 2 pregnant women. Gestational diabetes was
present in 4 women, of whom 3 were treated with diet restrictions
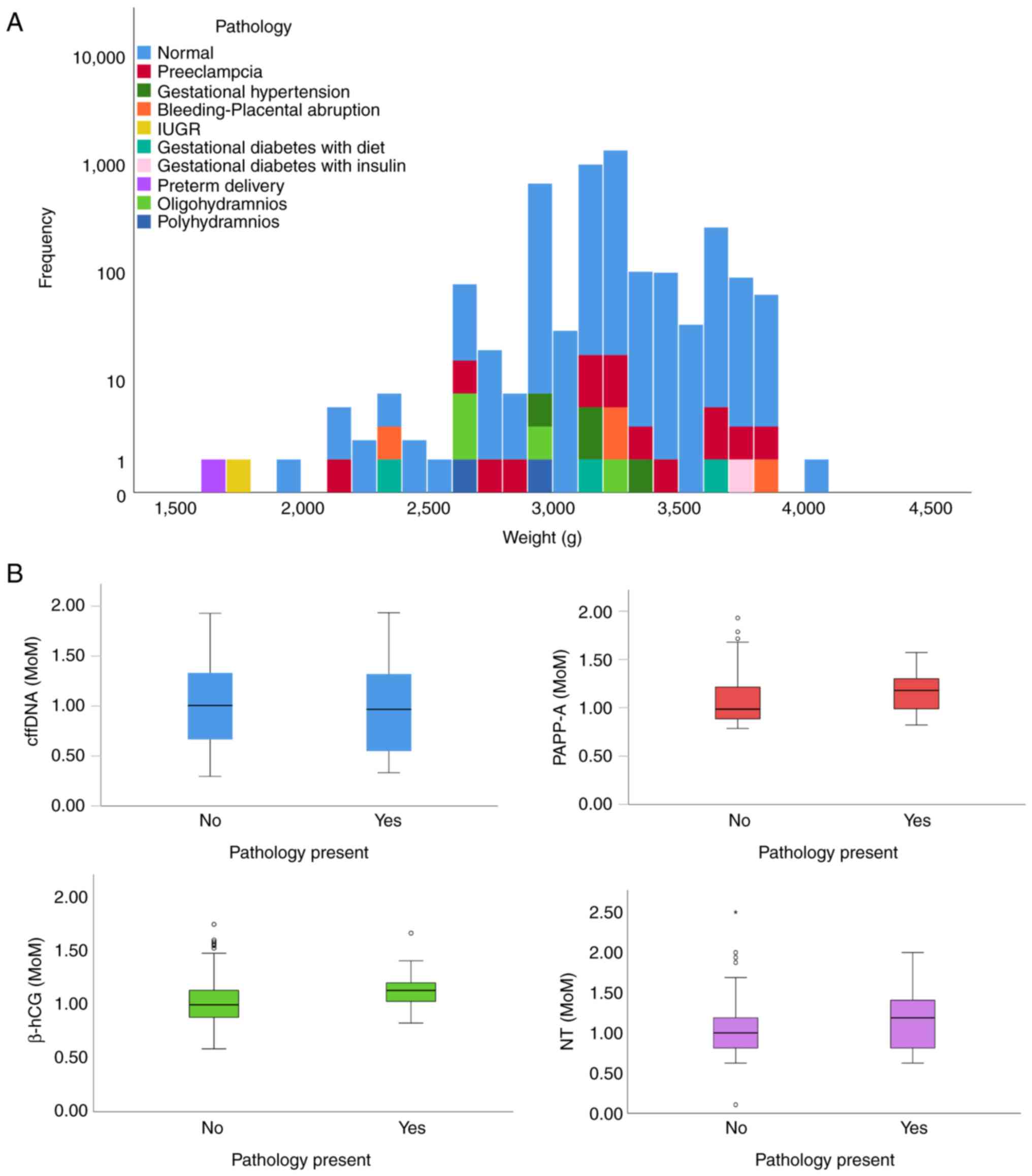
and 1 with insulin. The histogram of weight distribution was
prepared according to the pathological conditions present (Fig. 1A). No significant differences were
noted following comparison of the sex with the pathological or
demographical parameters.
‘MoM’ is a measure of the deviation of an individual
result from the median and is commonly used to report the results
of medical screening tests, particularly where the results of
individual tests are highly variable. The mean MoM values of
PAPP-A, β-hCG and cervical transparency measurement were used to
perform comparisons between samples derived from pregnancies with
pathological features in the fetus and those derived from normal
pregnancies. The MoM distributions for PAPP-A and β-hCG, NT and
cffDNA between the two groups of pregnancies are revealed in
Fig. 1B. The expression levels of
PAPP-A, β-hCG and the NT mean MoM values were significantly
different between these two groups (P=0.005, P<0.001 and
P=0.007, respectively). However, the expression levels of cffDNA
and the mean MoM values were not significantly different between
these two groups (P=0.687).
By comparing all pathological conditions
individually, the data indicated that β-hCG levels and NT mean MoM
values were significantly different despite the limited number of
samples used. In contrast to these findings, the levels of cffDNA
and the mean MoM values did not demonstrate a significant
difference in IUGR and gestational diabetes cases, as well as in
preeclampsia and preterm delivery cases, although their values were
somewhat different. The MoM values for all the biochemical markers
investigated according to each pathological condition are revealed
in Table III.
 | Table IIIMoM values for the cffDNA, the
biochemical markers (PAPP-A and β-hCG) and NT, investigated
according to each pathological condition. |
Table III
MoM values for the cffDNA, the
biochemical markers (PAPP-A and β-hCG) and NT, investigated
according to each pathological condition.
| |
Pathological conditions |
|---|
| MoM | | Normal | Preeclampsia | Gestational
hypertension | Bleeding-placental
abruption | IUGR | Gestational
diabetes with diet | Gestational
diabetes with insulin | Preterm
delivery |
Oligohydramnios | Polyhydramnios | Kruskal Wallis
test |
|---|
| cffDNA | Ν | 459 | 14 | 4 | 4 | 1 | 3 | 1 | 1 | 5 | 2 | 0.293 |
| | Mean | 1.01 | 0.88 | 1.08 | 0.94 | 0.40 | 1.20 | 1.79 | 0.60 | 1.00 | 1.47 | |
| | SD | 0.42 | 0.42 | 0.34 | 0.37 | 0 | 0.77 | 0 | 0 | 0.44 | 0.20 | |
| PAPP-A | Ν | 459 | 14 | 4 | 4 | 1 | 3 | 1 | 1 | 5 | 2 | 0.06 |
| | Mean | 1.07 | 1.14 | 1.24 | 1.14 | 0.87 | 1.44 | 1.37 | 0.89 | 1.10 | 1.24 | |
| | SD | 0.23 | 0.23 | 0.22 | 0.21 | 0 | 0.18 | 0 | 0 | 0.16 | 0.10 | |
| β-hCG | Ν | 459 | 14 | 4 | 4 | 1 | 3 | 1 | 1 | 5 | 2 | 0.032 |
| | Mean | 1.03 | 1.04 | 1.15 | 1.15 | 1.08 | 1.23 | 1.24 | 1.11 | 1.18 | 1.41 | |
| | SD | 0.20 | 0.15 | 0.17 | 0.13 | 0 | 0.16 | 0 | 0 | 0.10 | 0.36 | |
| NT | Ν | 459 | 14 | 4 | 4 | 1 | 3 | 1 | 1 | 5 | 2 | 0.015 |
| | Mean | 1.01 | 1.07 | 1.61 | 1.30 | 0.69 | 1.17 | 1.88 | 1.06 | 1.30 | 1.06 | |
| | SD | 0.26 | 0.34 | 0.31 | 0.56 | 0 | 0.61 | 0 | 0 | 0.37 | 0.35 | |
Discussion
cffDNA can serve as a pathological marker or be used
to provide genetic material for personalized medicine (30). The utilization of cffDNA, which is
present in the blood circulation of pregnant women (5), has modernized prenatal care for
genetic disorders and aneuploidies (31,32);
cffDNA has also been used for >20 years for fetal blood group
prediction (9,33). It is considered that non-invasive
prenatal diagnosis using fetal DNA in maternal blood may play an
increasingly important role in the future practice of prenatal
testing. However, it is important to address the ethical, legal and
social issues regarding this application. The advantage of
non-invasive testing compared with invasive testing is to avoid
harming the fetus. However, this method offers limited precision,
compared with that of specific diagnostic tests, such as chorionic
villus sampling or amniocentesis (10,34).
In addition, the false-positives and false-negatives in NIPT,
possibly related to the placental origin of fetal DNA, remain an
important issue to be addressed (35). Moreover, this method of testing does
not offer additional genetic information (10,34).
The main objective of the present study was to
investigate the predictive value of the increased levels of cffDNA
in the maternal circulation during the first trimester of pregnancy
with the subsequent onset of a number of serious pathological
conditions of pregnancy.
The findings indicated a lower C-section rate of
pregnant women, which was in stark contrast with the results
reported by Antoniou et al and the cited WHO report
published in 2020 (36,37). This further illustrated the
requirement for a more formal information source pertinent to WHO,
which can provide official data for C-sections in an effort to
limit unwarranted use. The mean MoM values of PAPP-A, β-hCG and NT,
reported in the present study, were affected in samples derived
from cases that presented with pathological conditions, which is in
agreement with previous studies (38-44).
By contrast, the mean MoM values of cffDNA were somewhat different
between the IUGR and the gestational diabetes groups, as well as
between the preeclampsia and the preterm delivery groups (14,45-48);
however, these differences did not reach statistical significance
in the present study. cffDNA levels are increased throughout the
course of normal pregnancy as well as in certain pathological
conditions (6,49) making it hard to measure incremental
differences. Cell-free DNA is present in healthy individuals. The
‘pathogenic’ value cut-offs cannot be easily distinguished from the
corresponding normal value cut-offs. Specific values of 6 (low
range) and 650 (high range) ng/ml have been measured in healthy
men, indicating the potential weaknesses of the quantification
methods used (50-52).
A review article (12) that summarized previous findings on
preterm birth and other adverse pregnancy outcomes was inconclusive
as to the role of cffDNA in preterm births; this report described
the technical and standardization issues between the pertinent
studies that examined the role of cffDNA in preterm birth and
highlighted the requirement to establish a normal range cut-off
related to cffDNA levels. In addition, two different systematic
reviews have been published on the use of cffDNA and the prediction
of preeclampsia (53,54). These studies concluded that cffDNA
is indeed a marker that can be used from the beginning of the
second trimester and onwards; after this period, its predictive
value is reduced. The studies also proposed the limitation of the
heterogeneity of the published data regarding cffDNA levels and the
challenges encountered during the interpretation of the findings. A
large study (55) of 1,949
singleton pregnancies concluded that cffDNA concentration levels
were variable and that maternal weight was affecting cffDNA MoM
values; however, it was not significantly altered in pregnancies
with pathological findings, such as preeclampsia.
In the present study, cffDNA was unaffected by
weight or any of the related factors. Thurik et al performed
a nested first-trimester case-control study investigating
preeclampsia, hypertension, gestational diabetes and preterm birth
(48). This study converted
first-trimester cffDNA values to MoM values, failing to present
predictive values for preeclampsia. Based on this evidence, it is
unknown whether cffDNA is actually a valid marker for the
identification of pathologies during pregnancies (24). Notably, for preeclampsia, it may not
provide added value to the existing screening methods (8,56);
this conclusion has also been claimed for preterm births (57). The latest review by Merriel et
al (24) is sceptical regarding
the use of cffDNA as a pathological marker since conflicting
results are presented in the reviewed studies. The authors of this
study also highlighted the lack of common guidelines, biochemical
tests and units, and the requirement for a normal concentration
range and specific time period of sample collection as factors that
pose serious challenges in the interpretation of the results.
Merriel et al (24) could
not identify a role for the use of cffDNA in clinical NIPT testing
for high-risk pregnancies. The present study reports similar
conclusions. A previous review (20) provided evidence and evaluated the
total cell-free DNA as a more appropriate alternative index,
especially for preeclampsia. Total cell-free DNA comprises
placental cffDNA and maternal cell-free DNA from maternal
leukocytes.
The evidence for the use of cffDNA in predicting
adverse pregnancy outcomes is controversial. However, this research
is vital for developing a better understanding of disease
processes. Currently cffDNA testing does not have any clinical
application for the prediction of pregnancy complications and
additional development is required for possible use in clinical
practice. Large-scale studies that will investigate the possible
alterations in cffDNA, in the process of the pregnancies that
exhibit severe complications, are required. Despite the small
number of cases, the present study revealed no alterations in the
first trimester; however, it would be interesting to address
whether the cffDNA is altered at the second or the third trimester
of complicated pregnancies, and, whether it may have any clinical
usefulness as a diagnostic or a predictive marker. The current use
of NIPT in prenatal diagnosis can be potentially added to more
novel technologies of personalized medicine, such as
next-generation sequencing and chromosomal microarray analysis
(58) if they can be applied in a
non-invasive manner.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets analyzed and/or generated during the
present study are available from the corresponding author on
reasonable request.
Authors' contributions
ZK, SS, EM and KR conceived and designed the study.
ZK, EP, EM, AP and SS acquired, analyzed and interpreted the data.
ZK, SS and IP confirmed the authenticity of the raw data. ZK, EP,
IP and SS drafted the manuscript. All authors critically reviewed
the manuscript for important intellectual content. All authors read
and approved the final version of the manuscript.
Ethics approval and consent to
participate
The present study protocol conformed to the globally
accepted regulations on clinical studies involving human data and
approval was conferred by the Ethics and Bioethics Scientific
Committee of the University Hospital of Heraklion-PAGNI (Crete,
Greece) (approval no. 822, 20/29-10-2014; Heraklion, Greece).
Written informed consent was obtained from all of the
participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Carlson LM and Vora NL: Prenatal
diagnosis: Screening and diagnostic tools. Obstet Gynecol Clin
North Am. 44:245–256. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
mLevy B and Stosic M: Traditional prenatal
diagnosis: Past to present. Methods Mol Biol. 1885:3–22.
2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Carbone L, Cariati F, Sarno L, Conforti A,
Bagnulo F, Strina I, Pastore L, Maruotti GM and Alviggi C:
Non-invasive prenatal testing: Current perspectives and future
challenges. Genes (Basel). 12(15)2021.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Sifakis S, Papantoniou N, Kappou D and
Antsaklis A: Noninvasive prenatal diagnosis of Down syndrome:
Current knowledge and novel insights. J Perinat Med. 40:319–327.
2012.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Lo YD, Corbetta N, Chamberlain PF, Rai V,
Sargent IL, Redman CW and Wainscoat JS: Presence of fetal DNA in
maternal plasma and serum. Lancet. 350:485–487. 1997.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Lo YD, Zhang J, Leung TN, Lau TK, Chang AM
and Hjelm NM: Rapid clearance of fetal DNA from maternal plasma. Am
J Hum Genet. 64:218–224. 1999.PubMed/NCBI View
Article : Google Scholar
|
|
7
|
Fan HC, Blumenfeld YJ, Chitkara U, Hudgins
L and Quake SR: Noninvasive diagnosis of fetal aneuploidy by
shotgun sequencing DNA from maternal blood. Proc Natl Acad Sci USA.
105:16266–16271. 2008.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Sifakis S, Zaravinos A, Maiz N, Spandidos
DA and Nicolaides KH: First-trimester maternal plasma cell-free
fetal DNA and preeclampsia. Am J Obstet Gynecol. 201:472.e1–e7.
2009.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Sifakis S, Koukou Z and Spandidos DA:
Cell-free fetal DNA and pregnancy-related complications (review).
Mol Med Rep. 11:2367–2372. 2015.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Liehr T, Harutyunyan T, Williams H and
Weise A: Non-invasive prenatal testing in Germany. Diagnostics
(Basel). 12(2816)2022.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Jakobsen TR, Clausen FB, Rode L, Dziegiel
MH and Tabor A: High levels of fetal DNA are associated with
increased risk of spontaneous preterm delivery. Prenat Diagn.
32:840–845. 2012.PubMed/NCBI View
Article : Google Scholar
|
|
12
|
van Boeckel SR, Davidson DJ, Norman JE and
Stock SJ: Cell-free fetal DNA and spontaneous preterm birth.
Reproduction. 155:R137–R145. 2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Sugito Y, Sekizawa A, Farina A, Yukimoto
Y, Saito H, Iwasaki M, Rizzo N and Okai T: Relationship between
severity of hyperemesis gravidarum and fetal DNA concentration in
maternal plasma. Clin Chem. 49:1667–1669. 2003.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Alberry MS, Maddocks DG, Hadi MA, Metawi
H, Hunt LP, Abdel-Fattah SA, Avent ND and Soothill PW:
Quantification of cell free fetal DNA in maternal plasma in normal
pregnancies and in pregnancies with placental dysfunction. Am J
Obstet Gynecol. 200:98.e91–e6. 2009.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Jimbo M, Sekizawa A, Sugito Y, Matsuoka R,
Ichizuka K, Saito H and Okai T: Placenta increta: Postpartum
monitoring of plasma cell-free fetal DNA. Clin Chem. 49:1540–1541.
2003.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Seval MM, Karabulut HG, Tükün A and Koç A:
Cell free fetal DNA in the plasma of pregnant women with
preeclampsia. Clin Exp Obstet Gynecol. 42:787–791. 2025.PubMed/NCBI
|
|
17
|
Yin A, Ng EH, Zhang X, He Y, Wu J and
Leung KY: Correlation of maternal plasma total cell-free DNA and
fetal DNA levels with short term outcome of first-trimester vaginal
bleeding. Hum Reprod. 22:1736–1743. 2007.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Sapantzoglou I, Gallardo Arozena M, Dragoi
V, Akolekar R, Nicolaides KH and Syngelaki A: Fetal fraction of
cell free DNA in screening for hypertensive disorders at 11-13
weeks. J Matern Fetal Neonatal Med. 35:5363–5368. 2022.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Wataganara T, Chen AY, LeShane ES,
Sullivan LM, Borgatta L, Bianchi DW and Johnson KL: Cell-free fetal
DNA levels in maternal plasma after elective first-trimester
termination of pregnancy. Fertil Steril. 81:638–644.
2004.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wu Y, Werlang A, Cheng W, Lanes A, Wen SW
and Walker M: Association between levels of total cell-free DNA and
development of preeclampsia-A literature review. AJP Rep.
11:e38–e48. 2021.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Desoye G, Gauster M and Wadsack C:
Placental transport in pregnancy pathologies. Am J Clin Nutr. 94 (6
Suppl):1896S–1902S. 2011.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Lazar L, Rigó J Jr, Nagy B, Balogh K, Makó
V, Cervenak L, Mézes M, Prohászka Z and Molvarec A: Relationship of
circulating cell-free DNA levels to cell-free fetal DNA levels,
clinical characteristics and laboratory parameters in preeclampsia.
BMC Med Genet. 10(120)2009.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Kolarova TR, Gammill HS, Nelson JL,
Lockwood CM and Shree R: At preeclampsia diagnosis, total cell-free
DNA concentration is elevated and correlates with disease severity.
J Am Heart Assoc. 10(e021477)2021.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Merriel A, Alberry M and Abdel-Fattah S:
Implications of non-invasive prenatal testing for identifying and
managing high-risk pregnancies. Eur J Obstet Gynecol Reprod Biol.
256:32–39. 2021.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Samango-Sprouse C, Banjevic M, Ryan A,
Sigurjonsson S, Zimmermann B, Hill M, Hall MP, Westemeyer M,
Saucier J, Demko Z and Rabinowitz M: SNP-based non-invasive
prenatal testing detects sex chromosome aneuploidies with high
accuracy. Prenat Diagn. 33:643–649. 2013.PubMed/NCBI View
Article : Google Scholar
|
|
26
|
Zimmermann B, Hill M, Gemelos G, Demko Z,
Banjevic M, Baner J, Ryan A, Sigurjonsson S, Chopra N, Dodd M, et
al: Noninvasive prenatal aneuploidy testing of chromosomes 13, 18,
21, X, and Y, using targeted sequencing of polymorphic loci. Prenat
Diagn. 32:1233–1241. 2012.PubMed/NCBI View
Article : Google Scholar
|
|
27
|
Ryan A, Hunkapiller N, Banjevic M,
Vankayalapati N, Fong N, Jinnett KN, Demko Z, Zimmermann B,
Sigurjonsson S, Gross SJ and Hill M: Validation of an enhanced
version of a SNP-Based noninvasive prenatal test for detection of
fetal chromosomal aneuploidies. Fetal Diagn Ther. 40:219–223.
2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Wald NJ, Cuckle H, Brock JH, Peto R,
Polani PE and Woodford FP: Maternal serum-alpha-fetoprotein
measurement in antenatal screening for anencephaly and spina bifida
in early pregnancy. Report of UK collaborative study on
alpha-fetoprotein in relation to neural-tube defects. Lancet.
1:1323–1332. 1977.PubMed/NCBI
|
|
29
|
Bishop JC, Dunstan FD, Nix BJ, Reynolds TM
and Swift A: All MoMs are not equal: Some statistical properties
associated with reporting results in the form of multiples of the
median. Am J Hum Genet. 52:425–430. 1993.PubMed/NCBI
|
|
30
|
Clausen FB: Cell-free fetal DNA and fetal
blood group genotyping: Non-invasive prenatal testing. ISBT Science
Series. 15:46–51. 2020.
|
|
31
|
Breveglieri G, D'Aversa E, Finotti A and
Borgatti M: Non-invasive prenatal testing using fetal DNA. Mol
Diagn Ther. 23:291–299. 2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Fiorentino F, Bono S, Pizzuti F, Duca S,
Polverari A, Faieta M, Baldi M, Diano L and Spinella F: The
clinical utility of genome-wide non invasive prenatal screening.
Prenat Diagn. 37:593–601. 2017.PubMed/NCBI View
Article : Google Scholar
|
|
33
|
van der Schoot CE, Winkelhorst D and
Clausen FB: Noninvasive fetal blood group typing. In: Noninvasive
Prenatal Testing (NIPT). Elsevier, pp125-156, 2018.
|
|
34
|
Radoi VE, Bohiltea CL, Bohiltea RE and
Albu DN: Cell free fetal DNA testing in maternal blood of Romanian
pregnant women. Iran J Reprod Med. 13:623–626. 2015.PubMed/NCBI
|
|
35
|
Liehr T: False-positives and
false-negatives in non-invasive prenatal testing (NIPT): What can
we learn from a meta-analyses on >750,000 tests? Mol Cytogenet.
15(36)2022.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Antoniou E, Orovou E, Sarella A, Iliadou
M, Palaska E, Sarantaki A, Iatrakis G and Dagla M: Is primary
cesarean section a cause of increasing cesarean section rates in
greece? Mater Sociomed. 32:287–293. 2020.PubMed/NCBI View Article : Google Scholar
|
|
37
|
World Health Organization (WHO): Caesarean
section rates continue to rise, amid growing inequalities in
access. WHO, Geneva, 2021. https://www.who.int/news/item/16-06-2021-caesarean-section-rates-continue-to-rise-amid-growing-inequalities-in-access.
Accessed June 16, 2021.
|
|
38
|
Yaron Y, Heifetz S, Ochshorn Y, Lehavi O
and Orr-Urtreger A: Decreased first trimester PAPP-A is a predictor
of adverse pregnancy outcome. Prenat Diagn. 22:778–782.
2002.PubMed/NCBI View
Article : Google Scholar
|
|
39
|
Cowans NJ, Stamatopoulou A, Maiz N,
Spencer K and Nicolaides KH: The impact of fetal gender on first
trimester nuchal translucency and maternal serum free β-hCG and
PAPP-A MoM in normal and trisomy 21 pregnancies. Prenat Diagn.
29:578–581. 2009.PubMed/NCBI View
Article : Google Scholar
|
|
40
|
Lee LC, Sheu BC, Shau WY, Liu DM, Lai TJ,
Lee YH and Huang SC: Mid-trimester β-hCG levels incorporated in a
multifactorial model for the prediction of severe pre-eclampsia.
Prenat Diagn. 20:738–743. 2000.PubMed/NCBI View Article : Google Scholar
|
|
41
|
D'Antonio F, Rijo C, Thilaganathan B,
Akolekar R, Khalil A, Papageourgiou A and Bhide A: Association
between first-trimester maternal serum pregnancy-associated plasma
protein-A and obstetric complications. Prenat Diagn. 33:839–847.
2013.PubMed/NCBI View
Article : Google Scholar
|
|
42
|
Nicolaides KH: Nuchal translucency and
other first-trimester sonographic markers of chromosomal
abnormalities. Am J Obstet Gynecol. 191:45–67. 2004.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Nicolaides KH, Azar G, Byrne D, Mansur C
and Marks K: Fetal nuchal translucency: Ultrasound screening for
chromosomal defects in first trimester of pregnancy. BMJ.
304:867–869. 1992.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Baer RJ, Norton ME, Shaw GM, Flessel MC,
Goldman S, Currier RJ and Jelliffe-Pawlowski LL: Risk of selected
structural abnormalities in infants after increased nuchal
translucency measurement. Am J Obstet Gynecol. 211:675.e1–19.
2014.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Sekizawa A, Jimbo M, Saito H, Iwasaki M,
Matsuoka R, Okai T and Farina A: Cell-free fetal DNA in the plasma
of pregnant women with severe fetal growth restriction. Am J Obstet
Gynecol. 188:480–484. 2003.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Smid M, Galbiati S, Lojacono A, Valsecchi
L, Platto C, Cavoretto P, Calza S, Ferrari A, Ferrari M and
Cremonesi L: Correlation of fetal DNA levels in maternal plasma
with Doppler status in pathological pregnancies. Prenat Diagn.
26:785–790. 2006.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Al Nakib M, Desbriere R, Bonello N,
Bretelle F, Boubli L, Gabert J and Levy-Mozziconacci A: Total and
fetal cell-free DNA analysis in maternal blood as markers of
placental insufficiency in intrauterine growth restriction. Fetal
Diagn Ther. 26:24–28. 2009.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Thurik FF, Lamain-de Ruiter M, Javadi A,
Kwee A, Woortmeijer H, Page-Christiaens GC, Franx A, van der Schoot
CE and Koster MP: Absolute first trimester cell-free DNA levels and
their associations with adverse pregnancy outcomes. Prenat Diagn.
36:1104–1111. 2016.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Birch L, English CA, O'Donoghue K, Barigye
O, Fisk NM and Keer JT: Accurate and robust quantification of
circulating fetal and total DNA in maternal plasma from 5 to 41
weeks of gestation. Clin Chem. 51:312–320. 2005.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Jung K, Fleischhacker M and Rabien A:
Cell-free DNA in the blood as a solid tumor biomarker-a critical
appraisal of the literature. Clin Chim Acta. 411:1611–1624.
2010.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Fernando MR, Chen K, Norton S,
Krzyzanowski G, Bourne D, Hunsley B, Ryan WL and Bassett C: A new
methodology to preserve the original proportion and integrity of
cell-free fetal DNA in maternal plasma during sample processing and
storage. Prenat Diagn. 30:418–424. 2010.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Manokhina I, Singh TK, Peñaherrera MS and
Robinson WP: Quantification of cell-free DNA in normal and
complicated pregnancies: Overcoming biological and technical
issues. PLoS One. 9(e101500)2014.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Contro E, Bernabini D and Farina A:
Cell-Free Fetal DNA for the prediction of pre-eclampsia at the
first and second trimesters: A systematic review and meta-analysis.
Mol Diagn Ther. 21:125–135. 2017.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Sarzynska-Nowacka U, Kosinski P and
Wielgos M: Is there a future for cell-free fetal dna tests in
screening for preeclampsia? Ginekol Pol. 90:55–60. 2019.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Poon LC, Musci T, Song K, Syngelaki A and
Nicolaides KH: Maternal plasma cell-free fetal and maternal DNA at
11-13 weeks' gestation: Relation to fetal and maternal
characteristics and pregnancy outcomes. Fetal Diagn Ther.
33:215–223. 2013.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Rolnik DL, da Silva Costa F, Lee TJ,
Schmid M and McLennan AC: Association between fetal fraction on
cell-free DNA testing and first-trimester markers for
pre-eclampsia. Ultrasound Obstet Gynecol. 52:722–727.
2018.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Quezada MS, Francisco C, Dumitrascu-Biris
D, Nicolaides KH and Poon LC: Fetal fraction of cell-free DNA in
maternal plasma in the prediction of spontaneous preterm delivery.
Ultrasound Obstet Gynecol. 45:101–105. 2015.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Ridnõi K, Muru K, Keernik M, Pajusalu S,
Ustav EL, Tammur P, Mölter-Väär T, Kahre T, Šamarina U, Asser K, et
al: A two-year prospective study assessing the performance of fetal
chromosomal microarray analysis and next-generation sequencing in
high-risk pregnancies. Mol Genet Genomic Med.
9(e1787)2021.PubMed/NCBI View Article : Google Scholar
|