Introduction
Lipogenic genes exert their biological effects
through transcriptional regulation of their target genes, several
of which are key regulatory genes involved in lipogenic metabolism
(1). Sterol regulatory
element-binding protein (SREBP)-1c is a key regulator of fatty acid
metabolism and plays a pivotal role in the transcriptional
regulation of different lipogenic genes that mediate lipid
synthesis (2,3). Nuclear SREBP-1c preferentially binds
to E-box motifs, thus enhancing transcription of genes required for
saturated and unsaturated fatty acid and triglyceride biosynthesis.
During this process, the lipogenic mRNAs for ATP citrate lyase
(ACLY) and fatty acid synthase (FASN) are elevated
(4,5), whereas the lipogenic mRNAs for
carnitine O-palmitoyltransferase type I (CPT-I) are
suppressed by SREBP-1c expression (6,7).
Cancer tissue proliferates by actively using the
energy supplied by fatty acid metabolism. In cancer as well as
normal cells, the upregulation of lipogenic enzymes is
indispensable for fatty acid metabolism and high lipogenic gene
expression is typical of a wide variety of cancers (8,9).
SREBP-1c is a key transcription factor that affects
cholesterol/lipid biosynthesis and uptake. miR-33b is
embedded in SREBP-1 introns and targets several key
regulators of cholesterol trafficking and of fatty
acid/triglyceride homeostasis for post-transcriptional repression
(10–12).
Although the mechanisms that underlie lipogenic gene
overexpression in gastric cancer have not been elucidated, part of
the lipogenic pathway is intensely expressed in metaplasia and in a
subset of gastric adenocarcinomas that are characterized by disease
progression, tumor aggressiveness and poor patient survival
(13,14).
The aim of the present study was to investigate
lipogenic gene expression in cancer and non-cancer tissues from
gastric cancer patients using quantitative PCR (qPCR) analysis.
Materials and methods
mRNA quantification
Samples were obtained during surgical resection of
gastric tissues from 34 Japanese gastric cancer patients (22 male
and 12 female; median age, 69.6±10.5 years). Cancer and non-cancer
tissues were investigated. Non-cancer tissue was sampled at >5
cm from the edge of each gastric cancer nodule. Samples were frozen
in RNase Later (Ambion, Foster City, CA, USA) immediately after
surgical resection and stored at −80°C until analysis. Written
informed consent was obtained from each patient.
Total RNA was extracted from 10 mg of tissue using
the Isogen II reagent (Nippon Gene, Tokyo, Japan) according to the
manufacturer’s instructions. Complementary DNA (cDNA) was prepared
by incubating DNase-treated total RNA (0.1 μg) with
PrimeScript® II First Strand cDNA Synthesis kit (Takara
Bio, Inc., Shiga, Japan). The qPCR reaction mixture was prepared
using FastStart TaqMan® Probe Master (Rox) (Roche
Applied Science, Mannheim, Germany) or Kapa Sybr® Fast
qPCR Master mix (Kapa Biosystems, Inc., Woburn, MA, USA). Primers
for amplifying the ACLY, FASN, CPT-I and
SREBP-1 mRNAs and for miR-33b, are presented in
Table I.
 | Table ISequences of primers and probes. |
Table I
Sequences of primers and probes.
| Gene
(primer/probe) | Sequence | Product size
(bp) |
|---|
| ACLY |
| Forward |
5′-GCTTGTTGGCGTGGATGAGAA-3′ | 138 |
| Reverse |
5′-ACTCCATCTTTGGTCACTACAA-3′ | |
| Probe |
5′-ACACCTGTTGGTCCACGCCCCTGA-3′ | |
| CPT-I |
| Forward |
5′-CGGCCGATGTTACGACAGGT-3′ | 190 |
| Reverse |
5′-GATGTCGCCTTTGCAGTGCC-3′ | |
| Probe |
5′-ACTCCTGGGCAGATGCGCCGATCG-3′ | |
| FASN |
| Forward |
5′-GTGTTTGAGGATGTGGTGCTGC-3′ | 185 |
| Reverse |
5′-CTTTCCGGGTGGTCGAAGAG-3′ | |
| Probe |
5′-TGTCAGAGAACGGCAACCTGGTAGTGAGTG-3′ | |
| SREBP-1 |
| Forward |
5′-GGTAGGAGCCATGGATTGCACTT-3′ | 164 |
| Reverse |
5′-GAGGTGGAGACAAGCTGCCTG-3′ | |
| miR-33b |
| Forward |
5′-TGTCAGGCAACCGTATTCACC-3′ | 74 |
| 1st reverse |
5′-CATGCAGTGAGTTAGATGTAGAACGTGCATTGCTGT-3′ | |
| 2nd reverse |
5′-CATGCAGTGAGTTAGATGTAGAACGTG-3′ | |
| Probe |
5′-CTGTTGCATTGCACCACTCACGG-3′ | |
PCR reactions comprised 45 cycles (at 95°C for 20
sec, at 60°C for 30 sec and at 72°C for 20 sec) with the CFX96
real-time PCR Detection system (Bio-Rad, Foster City, CA, USA). The
first PCR reaction comprised 7 cycles (at 95°C for 10 sec, at 60°C
for 15 sec and at 72°C for 10 sec) using FastStart TaqMan Probe
Master (Rox) (Roche Applied Science). The second reaction comprised
45 cycles (at 95°C for 10 sec, at 60°C for 15 sec and at 72°C for
10 sec) using the CFX96 real-time PCR Detection system (Bio-Rad).
The RNA samples were quantified by relating the PCR threshold
cycles obtained from the cell line samples to the amplicon-specific
standard curves. As normalization to the GAPDH housekeeping
gene was inaccurate, the RNA expression levels were presented as
the mRNA copy number per μg total RNA.
Statistical analysis
The samples in the experiments were tested in
triplicate or quadruplicate. Data are expressed as means ± standard
deviation. Differences between the mean values were evaluated using
the Student’s t-test. P<0.05 was considered to indicate a
statistically significant difference.
Results
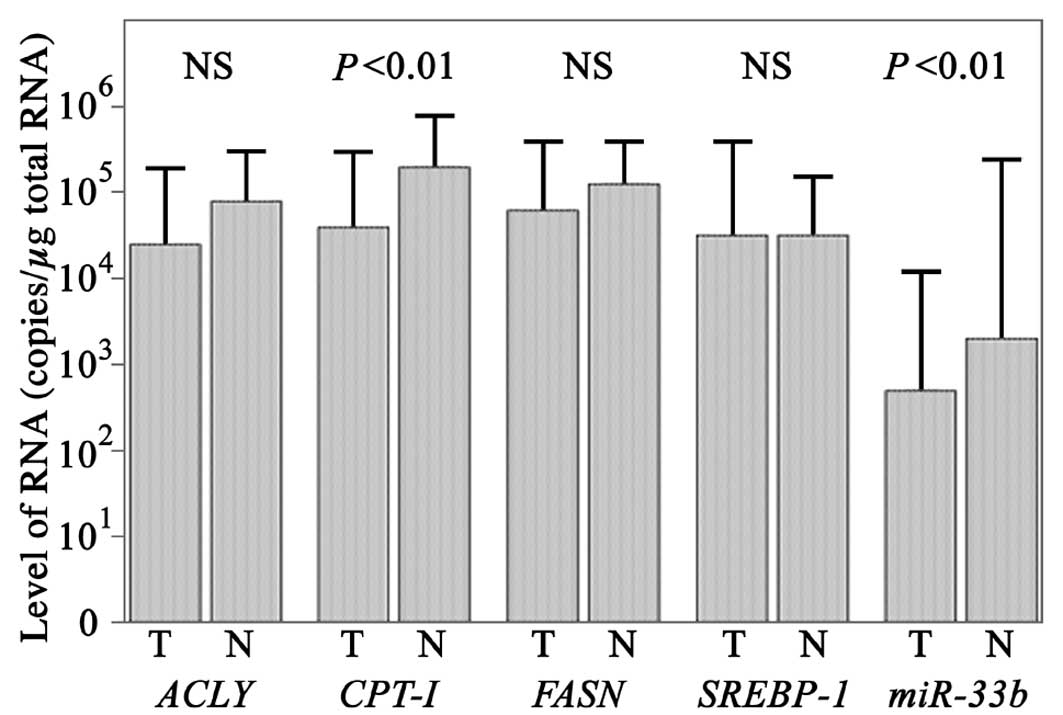
The levels of CPT-1 mRNA in cancerous tissues were
determined as 104.6±100.9 copies/μg total RNA
using qPCR (Fig. 1) and were
significantly higher in non-cancer tissues at
105.3±100.6 copies/μg total RNA (Student’s
t-test; P<0.01). The levels of miR-33b in cancerous
tissues were determined as 102.7±101.4
copies/μg total RNA and were significantly higher in non-cancerous
tissues at 103.3±102.1 copies/μg total RNA
(Student’s t-test; P<0.01).
The levels of SREBP-1 mRNA in cancer and
non-cancer tissues were determined as
104.5±101.1 and and
104.5±100.7 copies/μg total RNA, respectively
(Fig. 1), using qPCR.
The levels of ACLY mRNA in cancer and
non-cancer tissues were determined as
104.4±100.9 and
104.9±100.6 copies/μg total RNA,
respectively, while the levels of FASN mRNA in cancer and
non-cancer tissues were determined as
104.8±100.8 and
105.1±100.5 copies/μg total RNA,
respectively. The differences for SREBP-1, ACLY and
FASN were not statistically significant according to the
Student’s t-test.
Discussion
Although lipogenesis is negligible in the majority
of non-malignant adult tissues (15,16),
it is upregulated in several tumors, rendering the investigation of
endogenous lipogenesis a novel target for the prevention and/or
treatment of cancer. This limited-size study demonstrated no
statistically significant differences between the levels of mRNA
for lipogenic genes and other clinicopathological characteristics,
such as tumor size, degree of differentiation, tumor location,
stage TNM and p53 mutation. However, CPT-1 mRNA and
miR-33b expression were downregulated in the cancer tissues,
suggesting that the downregulation of CPT-1 mRNA may be part
of the mechanism responsible for SREBP upregulation and concords
with the increased lipogenesis and lipogenic enzyme expression
exhibited by a wide variety of cancers.
miR-33b mediates the transcription of its
target genes, several of which are critical to lipogenesis and
cholesterol metabolism (17,18),
including SREBP-1c, ACLY and FASN, which
increase fatty acid and triglyceride production (17,18).
SREBP-1 is synthesized as an integral protein of endoplasmic
reticulum membranes. At the nuclear level, mature SREBP-1 activates
genes that encode FAS and other lipogenic enzymes by interacting
with sterol response elements present in their promoter regions
(19). Consistent with this
hypothesis, SREBP expression was markedly stimulated by the
inhibition of miR-33b expression, which may also result in
increased fatty acid oxidation and reduced accumulation of fat in
the liver stores. Considering the promising results of the use of
anti-miRs in preclinical studies, miR-33b may become a
viable therapeutic target in the future.
Our results provide a basis for more detailed
studies on the regulation of SREBP activity and may assist in
further investigations of miR-33b as a target of gastric
cancer treatment. Although the reason for miR-33b
downregulation in cancerous tissues is uncertain, the elucidation
of its association with lipogenic genes may provide insight into
gastric carcinogenesis and lead to the development of novel
strategies for the genetic diagnosis of gastric cancer.
References
|
1
|
Li YY: Genetic and epigenetic variants
influencing the development of nonalcoholic fatty liver disease.
World J Gastroenterol. 18:6546–6551. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yokoyama C, Wang X, Briggs MR, et al:
SREBP-1, a basic-helix-loop-helix-leucine zipper protein that
controls transcription of the low density lipoprotein receptor
gene. Cell. 75:187–197. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jeon TI and Osborne TF: SREBPs: metabolic
integrators in physiology and metabolism. Trends Endocrinol Metab.
23:65–72. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lewis CA, Griffiths B, Santos CR, et al:
Genetic ablation of S6-kinase does not prevent processing of
SREBP1. Adv Enzyme Regul. 51:280–290. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rudolph MC, Monks J, Burns V, et al:
Sterol regulatory element binding protein and dietary lipid
regulation of fatty acid synthesis in the mammary epithelium. Am J
Physiol Endocrinol Metab. 299:E918–E927. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Matthews KA, Ozdemir C and Rawson RB:
Activation of sterol regulatory element binding proteins in the
absence of Scap in Drosophila melanogaster. Genetics.
185:189–198. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sampath H, Miyazaki M, Dobrzyn A, et al:
Stearoyl-CoA desaturase-1 mediates the pro-lipogenic effects of
dietary saturated fat. J Biol Chem. 282:2483–2493. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Israël M and Schwartz L: The metabolic
advantage of tumor cells. Mol Cancer. 10:702010.
|
|
9
|
Swinnen JV, Brusselmans K and Verhoeven G:
Increased lipogenesis in cancer cells: new players, novel targets.
Curr Opin Clin Nutr Metab Care. 9:358–365. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Horie T, Ono K, Horiguchi M, et al:
MicroRNA-33 encoded by an intron of sterol regulatory
element-binding protein 2 (Srebp2) regulates HDL in vivo. Proc Natl
Acad Sci USA. 107:17321–17326. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dávalos A, Goedeke L, Smibert P, et al:
miR-33a/b contribute to the regulation of fatty acid metabolism and
insulin signaling. Proc Natl Acad Sci USA. 108:9232–9237.
2011.PubMed/NCBI
|
|
12
|
Rayner KJ, Esau CC, Hussain FN, et al:
Inhibition of miR-33a/b in non-human primates raises plasma HDL and
lowers VLDL triglycerides. Nature. 478:404–407. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Perez-Tilve D, Heppner K, Kirchner H, et
al: Ghrelin-induced adiposity is independent of orexigenic effects.
FASEB J. 25:2814–2822. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Menendez JA and Lupu R: Fatty acid
synthase and the lipogenic phenotype in cancer pathogenesis. Nat
Rev Cancer. 7:763–777. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Weiss L, Hoffmann GE, Schreiber R, et al:
Fatty-acid biosynthesis in man, a pathway of minor importance.
Purification, optimal assay conditions, and organ distribution of
fatty-acid synthase. Biol Chem Hoppe Seyler. 367:905–912. 1986.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kusakabe T, Maeda M, Hoshi N, et al: Fatty
acid synthase is expressed mainly in adult hormone-sensitive cells
or cells with high lipid metabolism and in proliferating fetal
cells. J Histochem Cytochem. 48:613–622. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Najafi-Shoushtari SH, Kristo F, Li Y, et
al: MicroRNA-33 and the SREBP host genes cooperate to control
cholesterol homeostasis. Science. 328:1566–1569. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ramírez CM, Goedeke L and
Fernández-Hernando C: ‘Micromanaging’ metabolic syndrome. Cell
Cycle. 10:3249–3252. 2011.
|
|
19
|
Nohturfft A, DeBose-Boyd RA, Scheek S, et
al: Sterols regulate cycling of SREBP cleavage-activating protein
(SCAP) between endoplasmic reticulum and Golgi. Proc Natl Acad Sci
USA. 96:11235–11240. 1999. View Article : Google Scholar : PubMed/NCBI
|