Introduction
Colorectal cancer (CRC) is the third most common
gastrointestinal malignancy worldwide (1,2). In
China, with the improvement of living standards and changing of
dietary habits, the incidence of CRC exhibits a rising trend each
year, with the mortality rate rising to the fifth place (3). It has been demonstrated that
early-stage CRC may be cured by minimally invasive radical surgical
resection. However, a significant proportion of CRC patients are
diagnosed at an advanced stage, when conventional treatment options
are unavailable (4,5). Therefore, the early diagnosis of CRC
is crucial, as it may significantly reduce the mortality rate of
CRC. The pathogenesis of CRC is complicated. It was previously
reported that the occurrence of CRC is a process involving genetic
and environmental factors, including a variety of genetic
mutations, such as oncogene activation and tumor suppressor gene
inactivation (6). Therefore, the
mechanisms underlying the occurrence and the molecular pathogenesis
of CRC must be elucidated.
It was previously demonstrated that the expression
of peroxiredoxins (Prxs) is significantly upregulated in the
majority of tumors, suggesting that Prxs may play an important role
in tumor stage, invasion, recurrence and prognosis; it is also
hypothesized that the high expression of Prxs may be induced by
high levels of reactive oxygen species within the tumor cells and
may be associated with resistance to apoptosis (7). Therefore, Prxs may be used as tumor
markers.
As a member of the Prx family, the newly discovered
peroxiredoxin 4 (Prx4) has important biological functions (8,9),
through acting as an antioxidant as well as promoting cell
proliferation and differentiation and participating in cell signal
transduction. However, the prognostic significance of Prx4
expression in CRC has yet to be investigated. The aim of this study
was to elucidate the role of Prx4 in the development of CRC and the
association between Prx4 and the clinical characteristics of
CRC.
Patients and methods
Tissue samples
Fresh samples of CRC and adjacent normal tissues
were obtained from 15 patients who underwent surgical resection at
the Affiliated Hospital of Nantong University, Jiangsu, China. Upon
surgical removal, the samples were immediately snap-frozen in
liquid nitrogen. All the tissues were microdissected prior to RNA
extraction. A total of 59 CRC tissue samples were obtained from 59
patients between April, 2004 and February, 2008. The median
follow-up was 50 months. The patients comprised 36 men and 23
women, with a mean age of 65 years (range, 28–80 years). The tumors
were classified as 33 right hemicolon carcinomas and 26 left
hemicolon and rectal carcinomas, with 15 in Dukes’ stage A, 26 in
Dukes’ stage B, 14 in Dukes’ stage C and 4 in Dukes’ stage D. In
addition, 26 normal colorectal tissue samples were used as
controls. The main clinicopathological variables of the patients
are summarized in Table I. All the
specimens were fixed in 10% formalin, embedded in paraffin and cut
into 4-μm sections. All the patients had undergone surgical
resection at the Surgery Department of the Affiliated Hospital of
Nantong University. None of the patients had undergone chemotherapy
prior to surgery. The diagnoses were confirmed by pathological
examination.
 | Table IProtein expression of peroxiredoxin 4
in colorectal cancer (CRC) and adjacent normal tissues. |
Table I
Protein expression of peroxiredoxin 4
in colorectal cancer (CRC) and adjacent normal tissues.
| Tissue | Cases | Positive rate
(%) | − | + | ++ | +++ | Za | Pa |
|---|
| CRC | 26 | 88.5 | 3 | 1 | 11 | 11 | 4.237 | 0.0000 |
| Adjacent tissues | 26 | 65.4 | 9 | 10 | 7 | 0 | | |
Approval for the study was obtained from the Ethics
Committee of the Affiliated Hospital of Nantong University
according to the relevant provisions of the Declaration of Helsinki
and the people involved biomedical research ethics review method
(trial).
Quantitative polymerase chain reaction
(qPCR)
RNA was extracted from the fresh frozen cancer
tissues and adjacent normal tissues using the Biozol RNA extraction
kit (BioFlux, Tokyo, Japan). Total RNA (1 μg) was used to prepare
cDNA using random hexamers (Fermentas, Glen Burnie, MD, USA) and
was used as template in qPCR. Real-time amplifications, using
SYBR-Green detection chemistry, were run in triplicate on 96-well
reaction plates with the QuantStudio™ 7 FLex machine (Invitrogen
Life Technologies, Carlsbad, CA, USA). The reactions were prepared
in a total volume of 25 μl containing 2.0 μl cDNA, 1.0 μl of each
10 μM primer (Invitrogen Life Technologies), 12.5 μl of Maxima
SYBR-Green Master mix (Fermentas, Glen Bernie, MD, USA) and 8.5 μl
RNase/DNase-free sterile water. Blank controls were run in
triplicate for each Master mix. The cycle conditions were set as
follows: initial template denaturation at 95°C for 30 sec, followed
by 40 cycles of denaturation at 95°C for 5 sec and combined primer
annealing/elongation at 60°C for 30 sec. β-actin was used as the
endogenous control for quantitation. The forward and reverse
primers used for each molecule were as follows: Prx4: forward,
5′-TCTTTCAGATTTGACCCATCAG-3′ and reverse,
5′-AGGGCAGACTTCTCCGTGT-3′; β-actin: forward,
5′-AAGTACTCCGTGTGGATCGG-3′ and reverse, 5′-ATGCTATCACCTCCCCTGTG-3′.
This cycle was followed by a melting curve analysis, ranging from
56 to 95°C, with temperature increasing by steps of 0.5°C every 10
sec. The QuantStudio™ 7 FLex software (Life Technologies) was used
to determine baseline and threshold values automatically for all
plates. Raw Ct values were transformed to quantities using an Excel
spreadsheet generated by the authors, based on the comparative Ct
method (10).
Immunohistochemistry
The immunohistochemical staining of Prx4 was
performed using Polymer Detection system for immunohistolochemical
staining (Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd.,
Beijing, China) according to the manufacturer’s protocol. The
paraffin-embedded tissues were cut into 4-μm sections and placed on
glass slides. The sections were deparaffinized in xylene, hydrated
in descending concentrations of ethanol and rinsed with deionized
water. For each washing, 0.05 mol/l Tris-buffered saline (pH 7.4)
was used. To retrieve the antigen, the sections were boiled for 10
min in citrate buffer (pH 6.0). Endogenous peroxidase activity was
blocked for 30 min with 0.3% hydrogen peroxide. The sections were
then incubated overnight at 4°C with mouse monoclonal antibodies
against Prx4 (dilution 1:200; Abfrontier, Shanghai, China). All the
sections were sequentially incubated with PV9003 I (polymer helper)
for 20 min. After rinsing in phosphate-buffered saline, the
sections were incubated with PV9003 II (polyperoxidase-anti-mouse
IgG) for 30 min. Peroxidase activity was revealed by
3,3-diaminobenzidine staining for 10 min; counterstaining was
performed with hematoxylin and the sections were dehydrated and
coverslipped. Finally, the sections were dehydrated through graded
ethanols, cleared in xylene and mounted on glass slides. Sections
without primary antibodies were used as negative control.
Immunohistochemical evaluation
In Prx4-positive staining, the cytoplasm of the
cells was stained brown-yellow. All the immunostained sections were
evaluated by pathologists who were blinded to the clinical and
pathological variables of the patients. For Prx4 assessment,
staining intensity was scored as follows: 0, negative; 1, weak; 2,
medium; and 3, strong. The extent of the staining was scored as
follows: 0, 0%; 1, 1–25%; 2, 26–50%; 3, 51–75%; and 4, 76–100%,
according to the percentage of the positively-stained areas in
relation to the entire carcinomatous area. The sum of the staining
intensity and extent scores was used as the final staining score
for Prx4: −, 0–2; +, 3; ++, 4; and +++, ≥5. Tumors with a final
staining score of ≥3 were considered to be positive (11). In all the samples, staining was
repeated twice to avoid possible technical errors, but similar
results were obtained in these samples. The abovementioned
evaluation procedures were performed by two independent
investigators and a consensus was achieved.
Statistical analysis
The Stata v8.0 software (StataCorp LP, College
Station, TX, USA) and the SPSS v15.0 software (SPSS Inc., Chicago,
IL, USA) were used for statistical analysis. Materials with skewed
distribution were tested with the rank sum test and expressed as
median (measurement range). The survival time was estimated using
the Kaplan-Meier method and the log-rank test was used for testing
differences between groups. The multivariate Cox proportional
hazards model was applied to detect independent predictors of
survival. P<0.05 was considered to indicate a statistically
significant difference.
Results
Prx4 gene and protein expression
differences between CRC and adjacent normal tissues
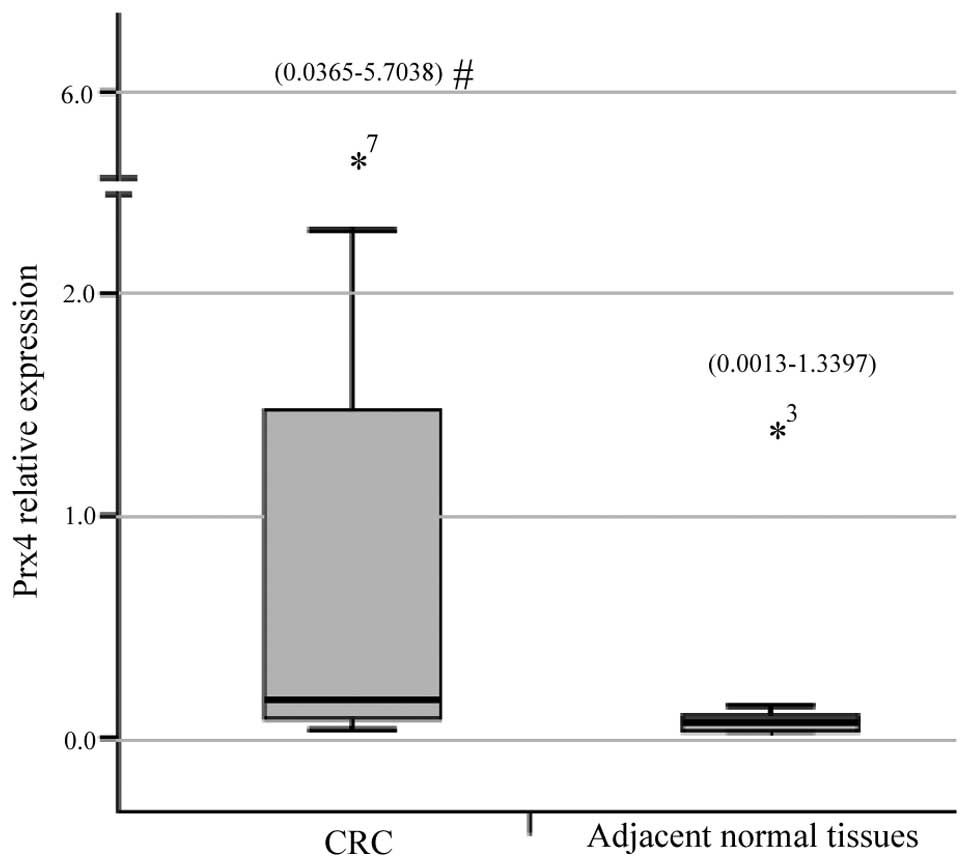
The qPCR demonstrated that the relative expression
measurements of the Prx4 gene exhibited skewed distribution and the
results for CRC tissues [0.1663 (0.0365–5.7038)] were significantly
higher compared to those for adjacent normal colorectal tissues
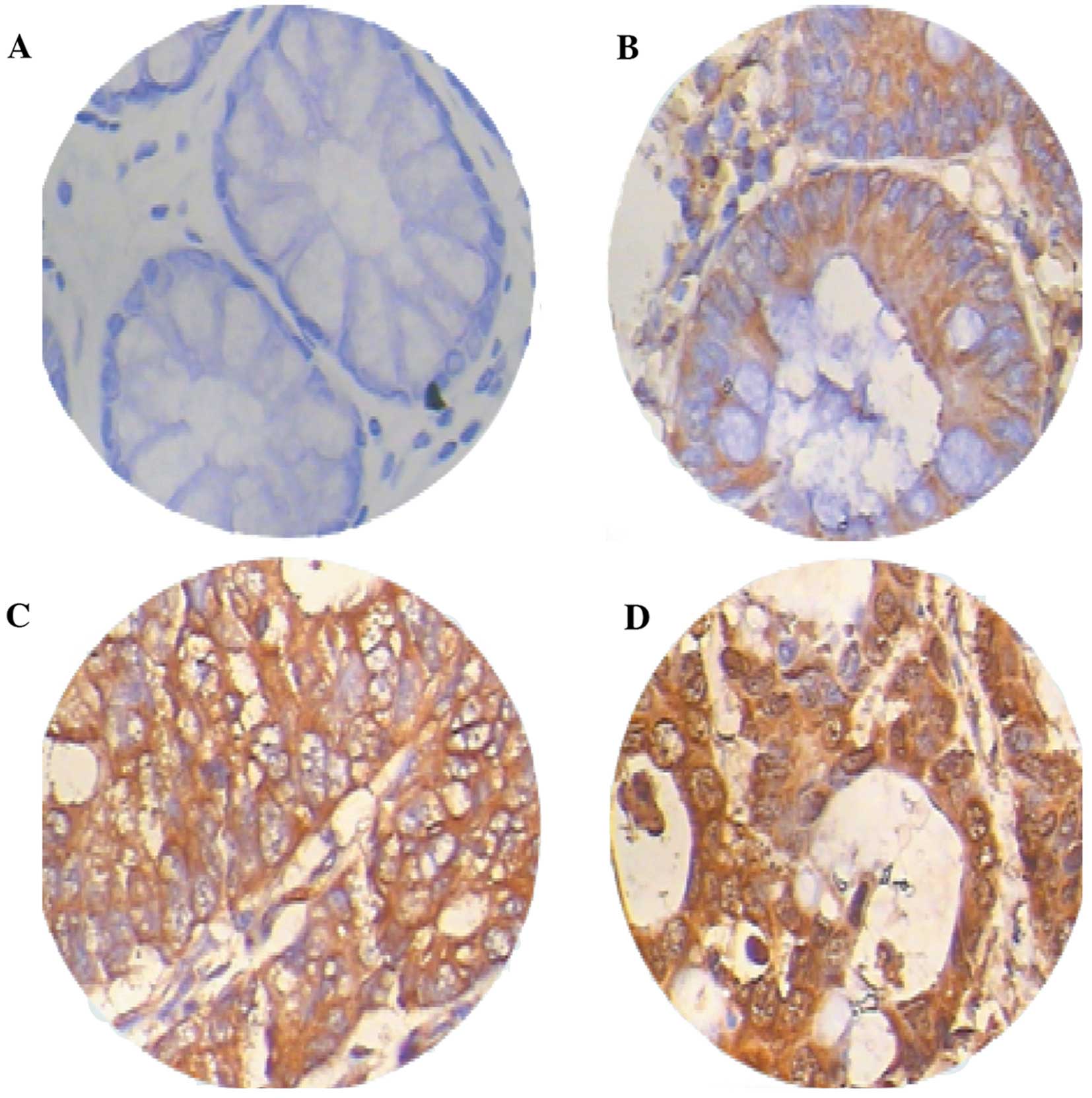
[0.0396 (0.0013–1.3397)] (Z=3.056, P=0.0022) (Fig. 1). The immunohistochemichal
examination revealed that the Prx4 protein was mainly expressed in
the endoplasmic reticulum and in the extracellular matrix,
occasionally in the nucleus, with different expression intensities.
Among the 26 samples of CRC and adjacent normal tissues, the
positive rate of Prx4 expression was 88.5% (23/26) and 65.4%
(17/26), respectively. The expression of Prx4 in the CRC tissues
(23/26) was significantly higher compared to that in the adjacent
normal tissues (17/26) (Z=4.237, P=0.0000) (Table I), with strongly positive
expression areas (++, +++) mainly concentrated in the CRC tissues
and weakly positive areas (+) mostly in the adjacent normal tissues
(Fig. 2, Table I).
Correlation of Prx4 expression with
clinicopathological variables in CRC
The clinicopathological data of the patients are
summarized in Table II. The
associations of Prx4 expression with clinicopathological variables
were evaluated. The Prx4 protein expression in CRC tissues was
significantly correlated with infiltration depth (P=0.0012), lymph
node metastasis (P=0.0061) and Dukes’ stage (P=0.0041). No
significant association was identified between overexpression of
Prx4 and gender, age, tumor location, tumor diameter, gross type,
tumor differentiation degree and distant metastasis.
 | Table IIAssociation between peroxiredoxin 4
expression and clinicopathological characteristics. |
Table II
Association between peroxiredoxin 4
expression and clinicopathological characteristics.
| Characteristics | No. (n=59) | Positive rate
(%) | − | + | ++ | +++ | Za | Pa |
|---|
| Gender | | | | | | | −0.108 | 0.9141 |
| Male | 36 | 31 (86.1) | 5 | 4 | 11 | 16 | | |
| Female | 23 | 22 (95.7) | 1 | 3 | 10 | 9 | | |
| Age, years | | | | | | | −1.660 | 0.0969 |
| <69 | 29 | 24 (82.8) | 5 | 4 | 10 | 10 | | |
| ≥69 | 30 | 29 (96.7) | 1 | 3 | 11 | 15 | | |
| Tumor location | | | | | | | −0.310 | 0.7567 |
| Right hemicolon | 33 | 31 (93.9) | 2 | 4 | 15 | 12 | | |
| Left hemicolon and
rectum | 26 | 22 (84.6) | 4 | 3 | 6 | 13 | | |
| Tumor diameter,
cm | | | | | | | −1.837 | 0.0663 |
| <5 | 32 | 26 (81.3) | 6 | 2 | 14 | 10 | | |
| ≥5 | 27 | 27 (100) | 0 | 5 | 7 | 15 | | |
| Gross type | | | | | | | 0.098 | 0.9223 |
| Massive | 27 | 25 (92.6) | 2 | 4 | 10 | 11 | | |
| Ulcerative | 32 | 28 (87.5) | 4 | 3 | 11 | 14 | | |
| Histological
differentiation | | | | | | | 1.028 | 0.5982 |
| High | 10 | 9 (90.0) | 1 | 1 | 3 | 5 | | |
| Moderate | 42 | 37 (88.1) | 5 | 5 | 16 | 16 | | |
| Poor | 7 | 7 (100) | 0 | 1 | 2 | 4 | | |
| Lymph node
metastasis | | | | | | | −2.744 | 0.0061 |
| Absent | 42 | 37 (88.1) | 5 | 7 | 17 | 13 | | |
| Present | 17 | 16 (94.1) | 1 | 0 | 4 | 12 | | |
| Distant
metastasis | | | | | | | −1.252 | 0.2106 |
| Absent | 52 | 46 (88.5) | 6 | 7 | 18 | 21 | | |
| Present | 7 | 7 (100) | 0 | 0 | 3 | 4 | | |
| Infiltration
depth | | | | | | | 15.844 | 0.0012 |
| T1 | 2 | 1 (50) | 1 | 1 | 0 | 0 | | |
| T2 | 12 | 10 (83.3) | 2 | 3 | 7 | 0 | | |
| T3 | 41 | 38 (92.7) | 3 | 3 | 14 | 21 | | |
| T4 | 4 | 4 (100) | 0 | 0 | 0 | 4 | | |
| Dukes’ stage | | | | | | | 13.248 | 0.0041 |
| A | 15 | 12 (80.0) | 3 | 4 | 7 | 1 | | |
| B | 26 | 24 (92.3) | 2 | 3 | 10 | 11 | | |
| C | 14 | 13 (92.9) | 1 | 0 | 3 | 10 | | |
| D | 4 | 4 (100) | 0 | 0 | 1 | 3 | | |
Univariate and multivariate analysis of
prognostic variables
At the end of the clinical follow-up, survival
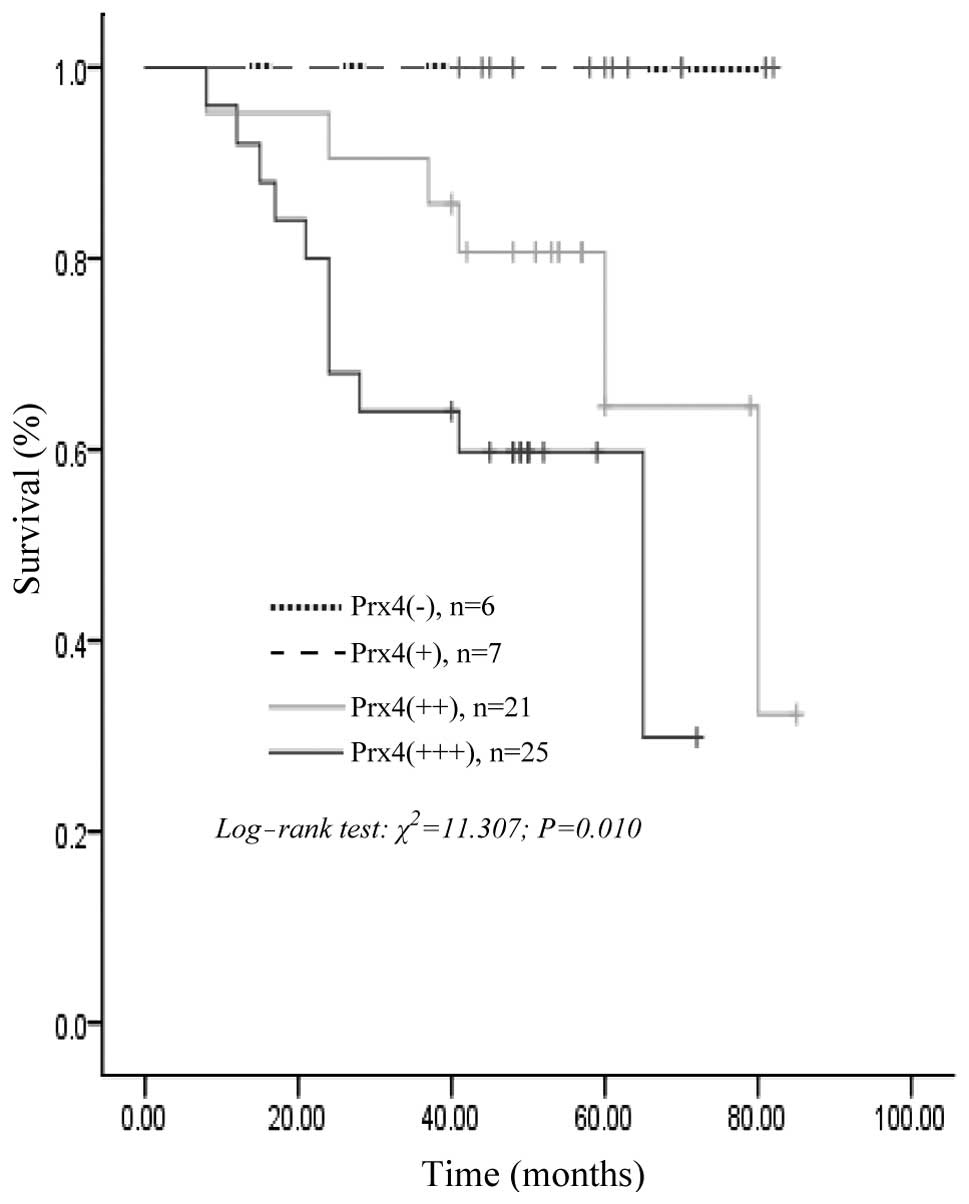
information was available for all the patients. The univariate
analysis for overall survival using the log-rank test identified
histological differentiation (P=0.015), lymph node metastasis
(P=0.000), distant metastasis (P=0.024), infiltration depth
(P=0.001), Dukes’ stage (P=0.000) and positive Prx4 expression
(P=0.010) as significant prognostic predictors. However, gender,
age, tumor location, tumor diameter and gross type were of no
prognostic value (Table III). In
59 patients with CRC, the survival rates for Prx4-positive patients
were significantly lower compared to those with negative expression
of Prx4 (log-rank test; P=0.010); the survival curve constructed
according to the Kaplan-Meier method is shown in Fig. 3. However, the multivariate Cox
regression analysis results revealed that Prx4 and other prognostic
markers by univariate analysis, including lymph node metastasis,
distant metastasis, histological differentiation, infiltration
depth and Dukes’ stage, were not independent unfavorable prognostic
factors for survival of CRC patients (P>0.05, Table III).
 | Table IIIUnivariate and multivariate analyses
of prognostic variables. |
Table III
Univariate and multivariate analyses
of prognostic variables.
| Univariatea |
Multivariateb |
|---|
|
|
|
|---|
| Variables | χ2 | P | Risk ratio | 95% CI | Z | P |
|---|
| Gender | 0.096 | 0.757 | | | | |
| Male/female | | | | | | |
| Age, years | 2.451 | 0.117 | | | | |
| <65/≥65 | | | | | | |
| Tumor location | −0.310 | 0.7567 | | | | |
| Right/left
hemicolon and rectum | | | | | | |
| Tumor diameter,
cm | 0.826 | 0.363 | | | | |
| <5/≥5 | | | | | | |
| Gross type | 0.389 | 0.533 | | | | |
|
Massive/ulcerative | | | | | | |
| Histological
differentiation | 8.418 | 0.015 | 1.375 | 0.410–4.605 | 0.266 | 0.606 |
|
High/moderate/poor | | | | | | |
| Lymph node
metastasis | 18.624 | 0.000 | 7.255 | 0.860–61.200 | 3.317 | 0.069 |
|
Absent/present | | | | | | |
| Distant
metastasis | 5.072 | 0.024 | 5.053 | 0.735–34.744 | 2.712 | 0.100 |
|
Absent/present | | | | | | |
| Infiltration
depth | 16.469 | 0.001 | 3.869 | 0.668–22.399 | 2.281 | 0.131 |
| T1/T2/T3/T4 | | | | | | |
| Dukes’ stage | 18.746 | 0.000 | 0.365 | 0.085–1.565 | 1.840 | 0.175 |
| A/B/C/D | | | | | | |
| Prx4 | 11.307 | 0.010 | 1.916 | 0.601–6.114 | 1.207 | 0.272 |
| −/+/++/+++ | | | | | | |
Discussion
Despite the advances in surgical techniques and
chemotherapy in the treatment of CRC, the cure rate for advanced
CRC remains low and the mortality rate remains high. Therefore, it
is crucial to elucidate the molecular mechanisms underlying the
development of this malignancy. The identification of the molecules
involved in the initiation and progression of CRC may be of value
for the diagnosis, targeted treatment and prediction of
prognosis.
Prxs belong to the antioxidant protein superfamily
and are abundant in prokaryotes and eukaryotes. All the Prx
proteins contain a conserved cysteine (Cys) residue in the
NH2-terminal portion of the molecule and the majority
contain an additional conserved Cys in the COOH-terminal region. A
small number of Prx proteins lack the COOH-terminal Cys. According
to the different number of conserved Cys residues, six Prx isoforms
in mammalian cells are divided into two subgroups, the 1-Cys Prx
and the 2-Cys Prx subgroups (8).
Prx6 belongs to 1-Cys Prx, whereas the other members belong to
2-Cys Prx. According to the different structures and mechanisms in
oxidation-reduction reactions, the 2-Cys Prx may be further
subdivided into two subgroups as follows: the designated 2-Cys
subgroup, including Prx1 through Prx4 and Prx5, the atypical 2-Cys
subgroup (12).
Prx4 (also referred to as AOE372 or TRANK) belongs
to the typical 2-Cys Prx that mainly localize to the endoplasmic
reticulum and extracellular matrix (13). Prx4 is overexpressed in the
pancreas, liver and cardiac tissues and its lowest expression was
found to be in brain tissue and blood granulocytes (14). Prx4 plays a key role in several
cellular functions, such as protein and lipid protection against
oxidative injury, cell proliferation, differentiation and cell
signaling transduction. Accumulating evidence also demonstrates
that Prx 4 is associated with the pathogenesis of tumors. Parkin
(15) proposed that ~20% of human
cancers are caused by infection or inflammatory conditions, such as
CRC caused by inflammatory bowel diseases (16), pancreatic cancer induced by chronic
pancreatitis (17) and liver
cancer generated from alcoholic liver disease (18). Under these conditions, the
activation of inflammatory cells may produce high levels of various
types of reactive oxygen species. Several studies reported that the
Prx4 protein is overexpressed in breast (19), ovarian (20), prostate (21) and other types of cancer (22) and Prx4 has been associated with
invasion, recurrence, prognosis and other characteristics of cancer
(23,24). It is generally believed that the
high expression of Prx proteins may be induced by high levels of
reactive oxygen species in tumor cells and is associated with its
anti-apoptotic function.
In this study, we used qPCR to investigate Prx4 mRNA
expression in CRC and adjacent normal tissues. The results
demonstrated that Prx4 mRNA expression in cancer tissues was
significantly higher compared to that in adjacent normal colorectal
tissues, suggesting that the upregulated Prx4 expression may play a
role in the occurrence and development of CRC. The
immunohistochemical examination revealed that the Prx4 protein
expression is higher in CRC compared to that in adjacent normal
tissues, with strongly positive staining areas concentrated in CRC
and weak expression areas mainly in adjacent normal tissues. This
finding suggested that a high expression of the Prx4 protein may be
associated with the pathogenesis and development of CRC. This study
also demonstrated that the expression of Prx4 was significantly
correlated with infiltration depth, lymph node metastasis and
Dukes’ stage. Although the results of the Cox regression analysis
revealed that Prx4, histological differentiation, lymph node
metastasis, distant metastasis, infiltration depth and Dukes’ stage
were not independent unfavorable prognostic factors in CRC, they
were consistently associated with survival time in CRC cases,
suggesting that Prx4 may play a role in cell proliferation,
infiltration and lymph node metastasis of CRC and its effect on
survival time may be synergistic with that of other
clinicopathological variables.
In summary, this study demonstrated that Prx4 is
overexpressed in CRC tissues and is correlated with the survival
time of postoperative CRC patients, indicating that Prx4 may be
associated with the pathogenesis and development of CRC and bears
potential as a prognostic marker and therapeutic target for
CRC.
Acknowledgements
This study was supported by grants from the
Foundation for Supporting Talents in Six Fields of Jiangsu Province
(no. 2012-WSN-065), the Health Project of Jiangsu Province (no.
H201318) and the Social Development Foundation of Nantong City
(nos. HS2011004 and BK2013069).
References
|
1
|
Esteban-Jurado C, Garre P, Vila M, et al:
New genes emerging for colorectal cancer predisposition. World J
Gastroenterol. 20:1961–1971. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gingras D and Beliveau R: Colorectal
cancer prevention through dietary and lifestyle modifications.
Cancer Microenviron. 4:133–139. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Zhang S, et al: Annual
report on status of cancer in China, 2010. Chin J Cancer Res.
26:48–58. 2014.
|
|
4
|
Mao Z, Sun J, Feng B, et al: The
metastasis suppressor, N-myc downregulated gene 1 (NDRG1), is a
prognostic biomarker for human colorectal cancer. PLoS One.
8:e682062013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Popa F, Bratucu M and Radu P: Present and
future tense in operable rectal cancer. Chirurgia (Bucur).
106:11–16. 2011.PubMed/NCBI
|
|
6
|
Cherry LM: The genetic etiology of
familial and nonfamilial colorectal cancer. Proc (Bayl Univ Med
Cent). 24:139–141. 2011.PubMed/NCBI
|
|
7
|
Hwang KE, Park DS, Kim YS, et al: Prx1
modulates the chemosensitivity of lung cancer to docetaxel through
suppression of FOXO1-induced apoptosis. Int J Oncol. 43:72–78.
2013.PubMed/NCBI
|
|
8
|
Fujii J and Ikeda Y: Advance in our
understanding of peroxiredoxin, a multifunctional, mammalian redox
protein. Redox Rep. 7:123–130. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sato Y, Kojima R, Okumura M, et al:
Synergistic cooperation of PDI family members in peroxiredoxin
4-driven oxidative protein folding. Sci Rep. 3:24562013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Spinsanti G, Zannolli R, Panti C, et al:
Quantitative real-time PCR detection of TRPV1-4 gene expression in
human leukocytes from healthy and hyposensitive subjects. Mol Pain.
4:512008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xie L, Ni WK, Chen XD, et al: The
expressions and clinical significances of tissue and serum
galectin-3 in pancreatic carcinoma. J Cancer Res Clin Oncol.
138:1035–1043. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chae HZ, Robison K, Poole LB, et al:
Cloning and sequencing of thiol-specific antioxidant from mammalian
brain: alkyl hydroperoxide reductase and thiol-specific antioxidant
define a large family of antioxidant enzymes. Proc Natl Acad Sci
USA. 91:7017–7021. 1994. View Article : Google Scholar
|
|
13
|
Tavender TJ and Bulleid NJ: Peroxiredoxin
IV protects cells from oxidative stress by removing
H2O2produced during disulphide formation. J
Cell Sci. 123:2672–2679. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Schulte J: Peroxiredoxin 4: a
multifunctional biomarker worthy of further exploration. BMC Med.
9:1372011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Parkin DM: The global health burden of
infection-associated cancers in the year 2002. Int J Cancer.
118:3030–3044. 2006.PubMed/NCBI
|
|
16
|
Lovasz BD, Lakatos L, Golovics PA, et al:
Risk of colorectal cancer in Crohn’s disease patients with colonic
involvement and stenosing disease in a population-based cohort from
Hungary. J Gastrointestin Liver Dis. 22:265–268. 2013.
|
|
17
|
Wu Y, Antony S, Hewitt SM, et al:
Functional activity and tumor-specific expression of dual oxidase 2
in pancreatic cancer cells and human malignancies characterized
with a novel monoclonal antibody. Int J Oncol. 42:1229–1238.
2013.
|
|
18
|
Machida K, Chen CL, Liu JC, et al: Cancer
stem cells generated by alcohol, diabetes, and hepatitis C virus. J
Gastroenterol Hepatol. 27(Suppl 2): 19–22. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Karihtala P, Kauppila S, Soini Y and
Arja-Jukkola-Vuorinen: Oxidative stress and counteracting
mechanisms in hormone receptor positive, triple-negative and
basal-like breast carcinomas. BMC Cancer. 11:2622011. View Article : Google Scholar
|
|
20
|
Karihtala P, Soini Y, Vaskivuo L, Bloigu R
and Puistola U: DNA adduct 8-hydroxydeoxyguanosine, a novel
putative marker of prognostic significance in ovarian carcinoma.
Int J Gynecol Cancer. 19:1047–1051. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Basu A, Banerjee H, Rojas H, et al:
Differential expression of peroxiredoxins in prostate cancer:
consistent upregulation of PRDX3 and PRDX4. Prostate. 71:755–765.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chang KP, Yu JS, Chien KY, et al:
Identification of PRDX4 and P4HA2 as metastasis-associated proteins
in oral cavity squamous cell carcinoma by comparative tissue
proteomics of microdissected specimens using iTRAQ technology. J
Proteome Res. 10:4935–4947. 2011. View Article : Google Scholar
|
|
23
|
Soini Y, Haapasaari KM, Vaarala MH,
Turpeenniemi-Hujanen T, Karja V and Karihtala P:
8-hydroxydeguanosine and nitrotyrosine are prognostic factors in
urinary bladder carcinoma. Int J Clin Exp Pathol. 4:267–275.
2011.PubMed/NCBI
|
|
24
|
Ishii T, Warabi E and Yanagawa T: Novel
roles of peroxiredoxins in inflammation, cancer and innate
immunity. J Clin Biochem Nutr. 50:91–105. 2012.PubMed/NCBI
|