Research has shown that events during pregnancy and
up to the first 2-3 years of life, influence biological functions,
such as the immune response and shape brain structure in ways that
affect well-being, health and productivity (1). Several factors that influence the
developmental environment have been found to be associated with the
risk of disease development during adult life. It appears that the
interaction between genes and environmental factors in the prenatal
to early postnatal period is critical for the development of
diseases in adulthood. Until recently, the aforementioned factors
were studied in a single effect-health result manner, an imprecise
approach, since it does not take fully into consideration the
numerous interactions between the environmental factors and genes
themselves. Thus, the concept of the exposome, i.e., the totality
of exposures from external and internal stimuli throughout the
lifespan of an organism, was proposed as a more inclusive method of
studying early-life environment-gene interactions (2). The mechanisms that consolidate the
effect of these numerous stimuli are epigenetic modifications, such
as DNA methylation, histone modifications and regulation by
non-coding RNAs (ncRNAs), while one of the main interfaces between
the external exposome and the human host is the gut microbiota
(3). Extracellular vesicles (EVs),
and particularly exosomes, can function as mediators of both
epigenetic modifications and mechanisms that alter gut microbiota
diversity (4,5). Breast milk is known to contain its own
microbiota and large quantities of EVs, particularly exosomes, and
plays an essential role in neonatal and infant health (6). Thus, the study of how exosomes may
influence epigenetic mechanisms and microbiota in early life may
further elucidate the specifics of the effects of breast milk on
the health of an infant and subsequently, its adulthood.
EVs are heterogeneous lipid bilayer-surrounded
vesicles that carry bioactive molecules and are secreted by
different cell types in the extracellular space (7,8). EV
secretion is an ubiquitous mechanism present in all domains of life
and occurs under a wide range of conditions, both physiological and
pathological (Table I) (9). Prokaryotes produce EVs termed membrane
vesicles. Pathogenic and non-pathogenic Gram-negative bacteria
secrete outer membrane vesicles, which are 10 to 500 nm in size and
originate from the outer membrane of a cell, while Gram-positive
bacteria secrete membrane vesicles which are 20 to 400 nm in size
and originate from the single cytoplasmic cell membrane. Bacterial
EVs are involved in numerous processes, including intercellular
communications, stress response and antibiotic resistance. Although
research on archaea is somewhat limited, the majority of species
appear to produce vesicles originating from the cytoplasmic cell
membrane with an average size of 50 to 250 nm (10). These EVs appear to promote
horizontal gene transfer and nutrient cycling under extreme
conditions (11). In eukaryotic
cells, and multicellular eukaryotic organisms such as humans,
vesicles are grouped into three subtypes based on their biogenesis,
size, content, release pathway and function (8). The three classical types of EVs in
eukaryotes are microvesicles, which are 40 nm to 1 µm in size and
emerge through direct outward budding and shedding from the plasma
membrane, apoptotic bodies which are 1 to 5 µm in size and are
created in the process of apoptosis, and exosomes which are 40 to
150 nm in size and originate from the endosome (12).
Exosomes, in particular, have garnered scientific
attention due their potential as diagnostic and therapeutic
molecules (13). These vesicles are
secreted by virtually any cell type and are found in numerous
biofluids including plasma, saliva, urine, semen, cerebrospinal
fluid and breast milk (8). Their
cargo depends on the cell of origin and consists-among others-of
nucleic acids, proteins, lipids and metabolites (14). The physiological state of the cell
of origin also influences exosomal cargo, with pathological
conditions leading to alterations in exosome content (15). Human exosomes are mainstays of
cellular communication and take part in both physiological
processes, such as the immune response, tissue repair and cell
programming, and pathological conditions, such as tumorigenesis,
chronic inflammation and neurodegeneration (16).
Epigenetics can aid in the determination of how
events in early life can influence adult health. Epigenetics study
the molecules and mechanisms that can extend alternative gene
activity states, while the DNA sequence remains the same (23). Epigenetic mechanisms include DNA
methylation, histone modifications and regulation by ncRNAs
(24). These mechanisms act as the
intermediate between genes and the environment, and influence
physiology and disease by altering gene expression and thus
regulating gene activity (25). DNA
methylation consists of a covalent attachment of a methyl group at
the C-5 position of cytosine, resulting in the formation of
5-methyl cytosine. This attachment mostly occurs in in
cytosine-guanosine (CpG) dinucleotide sequences. The catalyzation
of this process is achieved through the action of DNA
methyltransferases (DNMTs), while demethylation is achieved through
the action of ten-eleven translocation dioxygenases (26,27).
Gene promoters often showcase regions rich in such sequences, which
are termed CpG islands (28). DNA
methylation functions together with histone modifications to alter
chromatin structure and influence DNA accessibility to factors that
regulate gene expression (29).
Function-wise DNA methylation is considered a regulator of gene
silencing (30). Histone
modifications include the acetylation, methylation, ubiquitination,
sumoylation, deimination, phosphorylation and ADP ribosylation of
histone proteins. Similar to DNA methylation, histone modifications
influence the availability of DNA to the transcriptional machinery.
Generally, histone modifications are catalyzed by enzymes that
mostly act on the N-terminal tail of histones and involve amino
acids such as lysine, arginine, threonine, serine and tyrosine
(31,32). These modifications are catalyzed by
enzymes, such as histone acetyltransferases, histone
methyltransferases, histone deacetylases (HDACs) and histone
demethylases (33). The most
extensively studied histone modification is histone acetylation,
which usually results in a higher gene expression (31). ncRNAs are ribonucleic acid molecules
that do not encode proteins. These molecules are ubiquitous in the
human body and include ribosomal RNAs and transfer RNAs, plus RNAs
with a regulatory role in gene expression, such as miRNAs, long
ncRNAs (lncRNAs), small interfering RNAs, PIWI-interacting RNAs,
small nucleolar RNAs and circular RNAs. The expression of ncRNAs is
highly regulated and their expression profiles are distinct in each
developmental stage and tissue (34). The majority of studies on ncRNAs
have focused on the role of miRNAs and lncRNAs in different
pathologies (35). miRNAs, which
consist of 17 to 25 nucleotides, can recognize target messenger
RNAs (mRNAs) via sequence complementarity and regulate their
protein translation. The majority of miRNAs inhibit gene expression
by interfering with mRNA translation. In certain cases, such as the
binding of a miRNA to the promoter region of a gene, miRNAs may
promote gene expression (36).
lncRNAs, which consist of >200 nucleotides, appear to influence
gene expression via numerous mechanisms. Several lncRNAs influence
gene expression by creating scaffolds for protein-DNA complexes,
other lncRNAs recruit histone modification enzymes, while another
group bind neighboring genomic loci to promote gene imprinting.
Additionally, lncRNAs can interact with mRNAs or act as sponges
that inhibit the action of miRNAs (37).
Epigenetic modifications during early-life
development have a marked impact on both physiological and
pathological conditions in adult life (Fig. 1). Early-life environmental factors
significantly influence DNA methylation patterns across the human
lifespan. The effects of DNA methylation persist during the whole
human lifespan and appear stronger for heritable sites and sites
with play a role in gene expression (38). Some of the environmental factors
that can influence epigenetic mechanisms, such as DNA methylation
in early life include dietary habits during pregnancy, exposure to
metals, smoking, or alcohol consumption during the perinatal
period, and in particular, early-life stress (ELS) (39,40). A
prime example includes the altered methylation levels of monoamine
oxidase (MAO) A gene due to early-life stress which may be
associated with depression-like symptoms in adult mice. Monoamine
oxidases (MAOs) are enzymes that modulate the metabolism of
monoamine neurotransmitters, such as dopamine, epinephrine and
serotonin. These neurotransmitters act in tandem to modulate basic
emotions. Therefore, it is expected that their deregulated levels
have been associated with affective disorders, such as depression,
with MAO inhibitors being the first type of antidepressants
developed. Patients with depression showcase MAO A gene
hypomethylation, which may lead to increased MAO A activity and may
result in reduced monoamine utilization (40). Epigenetic mechanisms, such as
histone modifications help coordinate and fine-tune spatiotemporal
gene expression, while their dysregulation has been shown to be
associated with several disorders of the nervous system; indeed,
abnormal histone modification levels appear to promote
neuropsychiatric and neurodegenerative disorders (41). A previous study on rats found that
ELS appears to lead to the abnormal acetylation of the H3K9 histone
(42). Both hyperacetylation and
hypoacetylation have been implicated in anxiety-like behavior in
adult rats, with the conflicting results possibly arising due to
methodological differences (42).
miRNAs are main regulators of brain plasticity and higher brain
functions. ELS appears to influence miRNA levels well into
adulthood. Both human and animal studies have demonstrated that
miR-125-1-3p is particularly responsive to ELS and its low
expression levels are associated with schizophrenia in adulthood
(43). lncRNAs have been found to
be spatially and temporally restricted at certain developmental
stages, thus hinting at a potential role in proper organism
development. Indeed, lncRNAs, such as the CHD2 adjacent,
suppressive regulatory RNA (Chaserr) appear to play an integral
role in neurodevelopment in humans and has been associated with
neurodevelopmental delay in mice (44,45).
Nevertheless, the mechanisms through which early-life stressors
influence lncRNA levels and adult health remain poorly
characterized, with a study on mice showcasing that arsenic
exposure during embryonic development alters the expression of the
lncRNA growth arrest specific-5 in males (46). This lncRNA has been shown to be
associated with several pathological conditions in adult life, such
as cancer, type 2 diabetes mellitus and bone diseases (47).
The term microbiota refers to living organisms
located in a defined environment, with prime examples being the
human gut, oral, skin, respiratory and female genital tract
microbiota. Numerous studies on humans have highlighted the
mechanisms through which microbiota influence health and disease
(48,49). Specifically, human microbiota are
involved in essential biological processes, such as intercellular
communication and immune response, while dysbiosis, i.e., an
interruption in microbiota balance, has been found to be associated
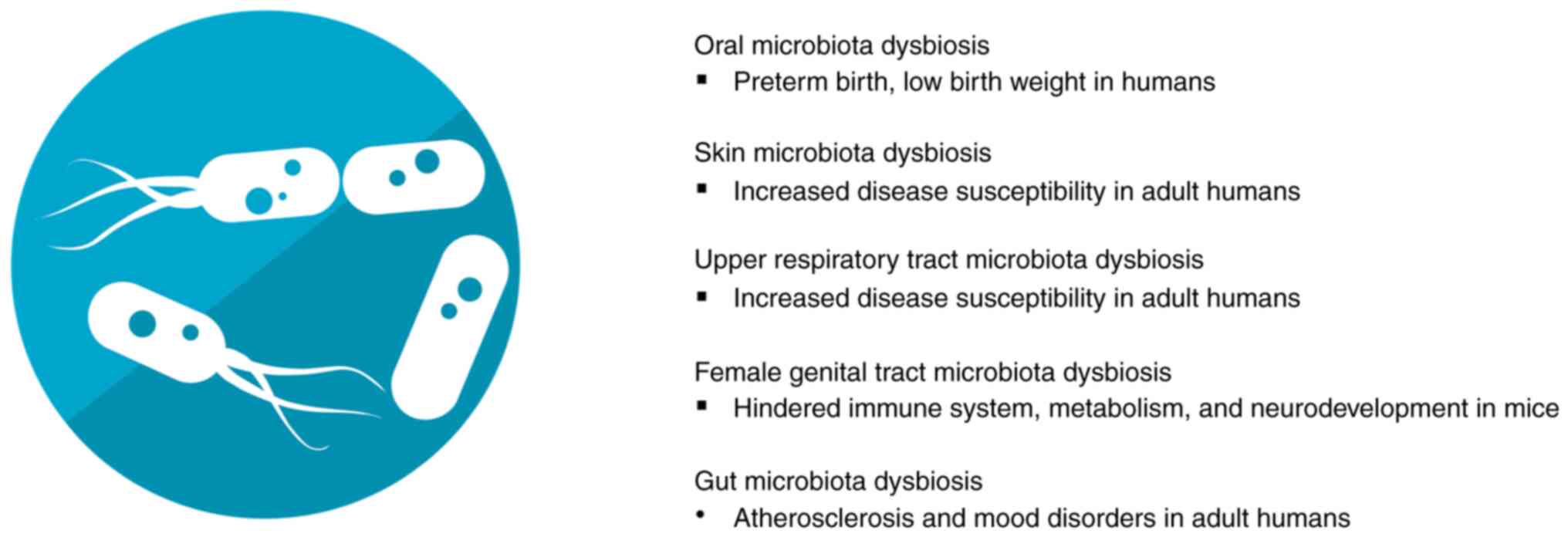
with numerous pathologies (Fig. 2).
Thus, it is evident that human microbiota have been hailed as
potential biomarkers and means of personalized medicine (48).
The oral microbiota interacts with the host to
communicate information regarding the status of immunity and
metabolism via a two-way communication between the oral cavity and
the systemic organs. Oral microbiota dysbiosis is associated with
diseases, such as periodontal diseases, as well as systemic
diseases, such as inflammatory bowel syndrome, diabetes and
pregnancy complications. As regards the latter, studies on mice
have suggested that oral symbiotic bacteria can colonize the
placenta and influence pregnancy (50). The skin is the largest organ of the
human body and carries a complex set of microorganisms, featuring
bacteria, archaea, eukaryotes, fungi, viruses and phages. The skin
microbiome is a main regulator of the skin immune system and
dysbiosis has been associated with atopic dermatitis, acne and
dandruff (51). The upper
respiratory tract is colonized by bacterial, fungal and viral
organisms. These respiratory microbiota are thought to hinder
potential pathogens from overgrowing and spreading towards the
lungs and dysbiosis has been associated with chronic respiratory
diseases (52). The female genital
tract microbiota play a critical role in the prevention of pathogen
invasion and growth, with dysbiosis being associated with
gynecological disorders and infertility (53). Gut microbiota are considered one of
the most significant mediators of human health. The gut microbiota
mainly consists of prokaryotic organisms and to a lesser extent,
fungi and viruses (54). These
organisms comprise a highly dynamic system that is influenced by
both exogenous factors, such as diet or environmental alterations
and endogenous factors, such as the genetic composition of the host
(55). The gut microbiota, and
particularly bacteria, function as a main regulator of the body
homeostasis, thus influencing several processes such as metabolism,
inflammation and hematopoiesis (49). It is then not surprising that gut
microbiota disorders have been found to be associated with several
diseases, such type 2 diabetes mellitus, atherosclerosis and even
neurodegenerative disorders, such as Alzheimer's disease and
Parkinson's disease (56).
Periodontal disease in mothers and the resulting
dysbiosis of their oral microbiota have been linked to pregnancy
complications, such as preterm birth and a low birth weight. Some
of the above may be a result of the existence of Gram-negative
anaerobic bacteremia, which originate from the gingival biofilm or
the entrance of pro-inflammatory factors that originate from the
gingival submucosa into the bloodstream (57). Microbial exposure via barrier
tissues such as skin and lungs in early life help imprint a healthy
immune response up to adult life in humans (58). Experiments on mice have also
highlighted that the vaginal microbiota can influence offspring
metabolism, immune response and even brain development (59). Research has indicated that the
microbes that inhabit the human intestinal tract may influence
infant development and the maturation of the immune system. It is
quite possible that the risk of disease is affected by the gut
microbiota during fetal development and early life, thus rendering
gut microbiota development research imperative (60). Gut microbiota development during
early life can be affected by numerous factors, including host
genetics, gestational age, delivery mode, maternal health,
antibiotics exposure, ELS, early-life environment and early-life
diet (61,62). Indeed, the development of adult
atherosclerosis is influenced by specific conditions during
infancy, such as preterm birth, malnutrition and microbiota
colonization. Malnutrition affects the gut microbiota composition
and leads to an increase in the number of potentially pathogenic
bacteria. This increase leads to decreased nutrient absorption and
may cause epithelial damage. Such epithelial damage may promote an
altered gut barrier permeability, leading to the systemic
circulation of metabolic products and bacteria, which in turn
results in an increased systemic inflammation, an underlying cause
of atherosclerosis (62). Another
instance of early-life microbiota influencing adult health is
susceptibility to mood disorders. ELS in humans appears to
interfere with metabolites of the glutamate pathway and promote
changes in functional brain connectivity that have been found to be
associated with mood disorders. These metabolites are considered to
be regulated by the gut microbiota, with a prime example being
5-oxoproline (63). This metabolite
is significantly increased in patients with major depressive
disorder (MDD) (64). Studies on
animal models have shown that mice transplanted with microbiota
from MDD display high levels of 5-oxoproline, while treatment of
rats with antibiotics reduces metabolite levels, thus implicating
the gut microbiota in its regulation (65,66).
As aforementioned, EVs, and specifically exosomes,
can act as mediators of epigenetic mechanisms and
microbiota-regulated mechanisms. The exosome-mediated transfer of
epigenetic regulators is considered to be an essential mechanism of
epigenetic information exchange between cells (67). In the case of DNA methylation,
exosomes derived from bovine milk have been found to contain miRNAs
that target DNMTs and influence the demethylation of the promoter
region of metabolic regulators, such as the fat mass and
obesity-associated protein (FTO). Studies on mice have suggested
that FTO expression may play a pivotal role in postnatal growth and
energy expenditure (68,69), while FTO loss-of-function mutations
have been shown to be associated with postnatal growth retardation
in humans (70). In the case of
exosome-mediated histone modifications, information is somewhat
limited. An in vitro study on isolated mouse cells
demonstrated that exosomes derived from developing cortical neurons
were enriched in HDAC2(71). HDAC2
has been implicated in the epigenetic regulation of genes that
encode synaptic proteins and appears to control dendritic spine
development and synapse establishment. Both in vitro and
in vivo mouse studies have detected a decrease in HDAC2
levels as cortical neurons mature and acquire news synapses. The
in vitro treatment of mature neurons with exosomes derived
from HDAC2-rich neurons was shown to lead to an increased
expression of HDAC2 in the recipient cells. These results implicate
a role for exosomes in nervous system maturation via the transport
of histone modifying enzymes (71).
Exosomes contain a wide variety of ncRNA molecules, including
miRNAs which are the most extensively studied, with lncRNAs
steadily receiving an increase in scientific interest (67). Exosomal miRNAs participate in both
physiological and pathological processes (72). In vitro studies on human
mesenchymal stem cells (hMSCs) have indicated that hMSC-derived
exosomes contribute to osteoblastic differentiation via a
miRNA-dependent mechanism (73).
Such studies highlight the importance of exosomal miRNAs in proper
organism development. Apart from their role in cell
differentiation, exosomal miRNAs are also involved in pathological
conditions, such as cancer. Exosomal miRNAs take part in processes
that regulate the tumor microenvironment and interfere with cancer
immunity. Specifically, tumor-derived exosomes modulate cancer
cell-to-cell communication via their miRNA cargo and are heavily
associated with angiogenesis, tumor growth, metastasis and drug
resistance (74). The majority of
research on exosomal lncRNAs focuses on their severe effects on
cancer, where they influence cell proliferation, angiogenesis, drug
resistance and metastasis (75). In
particular, several studies have focused on the interaction between
cancer cells and immune cells that have infiltrated the tumor
microenvironment, and how exosomal lncRNAs mediate this process.
Tumor-derived exosomal lncRNAs appear to boost M2 macrophage
polarization and inhibit the function of natural killer cells in
vitro, thus supporting tumor progression (76). Exosomes can potentially provide a
link between early-life development and cancer in adult life.
Early-life exposure to adverse environmental factors lead to
exosomal cargo alterations, which in turn may influence cancer risk
in adults (77).
Research on the role of EVs secreted by microbiota
in host health and the effect of host exosomes on microbiota are
limited and focus mainly on the gut microbiota-host interaction.
EVs derived from both pathogenic and probiotic bacteria can travel
short and long distances, and modulate both bacteria-bacteria and
bacteria-host communications (78,79).
It should be mentioned that the term probiotics refers to live
bacteria with health benefits and several gut commensal bacteria
display probiotic effects by boosting host health (80). Bacterial communications via EVs
appears to play an essential role in the gut microenvironment and
have been implicated in bacterial colonization and growth,
protection from host defense peptides and bacterial population
behavior. Bacteria-derived vesicles are known to play an
immunomodulatory role in human physiology. Specifically, EVs
derived from pathogenic bacteria display proinflammatory abilities
and play a role in immune response escape, while probiotic-derived
EVs are characterized by anti-inflammatory effects and promote
immune tolerance (80).
Microbiota-derived EVs may also play a crucial role in several
processes in early-life development. A previous study on healthy
suckling rats highlighted that probiotic-derived EVs promoted
immune and intestinal system maturation (81). It has also been demonstrated that
microbiota-derived EVs from human children exerted bone protective
effects in animal models of osteoporosis, possibly suggesting an
essential role in skeletal system maturation (82,83).
Additionally, the amniotic fluid of pregnant women contains
bacterial-derived EVs similar to those found in the maternal gut
microbiota. Experiments on pregnant mice have revealed that EVs
secreted from human maternal microbiota can pass to into the
intra-amniotic space. These findings suggest a potential role of
maternal microbiota-derived vesicles in priming the prenatal immune
system for gut colonization (84).
Respectively, host miRNAs can directly interact with bacterial
genes in the gut, with exosomes being a possible transfer
mechanism. A combination of in vitro experiments and animal
studies have shown that host miRNAs can act on microbiota genes and
promote or inhibit bacterial growth. Since host-derived EVs have
been shown to affect bacteria in animal models of several
pathologies, it is quite possible that this effect is based on
miRNA transportation via exosomes (85). Studies focusing on exosomes found in
human breast milk (HBM) have demonstrated that such vesicles
contain ncRNAs that are associated with infant health, thus
implicating maternal exosomes in proper offspring development
(86).
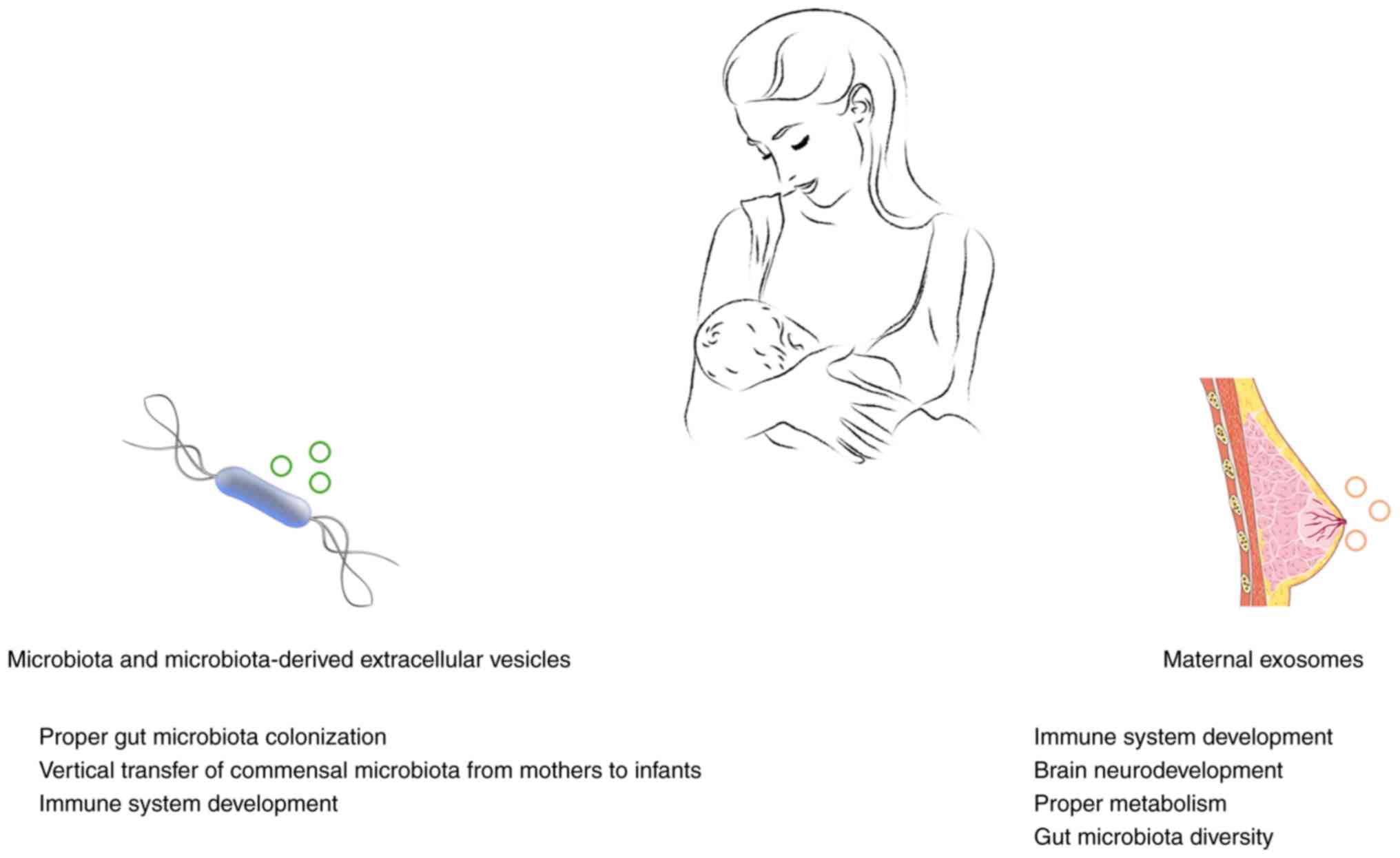
HBM is considered the optimal food source for
newborn infants and promotes infant health (Fig. 3). Indeed, breast feeding has been
shown to be associated with an improved infant health and immune
development, while breast-fed infants display lower mortality rates
than formula-fed infants (87). The
HBM consists of water, nutrients such as lactose and fat, plus
immune cells, hormones, miRNAs and immunomodulatory bioactive
molecules (88). Additionally, HBM
carries its own distinct microbiome with potentially probiotic
bacteria that participate in infant gut colonization (89). Moreover, during lactation the
composition of HBM microbiota alters in addition to immune and
nutritional composition. It is considered that these alterations in
the microbiota found in human breast milk play a role in the immune
system development of the infant (90). The HBM also contains bacterial EVs,
some of which originate from the gut microbiome, that promote the
vertical transfer of commensal microbiota from mothers to infants
(91).
All the aforementioned information suggests that EVs
play an essential role in early-life development.
Microbiota-derived EVs can influence infant gut microbiota
colonization and can affect maturation via their cargo. Moreover,
these types of EVs have been proposed as potential biomarkers of
metabolic diseases and various pathological conditions (104). Human exosomes can, through their
cargo, promote epigenetic changes that influence gene expression in
infant cells or alter gut microbiota diversity. Of note, it should
also be considered that the cargo found in HBM-derived exosomes is
influenced by maternal lifestyle and health (95,105).
For instance, maternal obesity is negatively associated with the
content of exosomal miR-148a and miR-30b in human breast milk.
These miRNAs are involved in glucose metabolism and adipogenesis
and appear to influence infant body composition (106). Hence, studying HBM-derived
exosomes could potentially provide a new direct link between
maternal health and infant development, highlighting their
potential role as intricate biomarkers and therapeutic agents.
Not applicable.
Funding: No funding was received.
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
DV conceived the study. TM, EP, KD, GPC and DV
wrote, drafted, revised, edited and reviewed the manuscript. All
authors have read and approved the final manuscript. Data
authentication is not applicable.
Not applicable.
Not applicable.
GPC is the Editor in Chief of the journal, and DV is
an Editor of the journal. However, they had no personal involvement
in the reviewing process, or any influence in terms of adjudicating
on the final decision, for this article. The other authors declare
that they have no competing interests.
|
1
|
Richter L, Black M, Britto P, Daelmans B,
Desmond C, Devercelli A, Dua T, Fink G, Heymann J, Lombardi J, et
al: Early childhood development: An imperative for action and
measurement at scale. BMJ Global Health. 4 (Suppl
4)(e001302)2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Ferrante G, Fasola S, Cilluffo G,
Piacentini G, Viegi G and La Grutta S: Addressing Exposome: An
innovative approach to environmental determinants in pediatric
respiratory health. Front Public Health. 10(871140)2022.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Fenga C: Gut microbiota modulation: A
tailored approach for the prevention of chronic diseases. Biomed
Rep. 16(23)2022.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Abeysinghe P, Turner N, Morean Garcia I,
Mosaad E, Peiris HN and Mitchell MD: The role of exosomal
epigenetic modifiers in cell communication and fertility of dairy
cows. Int J Mol Sci. 21(9106)2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Zhang B, Zhao J, Jiang M, Peng D, Dou X,
Song Y and Shi J: The potential role of gut microbial-derived
exosomes in metabolic-associated fatty liver disease: Implications
for treatment. Front Immunol. 13(893617)2022.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Galley JD and Besner GE: The therapeutic
potential of breast milk-derived extracellular vesicles. Nutrients.
12(745)2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Wang DR and Pan J: Extracellular vesicles:
Emerged as a promising strategy for regenerative medicine. World J
Stem Cells. 15:165–181. 2023.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Doyle LM and Wang MZ: Overview of
extracellular vesicles, their origin, composition, purpose, and
methods for exosome isolation and analysis. Cells.
8(727)2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Kumar MA, Baba SK, Sadida HQ, Marzooqi SA,
Jerobin J, Altemani FH, Algehainy N, Alanazi MA, Abou-Samra AB,
Kumar R, et al: Extracellular vesicles as tools and targets in
therapy for diseases. Signal Transduct Target Ther.
9(27)2024.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Mobarak H, Javid F, Narmi MT, Mardi N,
Sadeghsoltani F, Khanicheragh P, Narimani S, Mahdipour M, Sokullu
E, Valioglu F and Rahbarghazi R: Prokaryotic microvesicles Ortholog
of eukaryotic extracellular vesicles in biomedical fields. Cell
Commun Signal. 22(80)2024.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Liu J, Cvirkaite-Krupovic V, Commere PH,
Yang Y, Zhou F, Forterre P, Shen Y and Krupovic M: Archaeal
extracellular vesicles are produced in an ESCRT-dependent manner
and promote gene transfer and nutrient cycling in extreme
environments. ISME J. 15:2892–2905. 2021.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Sheta M, Taha EA, Lu Y and Eguchi T:
Extracellular vesicles: New classification and tumor
immunosuppression. Biology (Basel). 12(110)2023.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Zhang Y, Bi J, Huang J, Tang Y, Du S and
Li P: Exosome: A review of its classification, isolation
techniques, storage, diagnostic and targeted therapy applications.
Int J Nanomedicine. 15:6917–6934. 2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Duréndez-Sáez E, Calabuig-Fariñas S,
Torres-Martínez S, Moreno-Manuel A, Herreros-Pomares A, Escorihuela
E, Mosqueda M, Gallach S, Guijarro R, Serna E, et al: Analysis of
exosomal cargo provides accurate clinical, histologic and
mutational information in non-small cell lung cancer. Cancers
(Basel). 14(3216)2022.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Dimik M, Abeysinghe P, Logan J and
Mitchell M: The exosome: A review of current therapeutic roles and
capabilities in human reproduction. Drug Deliv Transl Res.
13:473–502. 2023.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Dilsiz N: Hallmarks of exosomes. Future
Sci OA. 8(FSO764)2022.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Zhang Y, Dou Y, Liu Y, Di M, Bian H, Sun X
and Yang Q: Advances in therapeutic applications of extracellular
vesicles. Int J Nanomedicine. 18:3285–3307. 2023.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Schiller EA, Cohen K, Lin X, El-Khawam R
and Hanna N: Extracellular Vesicle-microRNAs as diagnostic
biomarkers in preterm neonates. Int J Mol Sci.
24(2622)2023.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Lak NSM, van der Kooi EJ, Enciso-Martinez
A, Lozano-Andrés E, Otto C, Wauben MHM and Tytgat GAM:
Extracellular vesicles: A new source of biomarkers in pediatric
solid tumors? A systematic review. Front Oncol.
12(887210)2022.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Galardi A, Colletti M, Di Paolo V, Vitullo
P, Antonetti L, Russo I and Di Giannatale A: Exosomal MiRNAs in
pediatric cancers. Int J Mol Sci. 20(4600)2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Chaubey S, Thueson S, Ponnalagu D, Alam
MA, Gheorghe CP, Aghai Z, Singh H and Bhandari V: Early gestational
mesenchymal stem cell secretome attenuates experimental
bronchopulmonary dysplasia in part via exosome-associated factor
TSG-6. Stem Cell Res Ther. 9(173)2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Braun RK, Chetty C, Balasubramaniam V,
Centanni R, Haraldsdottir K, Hematti P and Eldridge MW:
Intraperitoneal injection of MSC-derived exosomes prevent
experimental bronchopulmonary dysplasia. Biochem Biophys Res
Commun. 503:2653–2658. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Cavalli G and Heard E: Advances in
epigenetics link genetics to the environment and disease. Nature.
571:489–499. 2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Bertogliat MJ, Morris-Blanco KC and
Vemuganti R: Epigenetic mechanisms of neurodegenerative diseases
and acute brain injury. Neurochem Int. 133(104642)2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Liang M: Epigenetic mechanisms and
hypertension. Hypertension. 72:1244–1254. 2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Nasrullah Hussain A, Ahmed S, Rasool M and
Shah AJ: DNA methylation across the tree of life, from micro to
macro-organism. Bioengineered. 13:1666–1685. 2022.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Shi J, Xu J, Chen YE, Li JS, Cui Y, Shen
L, Li JJ and Li W: The concurrence of DNA methylation and
demethylation is associated with transcription regulation. Nat
Commun. 12(5285)2021.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Uddin MG and Fandy TE: DNA methylation
inhibitors: Retrospective and perspective view. Adv Cancer Res.
152:205–223. 2021.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Sallustio F, Gesualdo L and Gallone A: New
findings showing how DNA methylation influences diseases. World J
Biol Chem. 10:1–6. 2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Tompkins JD: Discovering DNA methylation,
the history and future of the writing on DNA. J Hist Biol.
55:865–887. 2022.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Alaskhar Alhamwe B, Khalaila R, Wolf J,
von Bülow V, Harb H, Alhamdan F, Hii CS, Prescott SL, Ferrante A,
Renz H, et al: Histone modifications and their role in epigenetics
of atopy and allergic diseases. Allergy Asthma Clin Immunol.
14(39)2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Lee HT, Oh S, Ro DH, Yoo H and Kwon YW:
The Key Role of DNA Methylation and histone acetylation in
epigenetics of atherosclerosis. J Lipid Atheroscler. 9:419–434.
2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Liu R, Wu J, Guo H, Yao W, Li S, Lu Y, Jia
Y, Liang X, Tang J and Zhang H: Post-translational modifications of
histones: Mechanisms, biological functions, and therapeutic
targets. MedComm (2020). 4(e292)2023.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Zhang SF, Gao J and Liu CM: The role of
non-coding RNAs in neurodevelopmental disorders. Front Genet.
10(1033)2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Ratti M, Lampis A, Ghidini M, Salati M,
Mirchev MB, Valeri N and Hahne JC: MicroRNAs (miRNAs) and Long
Non-Coding RNAs (lncRNAs) as new tools for cancer therapy: First
steps from bench to bedside. Target Oncol. 15:261–278.
2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Jorge AL, Pereira ER, Oliveira CS,
Ferreira EDS, Menon ETN, Diniz SN and Pezuk JA: MicroRNAs:
Understanding their role in gene expression and cancer. Einstein
(Sao Paulo). 19(eRB5996)2021.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Borkiewicz L, Kalafut J, Dudziak K,
Przybyszewska-Podstawka A and Telejko I: Decoding LncRNAs. Cancers
(Basel). 13(2643)2021.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Li S, Ye Z, Mather KA, Nguyen TL, Dite GS,
Armstrong NJ, Wong EM, Thalamuthu A, Giles GG, Craig JM, et al:
Early life affects late-life health through determining DNA
methylation across the lifespan: A twin study. EBioMedicine.
77(103927)2022.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Schrott R, Song A and Ladd-Acosta C:
Epigenetics as a biomarker for early-life environmental exposure.
Curr Environ Health Rep. 9:604–624. 2022.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Xu Q, Jiang M, Gu S, Wang F and Yuan B:
Early life stress induced DNA methylation of monoamine oxidases
leads to depressive-like behavior. Front Cell Dev Biol.
8(582247)2020.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Park J, Lee K, Kim K and Yi SJ: The role
of histone modifications: From neurodevelopment to neurodiseases.
Signal Transduct Target Ther. 7(217)2022.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Guan L, Shi X, Tang Y, Yan Y, Chen L, Chen
Y, Gao G, Lin C and Chen A: Contribution of amygdala histone
acetylation in early life stress-induced visceral hypersensitivity
and emotional comorbidity. Front Neurosci.
16(843396)2022.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Allen L and Dwivedi Y: MicroRNA mediators
of early life stress vulnerability to depression and suicidal
behavior. Mol Psychiatry. 25:308–320. 2020.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Tsagakis I, Douka K, Birds I and Aspden
JL: Long non-coding RNAs in development and disease: Conservation
to mechanisms. J Pathol. 250:480–495. 2020.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Rom A, Melamed L, Gil N, Goldrich MJ,
Kadir R, Golan M, Biton I, Perry RB and Ulitsky I: Regulation of
CHD2 expression by the Chaserr long noncoding RNA gene is essential
for viability. Nat Commun. 10(5092)2019.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Caldwell KK, Hafez A, Solomon E,
Cunningham M and Allan AM: Arsenic exposure during embryonic
development alters the expression of the long noncoding RNA growth
arrest specific-5 (Gas5) in a sex-dependent manner. Neurotoxicol
Teratol. 66:102–112. 2018.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Zhou Z, Chen J, Huang Y, Liu D, Chen S and
Qin S: Long Noncoding RNA GAS5: A new factor involved in bone
diseases. Front Cell Dev Biol. 9(807419)2022.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Hou K, Wu ZX, Chen XY, Wang JQ, Zhang D,
Xiao C, Zhu D, Koya JB, Wei L, Li J and Chen ZS: Microbiota in
health and diseases. Signal Transduct Target Ther.
7(135)2022.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Afzaal M, Saeed F, Shah YA, Hussain M,
Rabail R, Socol CT, Hassoun A, Pateiro M, Lorenzo JM, Rusu AV and
Aadil RM: Human gut microbiota in health and disease: Unveiling the
relationship. Front Microbiol. 13(999001)2022.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Peng X, Cheng L, You Y, Tang C, Ren B, Li
Y, Xu X and Zhou X: Oral microbiota in human systematic diseases.
Int J Oral Sci. 14(14)2022.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Skowron K, Bauza-Kaszewska J, Kraszewska
Z, Wiktorczyk-Kapischke N, Grudlewska-Buda K, Kwiecińska-Piróg J,
Wałecka-Zacharska E, Radtke L and Gospodarek-Komkowska E: Human
skin microbiome: Impact of intrinsic and extrinsic factors on skin
microbiota. Microorganisms. 9(543)2021.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Man WH, de Steenhuijsen Piters WA and
Bogaert D: The microbiota of the respiratory tract: Gatekeeper to
respiratory health. Nat Rev Microbiol. 15:259–270. 2017.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Cocomazzi G, De Stefani S, Del Pup L,
Palini S, Buccheri M, Primiterra M, Sciannamè N, Faioli R, Maglione
A, Baldini GM, et al: The impact of the female genital microbiota
on the outcome of assisted reproduction treatments. Microorganisms.
11(1443)2023.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Al Bander Z, Nitert MD, Mousa A and
Naderpoor N: The gut microbiota and inflammation: An overview. Int
J Environ Res Public Health. 17(7618)2020.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Ferraris C, Elli M and Tagliabue A: Gut
microbiota for health: How can diet maintain a healthy Gut
Microbiota? Nutrients. 12(3596)2020.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Chen Y, Zhou J and Wang L: Role and
mechanism of gut microbiota in human disease. Front Cell Infect
Microbiol. 11(625913)2021.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Russo M, Calevo MG, D'Alessandro G,
Tantari M, Migliorati M, Piccardo I, Perucchin PP and Arioni C:
Influence of maternal oral microbiome on newborn oral microbiome in
healthy pregnancies. Ital J Pediatr. 49(140)2023.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Dhariwala MO and Scharschmidt TC: Baby's
skin bacteria: First impressions are long-lasting. Trends Immunol.
42:1088–1099. 2021.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Jašarević E, Hill EM, Kane PJ, Rutt L,
Gyles T, Folts L, Rock KD, Howard CD, Morrison KE, Ravel J and Bale
TL: The composition of human vaginal microbiota transferred at
birth affects offspring health in a mouse model. Nat Commun.
12(6289)2021.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Zhuang L, Chen H, Zhang S, Zhuang J, Li Q
and Feng Z: Intestinal microbiota in early life and its
implications on childhood health. Genomics Proteomics
Bioinformatics. 17:13–25. 2019.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Niu J, Xu L, Qian Y, Sun Z, Yu D, Huang J,
Zhou X, Wang Y, Zhang T, Ren R, et al: Evolution of the gut
microbiome in early childhood: A cross-sectional study of Chinese
children. Front Microbiol. 11(439)2020.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Sarkar A, Yoo JY, Valeria Ozorio Dutra S,
Morgan KH and Groer M: The Association between early-life gut
microbiota and long-term health and diseases. J Clin Med.
10(459)2021.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Coley EJL, Mayer EA, Osadchiy V, Chen Z,
Subramanyam V, Zhang Y, Hsiao EY, Gao K, Bhatt R, Dong T, et al:
Early life adversity predicts brain-gut alterations associated with
increased stress and mood. Neurobiol Stress.
15(100348)2021.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Erabi H, Okada G, Shibasaki C, Setoyama D,
Kang D, Takamura M, Yoshino A, Fuchikami M, Kurata A, Kato TA, et
al: Kynurenic acid is a potential overlapped biomarker between
diagnosis and treatment response for depression from metabolome
analysis. Sci Rep. 10(16822)2020.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Li B, Guo K, Zeng L, Zeng B, Huo R, Luo Y,
Wang H, Dong M, Zheng P, Zhou C, et al: Metabolite identification
in fecal microbiota transplantation mouse livers and combined
proteomics with chronic unpredictive mild stress mouse livers.
Transl Psychiatry. 8(34)2018.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Behr C, Kamp H, Fabian E, Krennrich G,
Mellert W, Peter E, Strauss V, Walk T, Rietjens IMCM and van
Ravenzwaay B: Gut microbiome-related metabolic changes in plasma of
antibiotic-treated rats. Arch Toxicol. 91:3439–3454.
2017.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Padmasekar M, Savai R, Seeger W and
Pullamsetti SS: Exposomes to exosomes: Exosomes as tools to study
epigenetic adaptive mechanisms in high-altitude humans. Int J
Environ Res Public Health. 18(8280)2021.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Fischer J, Koch L, Emmerling C, Vierkotten
J, Peters T, Brüning JC and Rüther U: Inactivation of the Fto gene
protects from obesity. Nature. 458:894–898. 2009.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Sachse G, Church C, Stewart M, Cater H,
Teboul L, Cox RD and Ashcroft FM: FTO demethylase activity is
essential for normal bone growth and bone mineralization in mice.
Biochim Biophys Acta Mol Basis Dis. 1864:843–850. 2018.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Melnik BC and Schmitz G: Milk's role as an
epigenetic regulator in health and disease. Diseases.
5(12)2017.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Zhang L, Lin TV, Yuan Q, Sadoul R, Lam TT
and Bordey A: Small extracellular vesicles control dendritic spine
development through regulation of HDAC2 signaling. J Neurosci.
41:3799–3807. 2021.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Schwarzenbach H and Gahan PB: MicroRNA
shuttle from cell-to-cell by exosomes and its impact in cancer.
Noncoding RNA. 5(28)2019.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Shirazi S, Huang CC, Kang M, Lu Y,
Ravindran S and Cooper LF: The importance of cellular and exosomal
miRNAs in mesenchymal stem cell osteoblastic differentiation. Sci
Rep. 11(5953)2021.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Li C, Zhou T, Chen J, Li R, Chen H, Luo S,
Chen D, Cai C and Li W: The role of Exosomal miRNAs in cancer. J
Transl Med. 20(6)2022.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Wang Y, Zhang M and Zhou F: Biological
functions and clinical applications of exosomal long non-coding
RNAs in cancer. J Cell Mol Med. 24:11656–11666. 2020.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Zhang W, Yan Y, Peng J, Thakur A, Bai N,
Yang K and Xu Z: Decoding roles of exosomal lncRNAs in tumor-immune
regulation and therapeutic potential. Cancers (Basel).
15(286)2022.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Yang Q, Diamond MP and Al-Hendy A: Early
life adverse environmental exposures increase the risk of uterine
fibroid development: role of epigenetic regulation. Front
Pharmacol. 7(40)2016.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Díez-Sainz E, Milagro FI, Riezu-Boj JI and
Lorente-Cebrián S: Effects of gut microbiota-derived extracellular
vesicles on obesity and diabetes and their potential modulation
through diet. J Physiol Biochem. 78:485–499. 2022.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Liang X, Dai N, Sheng K, Lu H, Wang J,
Chen L and Wang Y: Gut bacterial extracellular vesicles: Important
players in regulating intestinal microenvironment. Gut Microbes.
14(2134689)2022.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Macia L, Nanan R, Hosseini-Beheshti E and
Grau GE: Host- and Microbiota-derived extracellular vesicles,
immune function, and disease development. Int J Mol Sci.
21(107)2019.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Martínez-Ruiz S, Sáez-Fuertes L,
Casanova-Crespo S, Rodríguez-Lagunas MJ, Pérez-Cano FJ, Badia J and
Baldoma L: Microbiota-Derived extracellular vesicles promote
immunity and intestinal maturation in suckling rats. Nutrients.
15(4701)2023.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Liu H, Zhang Q, Wang S, Weng W, Jing Y and
Su J: Bacterial extracellular vesicles as bioactive nanocarriers
for drug delivery: Advances and perspectives. Bioact Mater.
14:169–181. 2021.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Liu JH, Chen CY, Liu ZZ, Luo ZW, Rao SS,
Jin L, Wan TF, Yue T, Tan YJ, Yin H, et al: Extracellular vesicles
from child gut microbiota enter into bone to preserve bone mass and
strength. Adv Sci (Weinh). 8(2004831)2021.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Kaisanlahti A, Turunen J, Byts N,
Samoylenko A, Bart G, Virtanen N, Tejesvi MV, Zhyvolozhnyi A,
Sarfraz S and Kumpula S: , et al: Maternal microbiota
communicates with the fetus through microbiota-derived
extracellular vesicles. Microbiome. 11(249)2023.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Du X, Ley R and Buck AH: MicroRNAs and
extracellular vesicles in the gut: New host modulators of the
microbiome? Microlife. 2(uqab010)2021.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Feng X, Chen X, Zheng X, Zhu H, Qi Q, Liu
S, Zhang H and Che J: Latest trend of milk derived exosomes:
Cargos, functions, and applications. Front Nutr.
8(747294)2021.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Lyons KE, Ryan CA, Dempsey EM, Ross RP and
Stanton C: Breast milk, a source of beneficial microbes and
associated benefits for infant health. Nutrients.
12(1039)2020.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Duale A, Singh P and Al Khodor S: Breast
milk: A meal worth having. Front Nutr. 8(800927)2022.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Yi DY and Kim SY: Human breast milk
composition and function in human health: From nutritional
components to microbiome and MicroRNAs. Nutrients.
13(3094)2021.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Banić M, Butorac K, Čuljak N, Leboš Pavunc
A, Novak J, Bellich B, Kazazić S, Kazazić S, Cescutti P, Šušković
J, et al: The human milk microbiota produces potential therapeutic
biomolecules and shapes the intestinal microbiota of infants. Int J
Mol Sci. 23(14382)2022.PubMed/NCBI View Article : Google Scholar
|
|
91
|
Notarbartolo V, Giuffrè M, Montante C,
Corsello G and Carta M: Composition of human breast milk microbiota
and its role in children's health. Pediatr Gastroenterol Hepatol
Nutr. 25:194–210. 2022.PubMed/NCBI View Article : Google Scholar
|
|
92
|
Kim KU, Kim WH, Jeong CH, Yi DY and Min H:
More than Nutrition: Therapeutic potential of breast milk-derived
exosomes in cancer. Int J Mol Sci. 21(7327)2020.PubMed/NCBI View Article : Google Scholar
|
|
93
|
Shah J, Sims B and Martin C: Therapeutic
potential of human breast milk derived exosomes. J Nanopart Res.
24(260)2022.
|
|
94
|
Admyre C, Johansson SM, Qazi KR, Filén
JJ, Lahesmaa R, Norman M, Neve EP, Scheynius A and Gabrielsson S:
Exosomes with immune modulatory features are present in human
breast milk1. J Immunol. 179:1969–1978. 2007.PubMed/NCBI View Article : Google Scholar
|
|
95
|
de la Torre Gomez C, Goreham RV, Bech
Serra JJ, Nann T and Kussmann M: ‘Exosomics’-A review of
biophysics, biology and biochemistry of exosomes with a focus on
human breast milk. Front Genet. 9(92)2018.PubMed/NCBI View Article : Google Scholar
|
|
96
|
Mirza AH, Kaur S, Nielsen LB, Størling J,
Yarani R, Roursgaard M, Mathiesen ER, Damm P, Svare J, Mortensen HB
and Pociot F: Breast milk-derived extracellular vesicles enriched
in exosomes from mothers with type 1 diabetes contain aberrant
levels of microRNAs. Front Immunol. 10(2543)2019.PubMed/NCBI View Article : Google Scholar
|
|
97
|
Kim KU, Han K, Kim J, Kwon DH, Ji YW, Yi
DY and Min H: The protective role of exosome-derived MicroRNAs and
proteins from human breast milk against infectious agents.
Metabolites. 13(635)2023.PubMed/NCBI View Article : Google Scholar
|
|
98
|
Chiurazzi M, Cozzolino M, Reinelt T,
Nguyen TD, Elke Chie S, Natalucci G and Miletta MC: Human milk and
brain development in infants. Reprod Med. 2:107–117. 2021.
|
|
99
|
Guo MM, Zhang K and Zhang JH: Human breast
milk-derived exosomal miR-148a-3p protects against necrotizing
enterocolitis by regulating p53 and Sirtuin 1. Inflammation.
45:1254–1268. 2022.PubMed/NCBI View Article : Google Scholar
|
|
100
|
Gialeli G, Panagopoulou O, Liosis G and
Siahanidou T: Potential epigenetic effects of human milk on
infants' neurodevelopment. Nutrients. 15(3614)2023.PubMed/NCBI View Article : Google Scholar
|
|
101
|
Cintio M, Polacchini G, Scarsella E,
Montanari T, Stefanon B and Colitti M: MicroRNA Milk Exosomes: From
cellular regulator to genomic marker. Animals (Basel).
10(1126)2020.PubMed/NCBI View Article : Google Scholar
|
|
102
|
Melnik BC, Stremmel W, Weiskirchen R, John
SM and Schmitz G: Exosome-Derived MicroRNAs of human milk and their
effects on infant health and development. Biomolecules.
11(851)2021.PubMed/NCBI View Article : Google Scholar
|
|
103
|
Zhou F, Paz HA, Sadri M, Cui J, Kachman
SD, Fernando SC and Zempleni J: Dietary bovine milk exosomes elicit
changes in bacterial communities in C57BL/6 mice. Am J Physiol
Gastrointest Liver Physiol. 317:G618–G624. 2019.PubMed/NCBI View Article : Google Scholar
|
|
104
|
Turunen J, Tejesvi MV, Suokas M, Virtanen
N, Paalanne N, Kaisanlahti A, Reunanen J and Tapiainen T: Bacterial
extracellular vesicles in the microbiome of first-pass meconium in
newborn infants. Pediatr Res. 93:887–896. 2023.PubMed/NCBI View Article : Google Scholar
|
|
105
|
Holzhausen EA, Kupsco A, Chalifour BN,
Patterson WB, Schmidt KA, Mokhtari P, Baccarelli AA, Goran MI and
Alderete TL: Influence of technical and maternal-infant factors on
the measurement and expression of extracellular miRNA in human
milk. Front Immunol. 14(1151870)2023.PubMed/NCBI View Article : Google Scholar
|
|
106
|
Shah KB, Chernausek SD, Garman LD, Pezant
NP, Plows JF, Kharoud HK, Demerath EW and Fields DA: Human milk
exosomal MicroRNA: Associations with maternal overweight/obesity
and infant body composition at 1 month of life. Nutrients.
13(1091)2021.PubMed/NCBI View Article : Google Scholar
|