Introduction
Hepatocellular carcinoma (HCC) is the fifth most
common cancer type, ranking as the third leading cause of
cancer-associated mortality due to its high invasive and metastatic
potential (1). The morbidity and
mortality of HCC is markedly higher in the China and Asian Pacific
region as a result of the prevalence of hepatitis B and C viral
infections (2). Due to the lack of an
early diagnostic system, the majority of patients with HCC are
diagnosed at advance stages. Recurrence and metastasis are the
leading causes of mortality in patients with HCC (3). Therefore, identifying novel diagnostic
and therapeutic targets of HCC is urgently required.
Human enhancer of filamentation 1 (HEF1), also
termed neural precursor cell-expressed developmentally
downregulated 9 or Crk-associated substrate lymphocyte type, was
first identified in neuronal precursor cells by Kumar et al
(4) in 1992. HEF1 is located on human
chromosome 6p25–24 and its translational product contains 843 amino
acids. According to previous studies, HEF1 is a scaffold protein
and is involved in a variety of cellular functions and behaviors,
including cell adhesion, migration, invasion, apoptosis and cell
cycle (5–8). Previous studies indicate that HEF1 is
important in the development of numerous cancer types, particularly
in the metastatic process (9–15). In prostate cancer, HEF1 promotes the
epithelial mesenchymal transition and bone invasion under the
regulation of microRNA-145 (16). In
pancreatic carcinoma, patients with a high expression of HEF1
exhibited a significantly poorer prognosis compared with those with
low expression of HEF1 (12).
However, the role of HEF1 and its clinical
significance in HCC remain to be elucidated. The aim of the present
study was to evaluate the expression of HEF1 in HCC and determine
its clinical significance. Immunohistochemistry (IHC) was performed
to examine the protein expression levels of HEF1 in 123 HCC tissues
and adjacent normal liver tissues. Correlation between the
expression of HEF1, and the clinicopathological characteristics and
patient survival were analyzed.
Materials and methods
Patient information
The present study was approved by the medical ethics
committee of Sun Yat-sen University Cancer Center (Guangdong,
China). Written informed consent was obtained from all patients
involved. In the present study, all paraffin-embedded pathological
specimens from 123 patients [107 (87.0%) men and 16 (13.0%) women;
mean age, 47.7 years] with HCC were collected from the archives of
the Department of Pathology, Sun Yat-Sen University Cancer Center.
Patients enrolled in the present study were diagnosed with HCC
between July 2005 and May 2008, undergoing primary and curative
resection for tumor without preoperative anticancer treatment. The
average follow-up time was 26.79 months (median, 28.0; range,
1.0–61 months). Clinicopathological characteristics of these
patients, including age, sex, hepatitis history, serum
α-fetoprotein (AFP), liver cirrhosis, tumor number, size,
differentiation, vascular invasion, relapse and tumor, node
metastasis (TNM) stage are summarized in Table I. Tumor differentiation was
determined, according to the criteria proposed by Edmonson and
Steiner (17). The histological grade
and clinical stage of the tumors were defined, according to the
International Union Against Cancer TNM classification system
(18).
 | Table I.Correlation between HEF1 expression
and the clinicopathological variables of primary hepatocellular
carcinoma cases. |
Table I.
Correlation between HEF1 expression
and the clinicopathological variables of primary hepatocellular
carcinoma cases.
|
|
| HEF1 expression, n
(%) |
|
|---|
|
|
|
|
|
|---|
| Variable | No. cases
(n=123) | Low | High | P-valuea |
|---|
| Age
(years)b |
|
|
| 0.647 |
|
≤48.3 | 60 | 28 (46.7) | 32 (53.3) |
|
|
>48.3 | 63 | 32 (50.8) | 31 (49.2) |
|
| Gender |
|
|
| 0.712 |
| Male | 106 | 51 (48.1) | 55 (51.9) |
|
|
Female | 17 | 9 (52.9) | 8 (47.1) |
|
| AFP (ng/ml) |
|
|
| 0.423 |
| ≤20 | 59 | 31 (52.5) | 28 (47.5) |
|
|
>20 | 64 | 29 (45.3) | 35 (54.7) |
|
| Liver cirrhosis |
|
|
| 0.443 |
| Yes | 86 | 40 (46.5) | 46 (53.5) |
|
| No | 37 | 20 (54.1) | 17 (45.9) |
|
| Tumor size (cm) |
|
|
| 0.103 |
| ≤5 | 75 | 41 (54.7) | 34 (45.3) |
|
|
>5 | 48 | 19 (39.6) | 29 (60.4) |
|
| Tumor
multiplicity |
|
|
| 0.077 |
|
Single | 85 | 46 (54.1) | 39 (45.9) |
|
|
Multiple | 38 | 14 (36.8) | 24 (63.2) |
|
| Differentiation |
|
|
| 0.439 |
|
Well | 15 | 8 (53.3) | 7 (46.7) |
|
|
Moderate | 71 | 36 (50.7) | 35 (49.3) |
|
|
Poor | 31 | 15 (48.4) | 16 (51.6) |
|
|
Undifferentiated | 6 | 1 (16.7) | 5 (83.3) |
|
| Stage |
|
|
| 0.019c |
| I | 10 | 8 (80.0) | 2 (20.0) |
|
| II | 54 | 29 (53.7) | 25 (46.3) |
|
|
III | 50 | 22 (44.0) | 28 (56.0) |
|
| IV | 9 | 1 (11.1) | 8 (88.9) |
|
| Vascular
invasion |
|
|
| 0.008c |
|
Yes | 58 | 21 (36.2) | 37 (63.8) |
|
| No | 65 | 39 (60.0) | 26 (40.0) |
|
| Relapse |
|
|
| 0.083 |
|
Yes | 42 | 16 (38.1) | 26 (61.9) |
|
| No | 81 | 44 (54.3) | 14 (45.7) |
|
Tissue microarray (TMA)
construction
TMA was constructed, as previously described
(19). Briefly, triplicates of the
core tissue biopsies (0.6 mm diameter) were obtained from the
representative tumor area and adjacent non-malignant liver tissue
of each donor tissue block, which was rearranged in recipient
paraffin blocks (tissue array blocks) using a tissue arraying
instrument (Beecher Instruments, Silver Spring, MD, USA).
IHC
IHC was performed to assess the altered protein
expression in 123 HCC tissues. Briefly, TMA slides were heated at
60°C for 2 h prior to deparaffinization with xylene (Guangzhou
Chemical Reagent Factory, Guangzhou, China) and rehydrated in
graded alcohol. The tissue sections were boiled in antigen
retrieval buffer for 15 min in a microwave oven to retrieve
antigen, followed by incubation with 3% hydrogen peroxide in
methanol to quench endogenous peroxidase activity. Following this,
1% bovine serum albumin (Sigma-Aldrich, St. Louis, MO, USA) was
used to block non-specific binding. Subsequently, the tissue
sections were incubated with rabbit anti-human anti-HEF1 (cat. no.
ab18056; 10 µg/ml; Abcam, Burlingame, CA, USA) at 4°C overnight.
Replacement of the primary antibody with phosphate buffered saline
was performed as a negative control. Following incubation with
rabbit anti-mouse peroxidase/DAB-conjugated secondary antibody
(Envision; Dako, Glostrup, Denmark) for 30 min at 37°C, the slides
were incubated with 3,3-diaminobenzidine solution at room
temperature for visualization. The tissue samples were subsequently
dehydrated and mounted using neutral balsam (Sigma-Aldrich).
IHC evaluation
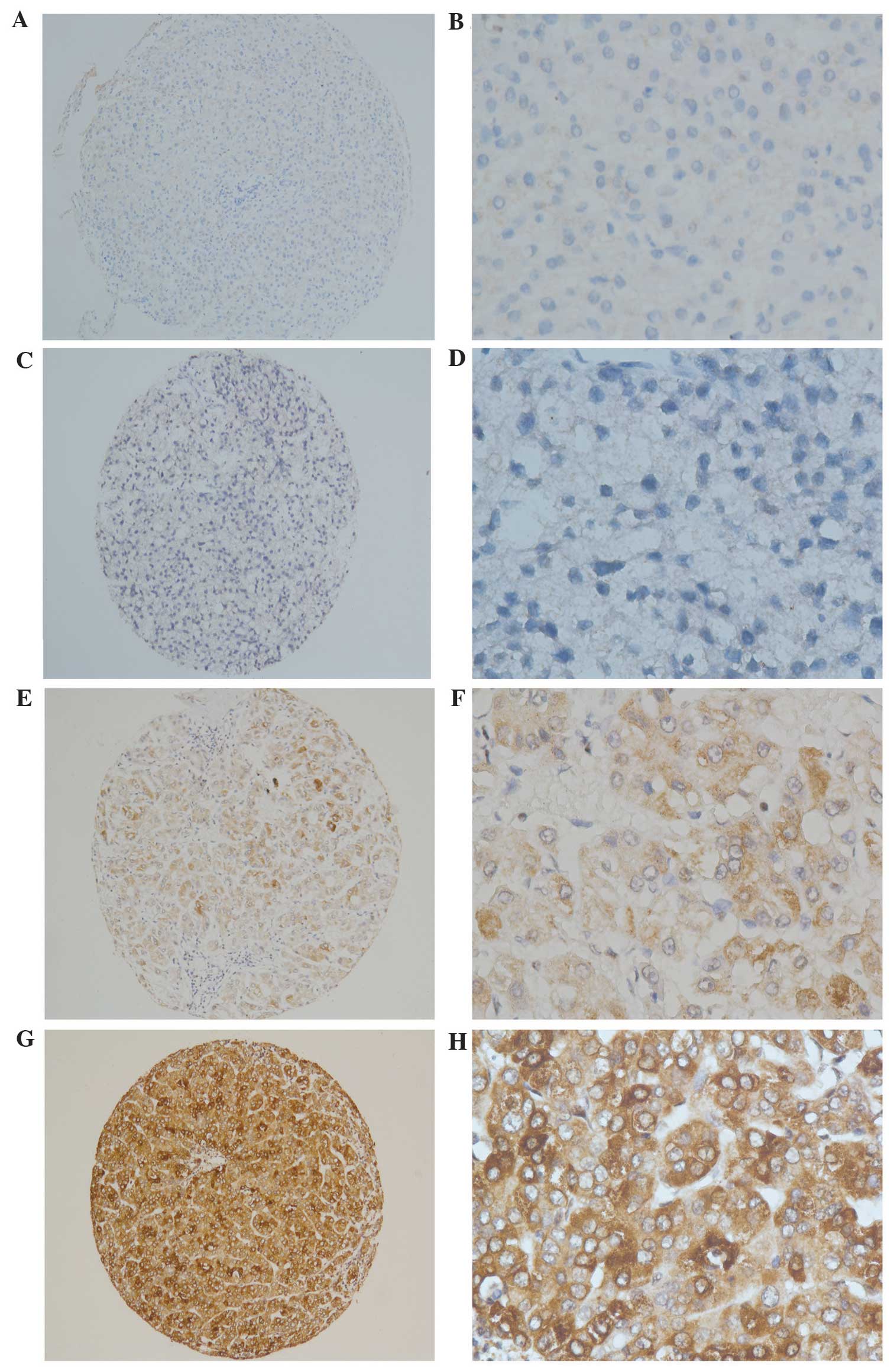
The immunoreactivity score (IRS) for HEF1 expression
were determined by two independent pathologists, in a blinded
manner (Fig. 1). The staining results
were scored based on the following criteria: i) Percentage of
positive tumor cells in the tumor tissue: 0 (0%), 1 (1–10%), 2
(11–50%), 3 (51–75%) and 4 (76–100%); ii) signal intensity: 0 (no
signal), 1 (weak), 2 (moderate) and 3 (marked). The IRS was
calculated by multiplying the score for the percentage of positive
cells by the intensity score (range, 0–12). The specimens were
rescored if the difference between the two pathologists was >3.
The definition of low and high expression levels of HEF1 was
determined by the median of the IRS results. Representative figures
are shown in Fig. 1.
Statistical analysis
Statistical analysis was performed using SPSS 17.0
software (SPSS, Inc., Chicago, IL, USA). The correlation between
the expression of HEF1 and clinicopathological variables were
assessed using Pearson's χ2 test. The overall survival
(OS) and disease free survival (DFS) were assessed by Kaplan-Meier
curve. The Cox regression model was used for multivariate survival
analysis. A two-sided P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression of HEF1 in HCC and adjacent
non-malignant liver tissues by IHC
The clinicopathological characteristics of the 123
patients are listed in Table I.
Immunostaining of HEF1 in HCC and normal hepatocyte tissues were
indicated as brown yellow granules in the cytoplasm (Fig. 1). The expression of HEF1 was divided
into two subgroups: Low and high HEF1 expression, as defined by IHC
evaluation. High expression of HEF1 was detected in 38.2% (47/123)
adjacent non-malignant normal tissues, whereas high expression of
HEF1 was detected in 51.2% (63/123) of HCC cases. Paired sample
tests revealed that the positive rate of HEF1 expression between
normal and HCC cases was statistically significant (Table II; P<0.05).
 | Table II.Comparison of HEF1 expression in 123
paired HCC and adjacent non-malignant normal tissues. |
Table II.
Comparison of HEF1 expression in 123
paired HCC and adjacent non-malignant normal tissues.
|
|
| HEF1 expression,
n |
|
|---|
|
|
|
|
|
|---|
| Tissue | Number | Low | High | % high
expression |
|---|
| Normala | 123 | 76 | 47 | 38.2 |
| HCCa | 123 | 60 | 63 | 51.2 |
Association between HEF1 expression
and clinicopathological variables
The association between immunohistochemical HEF1
expression in HCC tissues and various clinicopathological features
of patients with HCC were analyzed by Pearson's χ2 test
and are listed in Table I. The
expression of HEF1 correlated closely with TNM stage (P=0.019) and
vascular invasion status (P=0.008). However, no statistical
correlations were observed between HEF1 expression and age, sex,
AFP levels, liver cirrhosis, tumor size, tumor multiplicity,
differentiation and cancer relapse (P=0.647, 0.712, 0.423, 0.443,
0.103, 0.077, 0.439 and 0.083 respectively).
HEF1 expression and survival
All 123 hepatocellular carcinoma patients underwent
5 year follow-ups. The OS was defined as the duration between the
date of initial surgery and mortality, or the most recent
follow-up. The 5 year OS indicated by the Kaplan-Meier survival
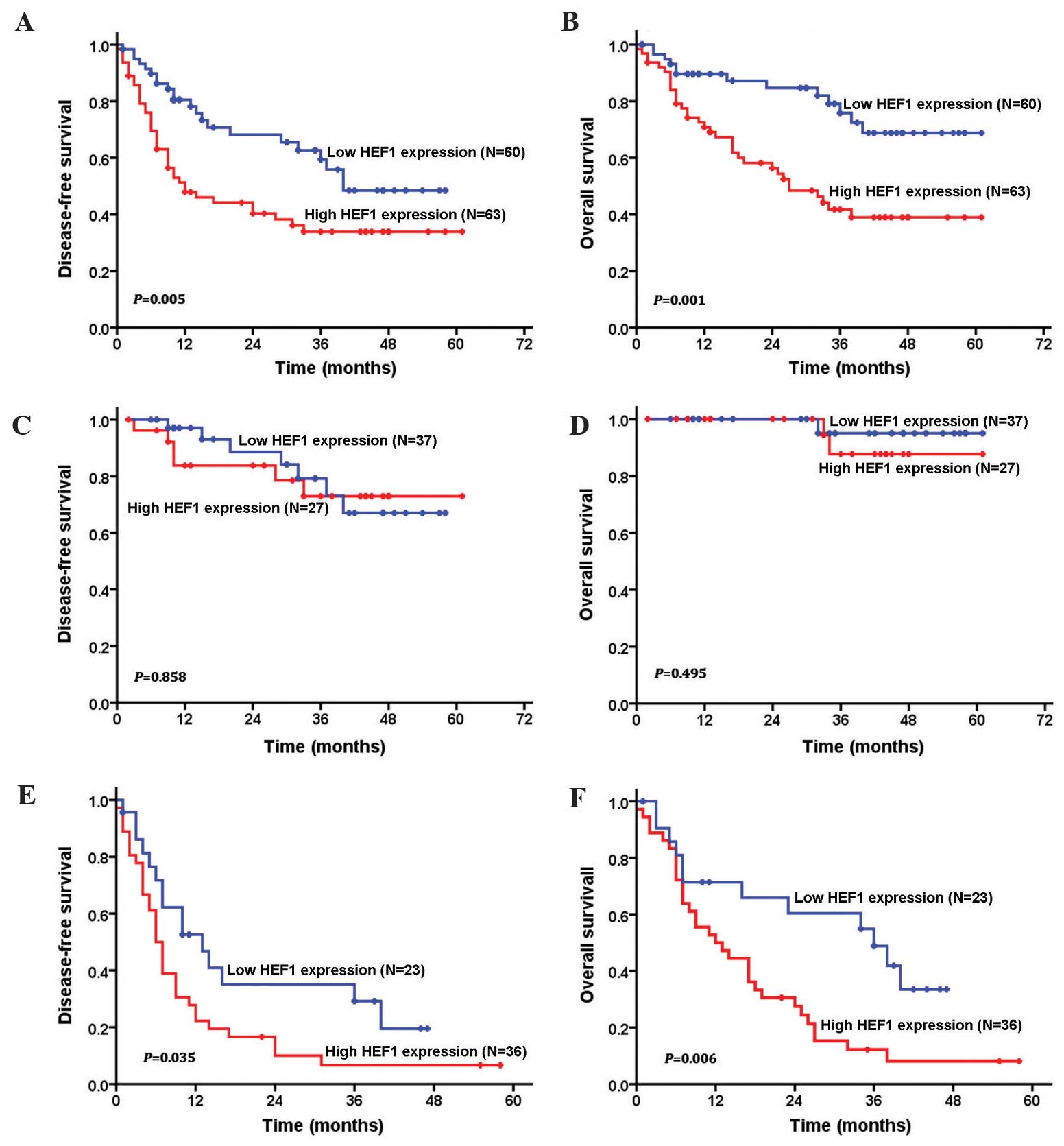
curve of low and high HEF1 expression is shown in Fig. 2. Patients with low expression of HEF1
had a significantly longer OS and DFS when compared with the
patients exhibiting high expression of HEF1 (P=0.001). The 5 year
survival rates for low and high HEF1 expression patients were 68.8
and 38.9%, respectively. The mean survival duration of patients
with low expression levels of HEF1 was 49.2 months, whereas the
mean survival time of patients with high expression levels of HEF1
was 33.4 months.
In addition, the present study also examined the
prognostic value of immunohistochemical HEF1 expression in
different subgroups of patients with HCC, according to the
classical TNM stage classification. Significant correlations
between high expression of HEF1, and shorter OS and DFS time were
observed in advanced TNM (stage III+IV) subgroups (P<0.05;
Fig. 2), which suggested that HEF1
expression is an effective prognostic marker for HCC patients in
both early and advanced stages.
Furthermore, univariate and multivariate analyses
were used to assess the prognostic value of HEF1 expression in HCC.
The results revealed that the following variables correlated
significantly with OS: AFP levels, tumor size, tumor multiplicity,
TNM stage, vascular invasion, cancer relapse and HEF1 expression
(Table III). Multivariate analysis
indicated that only HEF1 expression, serum AFP level and TNM stages
were independent factors, which affected the OS (P<0.05;
Table III).
 | Table III.Univariate and multivariate analysis
of different prognostic factors in 123 patients with hepatocellular
carcinoma using Cox Proportional Hazards Regression. |
Table III.
Univariate and multivariate analysis
of different prognostic factors in 123 patients with hepatocellular
carcinoma using Cox Proportional Hazards Regression.
|
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|
|---|
| Variable | All cases | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age
(years)a |
|
| 0.720 |
|
|
|
≤48.3 | 60 | 1.0 |
|
|
|
|
>48.3 | 63 | 1.111
(0.626–1.971) |
|
|
|
| Gender |
|
| 0.519 |
|
|
|
Male | 106 | 1.327
(0.562–3.133) |
|
|
|
|
Female | 17 | 1.0 |
|
|
|
| AFP (ng/ml) |
|
| <0.001 |
| 0.005 |
|
≤20 | 59 | 1.0 |
| 1.0 |
|
|
>20 | 64 | 3.363
(1.790–6.319) |
| 2.694
(1.359–5.337) |
|
| Liver
cirrhosis |
|
| 0.754 |
|
|
|
Yes | 86 | 1.106
(0.590–2.071) |
|
|
|
| No | 37 | 1.0 |
|
|
|
| Tumor size
(cm) |
|
| <0.001 |
| 0.956 |
| ≤5 | 75 | 1.0 |
| 1.0 |
|
|
>5 | 48 | 2.911
(1.619–5.234) |
| 0.981
(0.492–1.956) |
|
| Tumor
multiplicity |
|
| <0.001 |
| 0.369 |
|
Single | 85 | 1.0 |
| 1.0 |
|
|
Multiple | 38 | 3.749
(2.099–6.696) |
| 1.347
(0.703–2.581) |
|
|
Differentiation |
|
| 0.103 |
|
|
|
Well-moderate | 86 | 1.0 |
|
|
|
|
Poor-undifferentiated | 37 | 1.642
(0.905–2.980) |
|
|
|
| Stage |
|
| <0.001 |
| 0.007 |
|
I–II | 64 | 1.0 |
| 1.0 |
|
|
III–IV | 59 | 25.241
(7.812–81.551) |
| 11.406
(1.933–67.300) |
|
| Vascular
invasion |
|
| <0.001 |
| 0.314 |
|
Yes | 58 | 18.394
(6.554–51.619) |
| 2.172
(0.479–9.839) |
|
| No | 65 | 1.0 |
| 1.0 |
|
| Relapse |
|
| <0.001 |
| 0.653 |
|
Yes | 42 | 2.925
(1.630–5.250) |
| 0.860
(0.445–1.662) |
|
| No | 81 | 1.0 |
| 1.0 |
|
| HEF1
expression |
|
| 0.001 |
| 0.028 |
|
Low | 60 | 1.0 |
| 1.0 |
|
|
High | 63 | 2.876
(1.515–5.460) |
| 2.212
(1.091–4.484) |
|
Discussion
HEF1, also termed neural precursor cell-expressed
developmentally downregulated 9 or Crk-associated substrate
lymphocyte type, is identified as a member of the Crk-associated
substrate protein family (4).
Although no enzymatic domains have been determined in the amino
acid sequence of HEF1, its cytoplasmic location and numerous
tyrosine residues for phosphorylation by tyrosine kinases enable it
to function as a scaffold protein for intracellular signal
transduction (20). Previous studies
indicate that HEF1 is involved in a diversity of cellular functions
and behaviors, including cell adhesion, migration, invasion,
apoptosis and cell cycle (5–8).
In particular, overexpression of HEF1 and its
association with unfavorable prognosis have been revealed in
diverse cancer types, including melanoma, bladder cancer, ovarian
cancer, gastric cancer and colorectal cancer (11,21–24). In
addition, Cox analysis and the Kaplan-Meier method revealed that
HEF1 expression is an independent prognostic factor in these cancer
types and may serve as a predictive biomarker for these patients.
The present study of HEF1 in HCC is consistent with these findings.
A significantly upregulated HEF1 expression was observed in HCC
tissues compared with in normal non-malignant liver tissues, as
shown in Table II. Correlation
analysis between the expression of HEF1 and the clinicopathological
parameters of patients with HCC revealed that HEF1 overexpression
was associated with advanced TNM stage and vascular invasion,
indicating that HEF1 expression may be responsible for tumor
progression and metastasis in HCC. Further analysis revealed that
HEF1 expression is an independent prognostic factor for patients
with HCC, suggesting its potential to serve as an effective
biomarker for prognosis prediction and therapeutic target.
In terms of the mechanisms by which HEF1 regulates
cancer cell growth and metastasis, certain early studies showed
that HEF1 level is decreased in metastatic samples when compared
with primary tumors, indicating that it may serve a role in the
initial invasion process from the primary site (25). Numerous possible mechanisms have been
proposed in different cancer types (11,26).
McLaughlin et al (27)
reported that HEF1 expression is more frequently observed in
invasive phenotypes of breast cancer cells and is crucial for the
protease-dependent mesenchymal invasion process through the
regulation of matrix metalloproteinase 14, a key enzyme in
extracellular matrix degradation and tumor invasion. Other evidence
includes the discovery of the association between HEF1 and Aurora A
kinase (AURKA) in cancer cells. AURKA is an oncoprotein, which is
tightly associated with decreased survival and tumor metastasis.
Ice et al (28) showed that
binding of HEF1 with AURKA is critical for Aurora stabilization
and, therefore, protein level. A combination therapy with HEF1 RNA
interference and AURKA inhibitors revealed significant effects on
inhibiting growth and distant metastasis of xenografts of breast
tumors in mice. Another line of evidence for the role of HEF1 in
the growth of breast cancer cells is about cell surface-associated
glycan structure. Iida et al (29) demonstrated that HEF1 in breast cancer
cells enhances the expression of Chondroitin sulfate-E, which is a
key progression and metastasis regulator in breast cancer. This
provided a novel mechanism by which HEF1 promotes the malignant
phenotypes of breast cancer cells. Consistently, in the present
study, high expression of HEF1 was markedly associated with
vascular invasion, TNM stages and unfavorable prognosis in HCC.
However, the specific mechanism by which HEF1 enables HCC cells to
acquire growth and metastasis advantages remains to be
elucidated.
Staging of patients with HCC is critical for the
prognosis prediction and selection of therapy. HCC is distinctive
from other solid malignancies since its prognosis not only depends
upon the tumor stage, but also on the liver function impairment,
due to the accompanying cirrhosis. Individual specificity and
treatment plans are also important in the prognosis of a particular
patient (30). Therefore, a
comprehensive and effective staging system for HCC must take
traditional TNM stage, liver function parameters and various
molecular markers, including VEGF, Ki67, p53, c-myc and E-adherin,
into consideration (31). Nowadays,
various staging systems for HCC, including the European systems,
Barcelona Clinic Liver Cancer staging system, Chinese University
Prognostic Index, Japan integrated Staging and the cancer of the
liver Italian program, are working towards this goal. Japan
integrated staging has recently added biomarkers (AFP, DCP, AFP-L3)
into its evaluation system and extensive studies are focused on the
discovery of novel biomarkers, including HEF1 (32,33). As
shown in the present study, the expression of HEF1 correlated with
tumor metastasis and can be used as an independent prognostic
factor for HCC patients. It is anticipated that it can be used in
combination with other critical parameters to assess prognosis and
guide treatment.
In conclusion, the present study showed that
increased HEF1 expression is markedly associated with advanced
tumor stage, vascular invasion and poorer clinical outcomes of
patients with HCC. In addition, HEF1 expression was revealed to be
an independent prognostic factor for HCC patients, suggesting that
it may serve as an effective biomarker for prognosis prediction.
However, the detailed mechanisms by which HEF1 promotes tumor
invasion and metastasis in HCC requires further studies.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wei KR, Yu X, Zheng RS, Peng XB, Zhang SW,
Ji MF, Liang ZH, Ou ZX and Chen WQ: Incidence and mortality of
liver cancer in China, 2010. Chin J Cancer. 33:388–394.
2014.PubMed/NCBI
|
|
3
|
Fan JH, Wang JB, Jiang Y, Xiang W, Liang
H, Wei WQ, Qiao YL and Boffetta P: Attributable causes of liver
cancer mortality and incidence in China. Asian Pac J Cancer Prev.
14:7251–7256. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kumar S, Tomooka Y and Noda M:
Identification of a set of genes with developmentally
down-regulated expression in the mouse brain. Biochem Biophys Res
Commun. 185:1155–1161. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Feng Y, Wang Y, Wang Z, Fang Z, Li F, Gao
Y, Liu H, Xiao T, Li F, Zhou Y, et al: The CRTC1-NEDD9 signaling
axis mediates lung cancer progression caused by LKB1 loss. Cancer
Res. 72:6502–6511. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Guerrero MS, Parsons JT and Bouton AH: Cas
and NEDD9 contribute to tumor progression through dynamic
regulation of the cytoskeleton. Genes Cancer. 3:371–381. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Han T, Yi XP, Liu B, Ke MJ and Li YX:
MicroRNA-145 suppresses cell proliferation, invasion and migration
in pancreatic cancer cells by targeting NEDD9. Mol Med Rep.
11:4115–4120. 2015.PubMed/NCBI
|
|
8
|
Morimoto K, Tanaka T, Nitta Y, Ohnishi K,
Kawashima H and Nakatani T: NEDD9 crucially regulates
TGF-β-triggered epithelial-mesenchymal transition and cell invasion
in prostate cancer cells, Involvement in cancer progressiveness.
Prostate. 74:901–910. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Feng J, Zhao J, Xie H, Yin Y, Luo G, Zhang
J, Feng Y and Li Z: Involvement of NEDD9 in the invasion and
migration of gastric cancer. Tumour Biol. 36:3621–3628. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jin Y, Li F, Zheng C, Wang Y, Fang Z, Guo
C, Wang X, Liu H, Deng L, Li C, et al: NEDD9 promotes lung cancer
metastasis through epithelial-mesenchymal transition. Int J Cancer.
134:2294–2304. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Štajduhar E, Sedić M, Leniček T, Radulović
P, Kerenji A, Krušlin B, Pavelić K and Kraljević Pavelić S:
Expression of growth hormone receptor, plakoglobin and NEDD9
protein in association with tumour progression and metastasis in
human breast cancer. Tumour Biol. 35:6425–6434. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xue YZ, Sheng YY, Liu ZL, Wei ZQ, Cao HY,
Wu YM, Lu YF, Yu LH, Li JP and Li ZS: Expression of NEDD9 in
pancreatic ductal adenocarcinoma and its clinical significance.
Tumour Biol. 34:895–899. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li P, Zhou H, Zhu X, Ma G, Liu C, Lin B
and Mao W: High expression of NEDD9 predicts adverse outcomes of
colorectal cancer patients. Int J Clin Exp Pathol. 7:2565–2570.
2014.PubMed/NCBI
|
|
14
|
Kondo S, Iwata S, Yamada T, Inoue Y,
Ichihara H, Kichikawa Y, Katayose T, Souta-Kuribara A, Yamazaki H,
Hosono O, et al: Impact of the integrin signaling adaptor protein
NEDD9 on prognosis and metastatic behavior of human lung cancer.
Clin Cancer Res. 18:6326–6338. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sasaki T, Iwata S, Okano HJ, Urasaki Y,
Hamada J, Tanaka H, Dang NH, Okano H and Morimoto C: Nedd9 protein,
a Cas-L homologue, is upregulated after transient global ischemia
in rats, Possible involvement of Nedd9 in the differentiation of
neurons after ischemia. Stroke. 36:2457–2462. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Guo W, Ren D, Chen X, Tu X, Huang S, Wang
M, Song L, Zou X and Peng X: HEF1 promotes epithelial mesenchymal
transition and bone invasion in prostate cancer under the
regulation of microRNA-145. J Cell Biochem. 114:1606–1615. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Edmonson H and Steiner PE: Primary
carcinoma of the liver: A study of 100 cases among 48,900
necropsies. Cancer. 7:4621954. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sobin LH and Fleming ID: TNM
classification of malignant tumors fifth edition (1997).
Histopathology. Cancer. 80:1803–1804. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cai MY, Zhang B, He WP, Yang GF, Rao HL,
Rao ZY, Wu QL, Guan XY, Kung HF, Zeng YX and Xie D: Decreased
expression of PinX1 protein is correlated with tumor development
and is a new independent poor prognostic factor in ovarian
carcinoma. Cancer Sci. 101:1543–1549. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
O'Neill GM, Seo S, Serebriiskii IG, Lessin
SR and Golemis EA: A new central scaffold for metastasis, Parsing
HEF1/Cas-L/NEDD9. Cancer Res. 67:8975–8979. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang Q, Wang HJ, Zhang DH, Ru GQ, He XJ
and Ma YY: High expression of HEF1 is associated with poor
prognosis in urinary bladder carcinoma. Onco Targets Ther.
7:1319–1326. 2014.PubMed/NCBI
|
|
22
|
Wang H, Mu X, Zhou S, Zhang J, Dai J, Tang
L, Xiao L, Duan Z, Jia L and Chen S: NEDD9 overexpression is
associated with the progression of and an unfavorable prognosis in
epithelial ovarian cancer. Hum Pathol. 45:401–408. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang SS, Wu LH, Liu Q, Chen KS and Zhang
XF: Elevated expression of NEDD9 is associated with metastatic
activity in gastric cancer. Onco Targets Ther. 8:633–640. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xia D, Holla VR, Wang D, Menter DG and
DuBois RN: HEF1 is a crucial mediator of the proliferative effects
of prostaglandin E(2) on colon cancer cells. Cancer Res.
70:824–831. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Izumchenko E, Singh MK, Plotnikova OV,
Tikhmyanova N, Little JL, Serebriiskii IG, Seo S, Kurokawa M,
Egleston BL, Klein-Szanto A, et al: NEDD9 promotes oncogenic
signaling in mammary tumor development. Cancer Res. 69:7198–7206.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chang JX, Gao F, Zhao GQ and Zhang GJ:
Role of NEDD9 in invasion and metastasis of lung adenocarcinoma.
Exp Ther Med. 4:795–800. 2012.PubMed/NCBI
|
|
27
|
McLaughlin SL, Ice RJ, Rajulapati A,
Kozyulina PY, Livengood RH, Kozyreva VK, Loskutov YV, Culp MV, Weed
SA, Ivanov AV and Pugacheva EN: NEDD9 depletion leads to MMP14
inactivation by TIMP2 and prevents invasion and metastasis. Mol
Cancer Res. 12:69–81. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ice RJ, McLaughlin SL, Livengood RH, Culp
MV, Eddy ER, Ivanov AV and Pugacheva EN: NEDD9 depletion
destabilizes Aurora A kinase and heightens the efficacy of Aurora A
inhibitors: Implications for treatment of metastatic solid tumors.
Cancer Res. 73:3168–3180. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Iida J, Dorchak J, Clancy R, Slavik J,
Ellsworth R, Katagiri Y, Pugacheva EN, van Kuppevelt TH, Mural RJ,
Cutler ML and Shriver CD: Role for chondroitin sulfate
glycosaminoglycan in NEDD9-mediated breast cancer cell growth. Exp
Cell Res. 330:358–370. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bruix J and Llovet JM: Prognostic
prediction and treatment strategy in hepatocellular carcinoma.
Hepatology. 35:519–524. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Witjes CD, van Aalten SM, Steyerberg EW,
Borsboom GJ, de Man RA, Verhoef C and Ijzermans JN: Recently
introduced biomarkers for screening of hepatocellular carcinoma: A
systematic review and meta-analysis. Hepatol Int. 7:59–64. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kitai S, Kudo M, Minami Y, Ueshima K,
Chung H, Hagiwara S, Inoue T, Ishikawa E, Takahashi S, Asakuma Y,
et al: A new prognostic staging system for hepatocellular
carcinoma: Value of the biomarker combined Japan integrated staging
score. Intervirology. 51((Suppl 1)): S86–S94. 2008. View Article : Google Scholar
|
|
33
|
Nishikawa H, Kita R, Kimura T, Endo M,
Ohara Y, Sakamoto A, Saito S, Nishijima N, Nasu A, Komekado H and
Osaki Y: Proposal of the performance status combined Japan
integrated staging system in hepatocellular carcinoma complicated
with cirrhosis. Int J Oncol. 46:2371–2379. 2015.PubMed/NCBI
|