Introduction
Colorectal cancer is one of the most common
malignant diseases. In 2009, there were 146,970 new patients and
49,920 mortalities in the United States, with colorectal cancer
ranking third overall in terms of incidence and mortality in men
and women (1). Colorectal cancer is a
multifactorial disease, resulting from complex interactions between
environmental factors and genetic mutations. A number of studies
have revealed that diet may be involved in the development of
colorectal cancer (2,3). The abnormal intake of animal meat, fat,
vegetables and vitamins may increase the risk of development of
colorectal cancer. In addition, family history and genetic factors
are also closely associated with colorectal cancer; for example,
MutS homolog 2 (MSH2) and MutL homolog 1 (MLH1) gene mutations are
associated with hereditary non-polyposis colorectal cancer
(3).
Aberrant cellular proliferation is closely
associated with the development of cancers. The cyclin family is
considered to exert a key role in cell proliferation. Cyclin D1
(CCND1) is a major regulator protein, which fulfills a critical
role during transition from the growth (G)1 to the synthesis (S)
phase, promoting the progression of the cell cycle during cell
mitosis by adhering to cyclin-dependent kinases (CDKs) 4 and 6
(4,5).
The constitutively increased expression of CCND1 is observed in
numerous malignant cancers, and is associated with a poor prognosis
(6–8).
Single nucleotide polymorphisms (SNPs) are able to
change the structure of the genome and influence protein expression
and function, which leads to abnormal cell proliferation and an
increased risk of cancer (9). The
most common mutation locus of the CCND1 gene is at codon 242, with
a nucleotide change from guanine (G) to adenine (A) in exon 4. The
A allele increases the frequency of alternative splicing during
cell transcription, leading to an elevated level of CCND1, and
consequently resulting in abnormal cell proliferation and an escape
from apoptosis (10).
In 2009, Kong et al (11) reported the first case-control study on
the CCND1 G870A polymorphism and colorectal cancer risk, but no
significant differences in genotyping were observed in a population
in the United States. To date, several molecular epidemiological
investigations have been performed to evaluate the association
between the CCND1 G870A polymorphism and colorectal cancer
susceptibility, although the results were inconsistent. In the
present investigation, a meta-analysis of published case-control
studies was therefore performed to precisely assess the association
between the CCND1 G870A polymorphism and the risk of colorectal
cancer.
Materials and methods
This meta-analysis was designed according to the
guidelines of the Preferred Reporting Items for Systematic Reviews
and Meta-analyses (PRISMA Compliant) statement (12).
Search strategy
A comprehensive online search of the PubMed and
Embase databases was performed for studies published up to June 1
2015 using the following search terms: ‘CCND1’, ‘cyclin D1’,
‘colorectal cancer’, ‘colon cancer’, ‘rectum cancer’ and
‘polymorphism’. Additional studies were searched for from the
references of the retrieved studies, or from review articles on
this topic. The following criteria were used to include identified
studies in this meta-analysis: (i) a case-control study of the
CCND1 G870A polymorphism and colorectal cancer risk; and (ii)
sufficient data for estimating odds ratios (ORs) with 95%
confidence intervals (CIs). In cases of partly or completely
overlapping data, only the latest study, or the study with the
larger sample, was included (13,14).
Data extraction
The following data were extracted from all selected
studies independently by two investigators (Xiao-Ming Xu and
Xiao-Bing Ni): the first author's name, publication data, country
origin, racial descent of the study population (Asian, Caucasian or
mixed), sources of the controls, genotype distribution, genotyping
methods, adherence to the Hardy-Weinberg equilibrium (HWE), minor
allele frequency (MAF) in controls, and tumor subtypes.
Statistical analysis
Five genotype models were evaluated based on ORs and
95% CIs to assess the potential association between CCND1 G870A
polymorphisms and the risk of colorectal cancer: An allele contrast
model (A vs. G), a pair of co-dominant models (AA vs. GG and GA vs.
GG), a dominant model (GA+AA vs. GG) and a recessive model (AA vs.
GG+GA). Subgroup analyses were performed according to ethnicity and
control design. The study heterogeneity was assessed using
Cochran's Q statistic and the I2 statistic
(15). ORs were pooled using a random
effects model with the inverse variance (I–V) method (or the
DerSimonian and Laird method) when statistical heterogeneity was
identified to exist (P<0.10 or I2>50%)
(16); otherwise, a fixed effects
model (the Mantel-Haenszel method) was adopted (17).
Funnel plots and Egger's linear regression method
were used to assess any possible publication bias (18). Cumulative meta-analyses were also
performed to identify possible trends in the pooled estimate
according to the publication year (19).
All statistical analyses were performed using
Stata® version 11.0 (Stata Corporation, College Station,
TX, USA). A two-sided P<0.05 was considered to indicate a
statistically significant value.
Results
Study characteristics
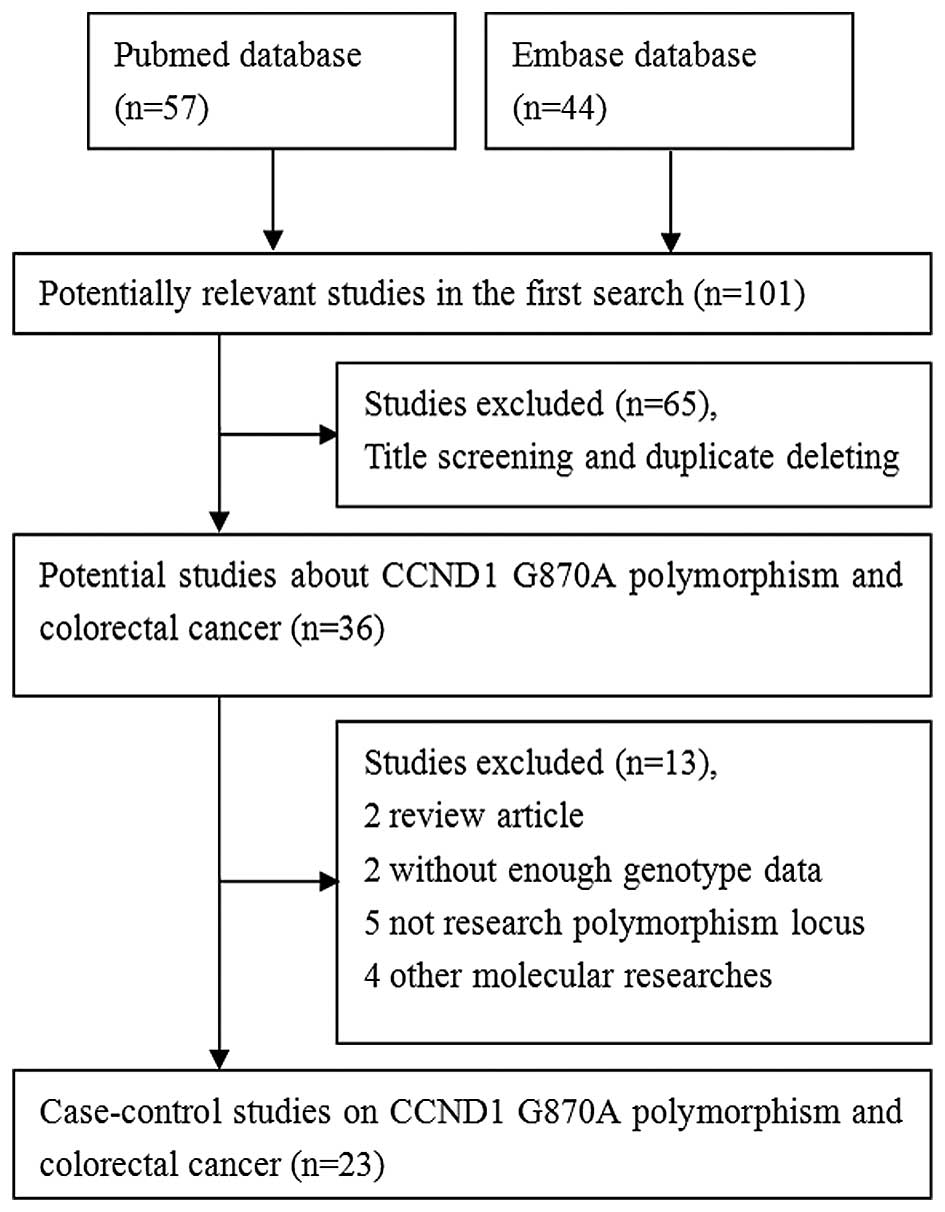
A total of 101 associated studies were searched.
During the first step, while screening the title and screening for
duplicates, 65 studies were excluded. Of the remaining 36 articles,
13 were excluded since two were reviews, two did not include
sufficient genotype data, five were not focused on the polymorphism
locus, and four were on other molecular studies. The flow chart of
study selection is shown in Fig. 1.
Ultimately, 23 published case-control studies met the inclusion
criteria, including 6,320 patients with colorectal cancer and 8,352
controls (11,20–41). The
genotype distribution in each study is shown in Table I. The diverse genotyping methods
included polymerase chain reaction-restriction fragment length
polymorphism (PCR-RFLP), PCR-single strand conformation
polymorphism, PCR Sequenase™ and TaqMan®. Overall, the
MAF in controls ranged between 0.412 and 0.614 in Caucasians, and
between 0.182 and 0.634 in Asians. In only one study did the
control population significantly deviate from the HWE (29).
 | Table I.Characteristics of case-control
studies included in the present meta-analysis. |
Table I.
Characteristics of case-control
studies included in the present meta-analysis.
|
| Genotype
distribution |
|
|---|
|
|
|
|
|---|
|
| Case | Control |
|
|---|
|
|
|
|
|
|---|
| Authors | Year | Country/region | Racial descent | Source of
controls | Case | Control | GG | GA | AA | GG | GA | AA | Genotyping
type | P-value for
HWEa | MAF | Type | Refs. |
|---|
| Kong et
al | 2000 | USA | Caucasian | Family-control | 49 | 37 | 9 | 36 | 4 | 10 | 21 | 6 | PCR-SSCP | 0.366 | 0.446 | HNPCC | (11) |
| McKay et
al | 2000 | UK | Caucasian |
Population-control | 100 | 101 | 25 | 58 | 17 | 34 | 50 | 17 | PCR-RFLP | 0.849 | 0.416 | sCRC | (20) |
| Kong et
al | 2001 | USA | Caucasian | Healthy
control | 156 | 152 | 36 | 71 | 49 | 45 | 84 | 23 | PCR-SSCP | 0.112 | 0.428 | sCRC | (21) |
| Bala et
al | 2001 | Finland | Caucasian | Family-control | 146 | 186 | 50 | 70 | 26 | 47 | 97 | 42 | PCR-SSCP | 0.551 | 0.487 | HNPCC | (22) |
| Porter et
al | 2002 | UK | Caucasian |
Hospital-control | 334 | 171 | 85 | 175 | 74 | 60 | 81 | 30 | PCR-RFLP | 0.768 | 0.412 | CRC | (23) |
|
|
|
| Caucasian |
Hospital-control | 99 | 171 | 30 | 47 | 22 | 60 | 81 | 30 | PCR-RFLP | 0.768 | 0.412 | HNPCC |
|
|
|
|
| Caucasian |
Hospital-control | 128 | 171 | 34 | 65 | 29 | 60 | 81 | 30 | PCR-RFLP | 0.768 | 0.412 | sCRC |
|
| Le Marchand et
al | 2003 | USA | Mixed |
Population-control | 504 | 624 | 109 | 253 | 142 | 164 | 315 | 145 | PCR-RFLP | 0.792 | 0.485 | CRC | (24) |
|
|
|
| Caucasian |
Population-control | 208 | 244 | 34 | 110 | 64 | 68 | 120 | 56 | PCR-RFLP | 0.792 | 0.475 | CRC |
|
|
|
|
| Asian |
Population-control | 296 | 380 | 75 | 143 | 78 | 96 | 195 | 89 | PCR-RFLP | 0.792 | 0.491 | CRC |
|
| Grieu et
al | 2003 | Australia | Caucasian |
Hospital-control | 569 | 327 | 142 | 313 | 114 | 90 | 158 | 79 | PCR-SSCP | 0.556 | 0.483 | sCRC | (25) |
| Hong et
al | 2005 | Singapore | Asian | Healthy
control | 254 | 101 | 55 | 128 | 71 | 12 | 50 | 39 | PCR-RFLP | 0.505 | 0.634 | sCRC | (26) |
| Jiang et
al | 2006 | India | Asian | Healthy
control | 301 | 291 | 46 | 130 | 125 | 56 | 145 | 90 | PCR-RFLP | 0.860 | 0.558 | CRC | (27) |
| Schernhammer et
al | 2006 | USA | Caucasian |
Population-control | 610 | 1,237 | 125 | 311 | 174 | 264 | 593 | 380 |
TaqMan® | 0.250 | 0.614 | CRC | (28) |
| Huang et
al | 2006 | China | Asian |
Hospital-control | 831 | 1,052 | 126 | 411 | 294 | 199 | 464 | 389 | PCR-RFLP | 0.004 | 0.590 | sCRC | (29) |
| Probst-Hensch et
al | 2006 | Singapore | Asian |
Population-control | 300 | 1,169 | 56 | 132 | 112 | 207 | 548 | 414 |
TaqMan® | 0.272 | 0.589 | CRC | (30) |
| Krüger et
al | 2006 | Germany | Caucasian |
Population-control | 406 | 245 | 141 | 188 | 77 | 73 | 121 | 51 | PCR Sequenase™ | 0.947 | 0.455 | HNPCC | (31) |
| Grünhage et
al | 2008 | Germany | Caucasian |
Hospital-control | 194 | 218 | 37 | 93 | 64 | 48 | 109 | 61 | PCR-RFLP | 0.958 | 0.530 | CRC | (32) |
|
|
|
| Caucasian |
Hospital-control | 98 | 218 | 13 | 50 | 35 | 48 | 109 | 61 | PCR-RFLP | 0.958 | 0.530 | HNPCC |
|
|
|
|
| Caucasian |
Hospital-control | 96 | 218 | 24 | 43 | 29 | 48 | 109 | 61 | PCR-RFLP | 0.958 | 0.530 | sCRC |
|
| Tan et
al | 2008 | Germany | Caucasian |
Population-control | 498 | 600 | 120 | 263 | 115 | 147 | 310 | 143 | PCR-RFLP | 0.414 | 0.497 | CRC | (33) |
| Forones et
al | 2008 | Brazil | Mixed |
Hospital-control | 123 | 120 | 36 | 66 | 21 | 34 | 67 | 19 | PCR-RFLP | 0.141 | 0.438 | CRC | (34) |
| Talseth et
al | 2008 | Australia | Caucasian |
Hospital-control | 157 | 153 | 34 | 78 | 45 | 42 | 80 | 31 |
TaqMan® | 0.527 | 0.464 | HNPCC | (39) |
| Liu et
al | 2010 | China | Asian |
Population-control | 373 | 838 | 66 | 187 | 120 | 160 | 429 | 249 | PCR-RFLP | 0.303 | 0.553 | CRC | (35) |
| Kanaan et
al | 2010 | USA | Caucasian |
Hospital-control | 75 | 93 | 19 | 39 | 17 | 24 | 48 | 21 |
TaqMan® | 0.748 | 0.484 | sCRC | (36) |
| Yaylim-Eraltan
et al | 2010 | Turkey | Caucasian |
Hospital-control | 57 | 117 | 9 | 28 | 20 | 29 | 60 | 28 | PCR-RFLP | 0.781 | 0.496 | CRC | (37) |
| Jelonek et
al | 2010 | Poland | Caucasian |
Population-control | 50 | 153 | 12 | 33 | 5 | 44 | 71 | 38 | PCR-RFLP | 0.383 | 0.480 | CRC | (38) |
| Sameer et
al | 2013 | India | Asian | Healthy
control | 130 | 160 | 19 | 70 | 41 | 41 | 76 | 43 | PCR-RFLP | 0.528 | 0.506 | CRC | (40) |
| Govatati et
al | 2014 | India | Asian | Healthy
control | 102 | 107 | 54 | 39 | 10 | 71 | 33 | 3 |
TaqMan® | 0.719 | 0.182 | CRC | (41) |
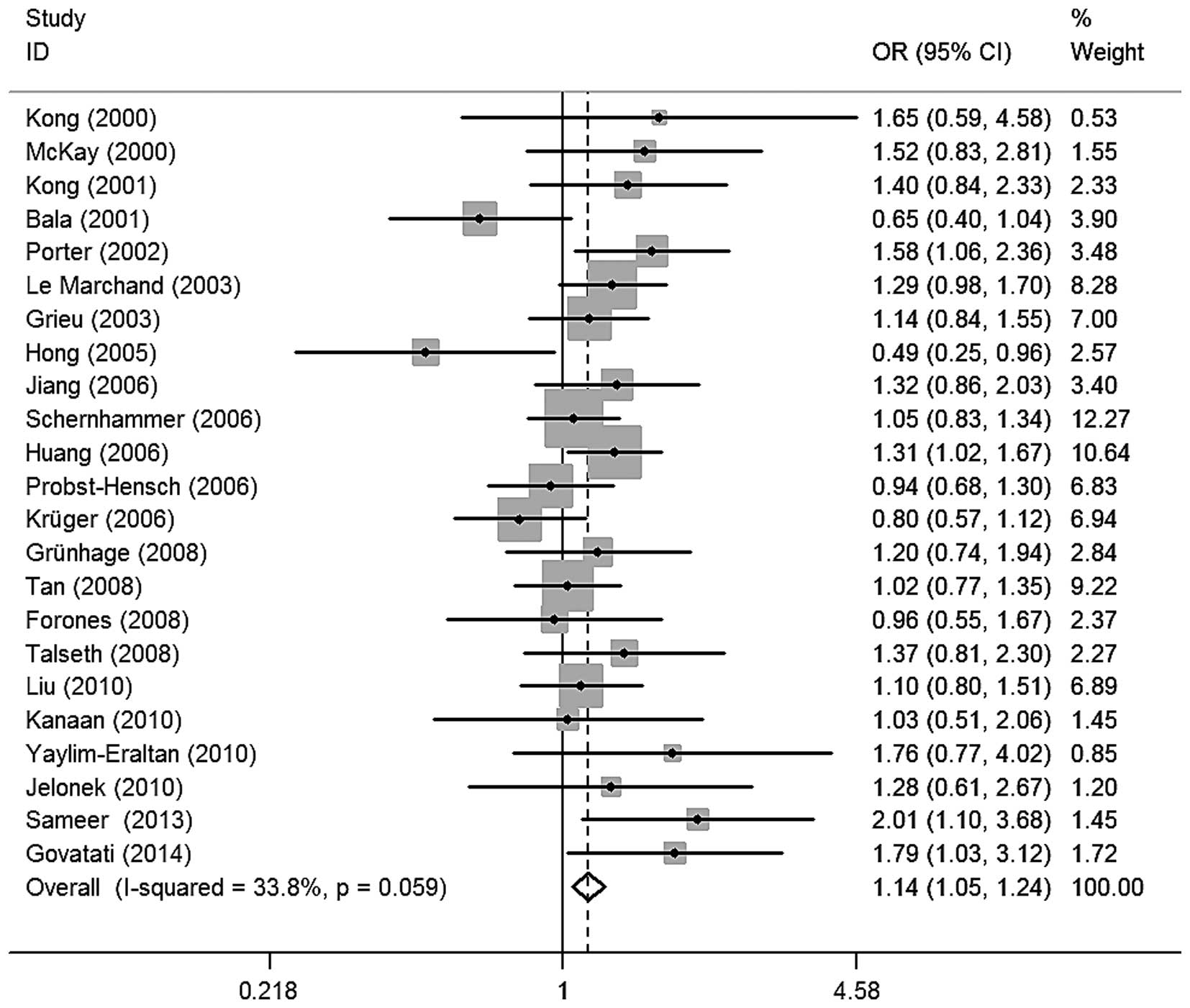
Meta-analysis
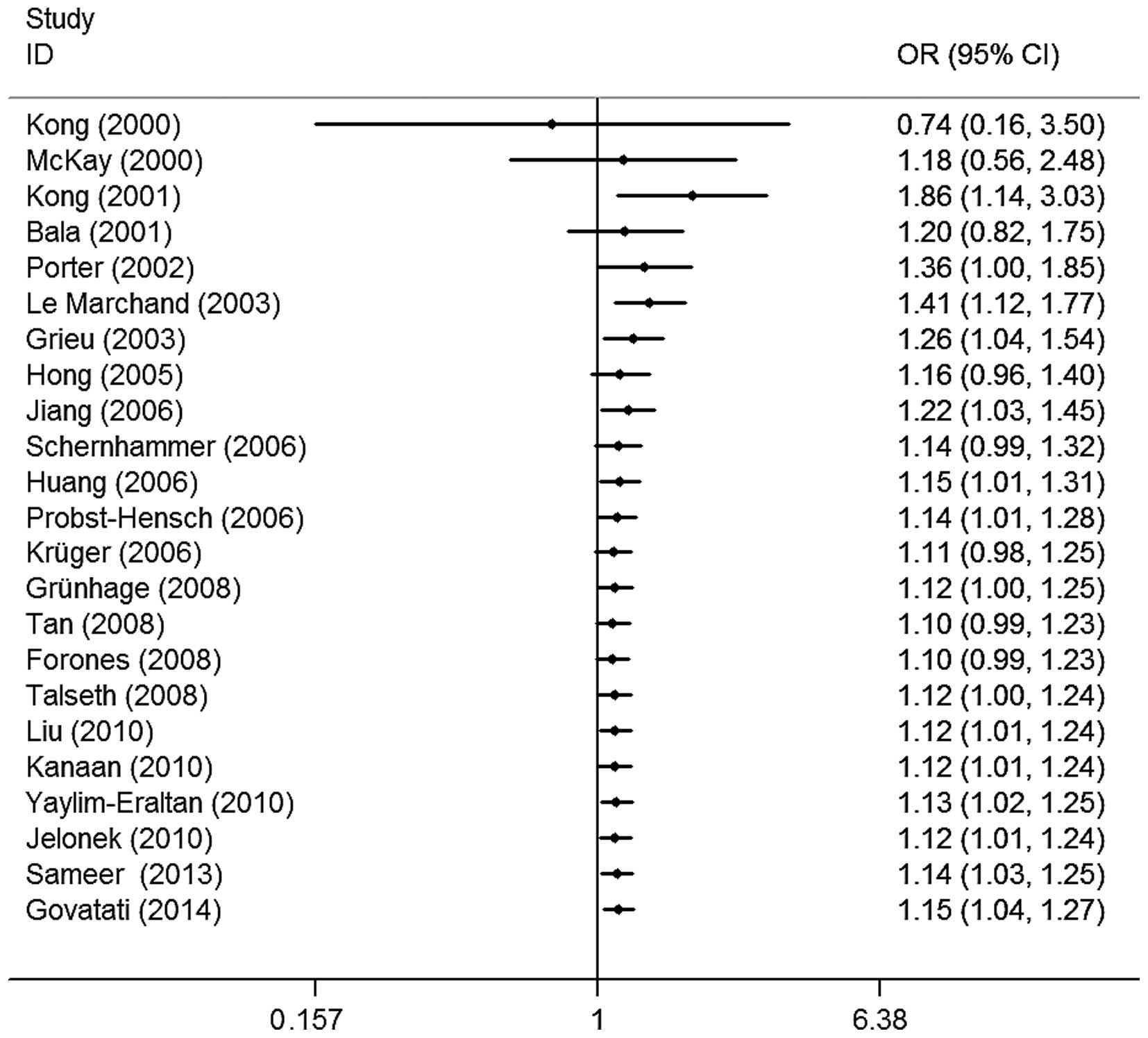
The evaluation of the association between CCND1
G870A polymorphisms and colorectal cancer risk is shown in Table II. Overall, four of the genetic
models revealed that the CCND1 G870A polymorphism was significantly
associated with an increased risk of colorectal cancer (A vs. G,
OR=1.09, 95% CI=1.00–1.18, I2=54.3%; GA vs. GG,
OR=1.13, 95% CI=1.04–1.24, I2=18.2%; AA vs. GG,
OR=1.17, 95% CI=1.00–1.38, I2=52.5%; and GA+AA
vs. GG, OR=1.14, 95% CI=1.05–1.24, I2=33.8%;
Fig. 2). Similarly, significant risk
effects were detected in the subgroup analysis of patients with
sporadic colorectal cancer (GA vs. GG, OR=1.21, 95% CI=1.04–1.42,
I2=24.1%; GA+AA vs. GG, OR=1.18, 95%
CI=1.02–1.37, I2=35.0%) and Caucasians (GA vs.
GG, OR=1.14, 95% CI=1.02–1.28, I2=19.8%; GA+AA
vs. GG, OR=1.14, 95% CI=1.02–1.27, I2=37.5%).
Significantly increased risks were also detected in the stratified
analysis of hospital-based studies and studies employing the
PCR-PRFLP genotyping method (Table
II).
 | Table II.Summary of the ORs and 95% CIs of the
CCND1 G870A polymorphism and colorectal cancer risk. |
Table II.
Summary of the ORs and 95% CIs of the
CCND1 G870A polymorphism and colorectal cancer risk.
|
|
| A vs. G | GA vs. GG | AA vs. GG | GA+AA vs. GG | AA vs. GG+GA |
|---|
|
|
|
|
|
|
|
|
|---|
| Parameter | Na | OR | 95% CI | P-value |
I2 | OR | 95% CI | P-value |
I2 | OR | 95% CI | P-value |
I2 | OR | 95% CI | P-value |
I2 | OR | 95% CI | P-value |
I2 |
|---|
| Total | 23 | 1.09 | 1.00–1.18 | <0.01 | 54.3 | 1.13 | 1.04–1.24 | <0.01 | 18.2 | 1.17 | 1.00–1.38 | 0.05 | 52.5 | 1.14 | 1.05–1.24 | <0.01 | 33.8 | 1.08 | 0.95–1.23 | 0.23 | 54.7 |
| HWE-yes | 22 | 1.09 | 1.00–1.19 | 0.05 | 56.3 | 1.10 | 1.01–1.21 | 0.04 | 12.8 | 1.17 | 0.99–1.40 | 0.07 | 54.6 | 1.12 | 1.03–1.22 | <0.01 | 34.2 | 1.10 | 0.95–1.26 | 0.20 | 55.1 |
| Subtype |
|
|
HNPCC | 6 | 1.06 | 0.86–1.31 | 0.59 | 59.5 | 0.98 | 0.79–1.22 | 0.88 | 37.3 | 1.13 | 0.72–1.76 | 0.60 | 58.9 | 1.08 | 0.78–1.51 | 0.63 | 54.8 | 1.08 | 0.87–1.35 | 0.48 | 37.1 |
|
sCRC | 8 | 1.06 | 0.91–1.24 | 0.46 | 59.0 | 1.21 | 1.04–1.42 | 0.02 | 24.1 | 1.13 | 0.82–1.56 | 0.45 | 59.9 | 1.18 | 1.02–1.37 | 0.03 | 35.0 | 1.04 | 0.80–1.35 | 0.77 | 62.2 |
| Ethnicity |
|
|
Caucasian | 15 | 1.09 | 0.98–1.22 | 0.10 | 56.1 | 1.14 | 1.02–1.28 | 0.02 | 19.8 | 1.19 | 0.95–1.90 | 0.13 | 56.7 | 1.14 | 1.02–1.27 | 0.02 | 37.5 | 1.08 | 0.89–1.30 | 0.43 | 57.2 |
|
Asian | 8 | 1.10 | 0.96–1.26 | 0.16 | 64.2 | 1.12 | 0.91–1.37 | 0.30 | 48.3 | 1.20 | 0.95–1.27 | 0.18 | 60.3 | 1.16 | 0.94–1.43 | 0.17 | 55.2 | 1.11 | 0.92–1.34 | 0.28 | 57.2 |
|
Mixed | 1 | 1.01 | 0.70–1.44 | 0.97 | NA | 0.93 | 0.52–1.66 | 0.81 | NA | 1.04 | 0.48–2.27 | 0.91 | NA | 0.96 | 0.55–1.67 | 0.87 | NA | 1.09 | 0.56–2.16 | 0.79 | NA |
| Design |
|
|
Fam-B | 2 | 0.80 | 0.61–1.06 | 0.12 | 0 | 1.02 | 0.38–2.75 | 0.97 | 66.9 | 0.60 | 0.34–1.08 | 0.09 | 0 | 0.92 | 0.38–2.23 | 0.86 | 616 | 0.69 | 0.42–1.15 | 0.16 | 0 |
|
Heal-B | 5 | 1.27 | 0.92–1.75 | 0.14 | 79.5 | 1.16 | 0.81–1.68 | 0.42 | 50.3 | 1.64 | 0.82–3.27 | 0.16 | 78.7 | 1.30 | 0.86–1.96 | 0.22 | 653 | 1.47 | 0.88–2.47 | 0.15 | 78.2 |
|
Hosp-B | 8 | 1.10 | 1.01–1.19 | 0.03 | 13.6 | 1.29 | 1.10–1.50 | <0.01 | 0 | 1.24 | 1.05–1.48 | <0.01 | 4.7 | 1.27 | 1.10–1.46 | <0.01 | 0 | 1.03 | 0.90–1.17 | 0.69 | 33.3 |
|
Pop-B | 8 | 1.03 | 0.96–1.19 | 0.43 | 17.5 | 1.06 | 0.94–1.20 | 0.32 | 0.6 | 1.05 | 0.92–1.21 | 0.46 | 18.7 | 1.06 | 0.95–1.19 | 0.30 | 0 | 1.02 | 0.81–1.13 | 0.77 | 33.5 |
| Genotyping
type |
|
|
PCR-RFLP | 13 | 1.10 | 1.04–1.18 | <0.01 | 48.7 | 1.21 | 1.08–1.36 | <0.01 | 13.0 | 1.24 | 1.09–1.41 | <0.01 | 45.5 | 1.22 | 1.09–1.36 | <0.01 | 25.1 | 1.08 | 0.98–1.19 | 0.11 | 49.2 |
|
PCR-SSCP | 4 | 1.04 | 0.76–1.41 | 0.81 | 73.1 | 1.08 | 0.85–1.37 | 0.53 | 42.7 | 1.05 | 0.54–2.05 | 0.87 | 74.1 | 1.07 | 0.75–1.54 | 0.71 | 51.4 | 0.99 | 0.52–1.91 | 0.98 | 80.5 |
|
Taqman | 5 | 1.11 | 0.94–1.32 | 0.22 | 54.1 | 1.09 | 0.91–1.30 | 0.36 | 0 | 1.09 | 0.89–1.33 | 0.42 | 45.9 | 1.10 | 0.93–1.30 | 0.27 | 14.9 | 1.04 | 0.89–1.21 | 0.65 | 49.5 |
|
Others | 1 | 0.87 | 0.70–1.09 | 0.23 | NA | 0.80 | 0.56–1.16 | 0.24 | NA | 0.78 | 0.50–1.23 | 0.28 | NA | 0.80 | 0.57–1.12 | 0.20 | NA | 0.89 | 0.60–1.32 | 0.57 | NA |
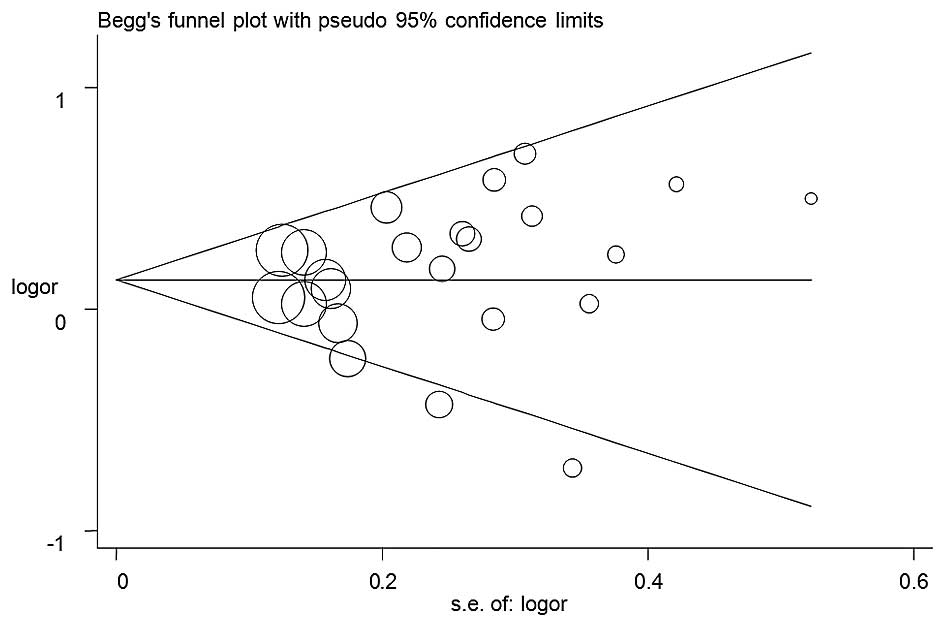
Publication bias
No publication bias was detected in any of the five
genetic models. The shape of the funnel plots appeared to be
symmetrical for all models (the GA+AA vs. GG model is shown in
Fig. 3), and Egger's test results
supported these findings (for A vs. G: P=0.34; for GA vs. GG:
P=0.67; for AA vs. GG: P=0.53; for AA+GA vs. GG: P=0.47; and for AA
vs. GG+GA: P=0.49).
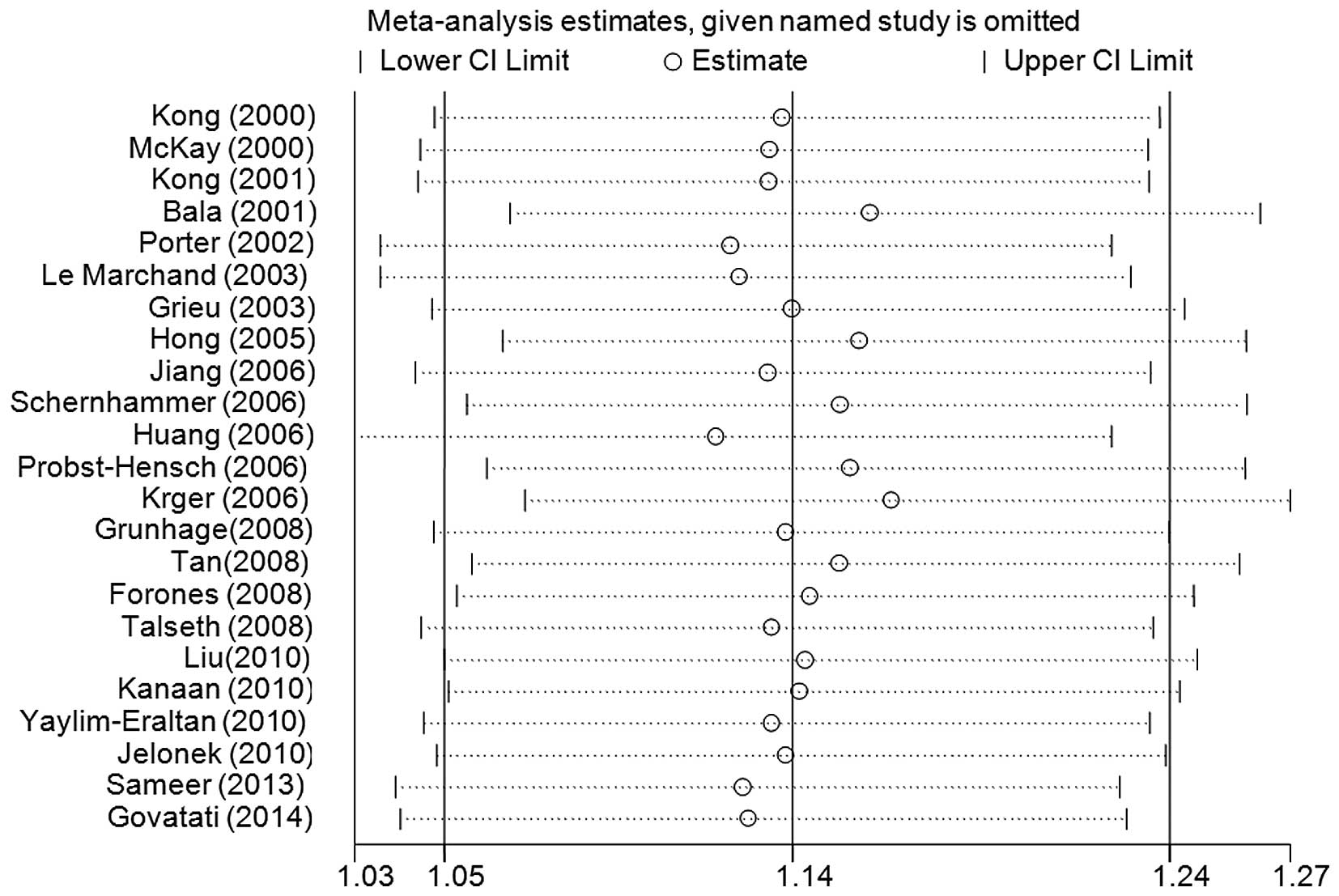
Sensitivity analysis and cumulative
analysis
Sensitivity analyses were performed, and the
exclusion of no single study qualitatively changed the pooled ORs,
indicating that the results of the present meta-analysis were
stable (the data for the GA+AA vs. GG model is shown in Fig. 4). Cumulative analyses according to the
publication date revealed that the cancer risk increased gradually,
and became positive, on inclusion of the study conducted by Talseth
et al (39) in 2008 (the GA+AA
vs. GG model is shown in Fig. 5).
Discussion
CCND1 is located on chromosome 11q13, and encodes a
critical cell cycle regulatory protein of 295 amino acids. CCND1
regulates the transition from the G1 to the S phase during cell
division. High levels of activity of CCND1 result in premature cell
passage through the G1-S transition, leading to an extension of
non-repaired DNA damage and the accumulation of genetic mistakes
(42). Overexpression of CCND1 has
been detected in several cancer types, which is also regarded as a
risk factor for cancer development. Of the SNPs in CCND1, the
G-to-A mutation is the most common, and does not result in an amino
acid alteration in the protein sequence. However, the A allele
change does lead to an alternatively spliced transcript of CCND1,
with a longer half-life compared with the G allele, which
facilitates the passage of the variant cell through the G1-S
checkpoint and rapid proliferation, ultimately resulting in cancer
development (43). Previous studies
have shown that the A allele may be associated with an increased
risk of breast, prostate, esophageal and other cancer types in
different ethnicities (44–46).
In 2000, the first case-control study performed by
McKay et al (20) failed to
reveal any significant association between the CCND1 G870A
polymorphism and the risk of colorectal cancer in Caucasians. To
date, conflicting data about the association between the CCND1
G870A polymorphism and colorectal cancer susceptibility exist. Kong
et al (21) identified that
the risk of developing colorectal cancer was 3-fold higher in
Caucasians with a homozygous A allele (OR=2.68, 95% CI=1.38–5.19).
The study by Jiang et al (27)
suggested that the AA genotype may increase the colorectal cancer
risk compared with the GG+AG genotype (OR=1.56, 95% CI=1.10–2.21)
in an Indian population. Huang et al (29) also identified a significant
association between the CCND1 G870A polymorphism and the risk of
colorectal cancer in young Chinese patients. Notably, the mechanism
of the G870A polymorphism differs according to ethnicity. Porter
et al (23) demonstrated an
increased risk of familiar colorectal cancer of almost 2-fold (for
GA+AA vs. GG: OR=1.7, 95% CI=1.1–2.7) although the authors did not
find any significant association with sporadic colorectal cancer in
Caucasians. Le Marchand et al (24) reported a significantly increased risk
in Hawaiian individuals with a heterozygous GA genotype (OR=3.9,
95% CI=1.2–13.2), and a marginal risk in Caucasians with a
homozygous AA genotype (OR=2.1, 95% CI=1.0–4.3), although the
authors did not find any significant association in a Japanese
population. However, the studies by Bala, Schernhammer, Krüger and
other research groups (22,25,28,31,33–36)
failed to identify any significant association between the CCND1
G870A polymorphism and colorectal cancer. By contrast, several
studies revealed that the A allele exerts a protective function in
the development of colorectal cancer (26,30,37,38).
The present meta-analysis, comprising 23 case control studies with
6,320 patients with colorectal cancer and 8,252 controls, explored
the association between an increased risk of colorectal cancer and
the CCND1 G870A polymorphism. The findings suggested that CCND1
exerts an important role in the development of colorectal cancer,
particularly in Caucasians and in the development of sporadic
colorectal cancer.
Several limitations of the present analysis should
be acknowledged. First, the results are based on the unadjusted
estimates, without the original data from the selected studies, and
lack information on certain co-variables, including diet, smoking,
drinking and other environmental factors. Secondly, small numbers
of patients were included in the cancer subgroups, including cancer
location and familiar hereditary, which prevented more precise
conclusions from being drawn. The actual association could
therefore be biased, and the analysis may not have enough
statistical power with the current small sample size. Thirdly, the
controls in several of the studies were hospital-based populations
with other diseases, which could also result in a certain selection
bias. Finally, the majority of the included studies were performed
in Caucasian and Asian populations, without any reported studies in
African populations; therefore, ethnicity may also result in a
certain bias. Despite these limitations, the present meta-analysis
included 23 published articles with the largest sample sizes and
latest data. A cumulative analysis also demonstrated that the
results of our meta-analysis were stable, which further confirm the
reliability and validity of the present study.
In conclusion, the present meta-analysis suggested
that the CCND1 G870A polymorphism may be associated with an
increased risk of the development of colorectal cancer. Considering
the limitations due to the small sample size, larger, well-designed
case-control studies are required to further validate these
findings.
Acknowledgements
The present study was supported by grant no.
D20142102 from the Ministry of Education of Hubei Province.
References
|
1
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J and
Thun MJ: Cancer statistics, 2009. CA Cancer J Clin. 59:225–249.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Brevik A, Joshi AD, Corral R, Onland-Moret
NC, Siegmund KD, Le Marchand L, Baron JA, Martinez ME, Haile RW,
Ahnen DJ, et al: Polymorphisms in base excision repair genes as
colorectal cancer risk factors and modifiers of the effect of diets
high in red meat. Cancer Epidemiol Biomarkers Prev. 19:3167–3173.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lynch PM: The hMSH2 and hMLH1 genes in
hereditary nonpolyposis colorectal cancer. Surg Oncol Clin N Am.
18:611–624. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shin MH, Holmes MD, Hankinson SE, Wu K,
Colditz GA and Willett WC: Intake of dairy products, calcium and
vitamin d and risk of breast cancer. J Natl Cancer Inst.
94:1301–1311. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sherr CJ: Cancer cell cycles. Science.
274:1672–1677. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kaminagakura E, da Werneck Cunha I, Soares
FA, Nishimoto IN and Kowalski LP: CCND1 amplification and protein
overexpression in oral squamous cell carcinoma of young patients.
Head Neck. 33:1413–1419. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Troncone G, Volante M, Iaccarino A, Zeppa
P, Cozzolino I, Malapelle U, Palmieri EA, Conzo G, Papotti M and
Palombini L: Cyclin D1 and D3 overexpression predicts malignant
behavior in thyroid fine-needle aspirates suspicious for Hurthle
cell neoplasms. Cancer. 117:522–529. 2009.PubMed/NCBI
|
|
8
|
Abramson VG, Troxel AB, Feldman M, Mies C,
Wang Y, Sherman L, McNally S, Diehl A and Demichele A: Cyclin D1b
in human breast carcinoma and coexpression with cyclin D1a is
associated with poor outcome. Anticancer Res. 30:1279–1285.
2010.PubMed/NCBI
|
|
9
|
Zhang L, Chen LM, Wang MN, Chen XJ, Li N,
Huang YD and Chen M: The G894t, T-786c and 4b/a polymorphisms in
Enos gene and cancer risk: A meta-analysis. J Evid Based Med.
7:263–269. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Betticher DC, Thatcher N, Altermatt HJ,
Hoban P, Ryder WD and Heighway J: Alternate splicing produces a
novel cyclin D1 transcript. Oncogene. 11:1005–1011. 1995.PubMed/NCBI
|
|
11
|
Kong S, Amos CI, Luthra R, Lynch PM, Levin
B and Frazier ML: Effects of cyclin D1 polymorphism on age of onset
of hereditary nonpolyposis colorectal cancer. Cancer Res.
60:249–252. 2000.PubMed/NCBI
|
|
12
|
Moher D, Liberati A, Tetzlaff J and Altman
DG: Preferred reporting items for systematic reviews and
meta-analyses: The PRISMA statement. BMJ. 339:b25352009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Little J, Bradley L, Bray MS, Clyne M,
Dorman J, Ellsworth DL, Hanson J, Khoury M, Lau J, O'Brien TR, et
al: Reporting, appraising and integrating data on genotype
prevalence and gene-disease associations. Am J Epidemiol.
156:300–310. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang Y, Jia Q, Xue P, Liu Y, Xiong T,
Yang J, Song C, He Q and Du L: The-786T>C polymorphism in the
NOS3 gene is associated with increased cancer risk. Tumour Biol.
35:3535–3540. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lau J, Ioannidis JP and Schmid CH:
Quantitative synthesis in systematic reviews. Ann Intern Med.
127:820–826. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
DerSimonian R and Laird N: Meta-analysis
in clinical trials. Control Clin Trials. 7:177–188. 1986.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mantel N and Haenszel W: Statistical
aspects of the analysis of data from retrospective studies of
disease. J Natl Cancer Inst. 22:719–748. 1959.PubMed/NCBI
|
|
18
|
Egger M, Davey Smith G, Schneider M and
Minder C: Bias in meta-analysis detected by a simple, graphical
test. BMJ. 315:629–634. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bagos PG and Nikolopoulos GK: Generalized
least squares for assessing trends in cumulative meta-analysis with
applications in genetic epidemiology. J Clin Epidemiol.
62:1037–1044. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
McKay JA, Douglas JJ, Ross VG, Curran S,
Murray GI, Cassidy J and McLeod HL: Cyclin D1 protein expression
and gene polymorphism in colorectal cancer. Aberdeen colorectal
initiative. Int J Cancer. 88:77–81. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kong S, Wei Q, Amos CI, Lynch PM, Levin B,
Zong J and Frazier ML: Cyclin D1 polymorphism and increased risk of
colorectal cancer at young age. J Natl Cancer Inst. 93:1106–1108.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bala S and Peltomäki P: Cyclin D1 as a
genetic modifier in hereditary nonpolyposis colorectal cancer.
Cancer Res. 61:6042–6045. 2001.PubMed/NCBI
|
|
23
|
Porter TR, Richards FM, Houlston RS, Evans
DG, Jankowski JA, Macdonald F, Norbury G, Payne SJ, Fisher SA,
Tomlinson I and Maher ER: Contribution of cyclin d1 (CCND1) and
E-cadherin (CDH1) polymorphisms to familial and sporadic colorectal
cancer. Oncogene. 21:1928–1933. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Le Marchand L, Seifried A, Lum-Jones A,
Donlon T and Wilkens LR: Association of the cyclin D1 A870G
polymorphism with advanced colorectal cancer. JAMA. 290:2843–2848.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Grieu F, Malaney S, Ward R, Joseph D and
Iacopetta B: Lack of association between CCND1 G870A polymorphism
and the risk of breast and colorectal cancers. Anticancer Res.
23:4257–4259. 2003.PubMed/NCBI
|
|
26
|
Hong Y, Eu KW, Seow-Choen F, Fook-Chong S
and Cheah PY: GG genotype of cyclin D1 G870A polymorphism is
associated with increased risk and advanced colorectal cancer in
patients in Singapore. Eur J Cancer. 41:1037–1044. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jiang J, Wang J, Suzuki S, Gajalakshmi V,
Kuriki K, Zhao Y, Nakamura S, Akasaka S, Ishikawa H and Tokudome S:
Elevated risk of colorectal cancer associated with the AA genotype
of the cyclin D1 A870G polymorphism in an Indian population. J
Cancer Res Clin Oncol. 132:193–199. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Schernhammer ES, Tranah GJ, Giovannucci E,
Chan AT, Ma J, Colditz GA, Hunter DJ, Willett WC and Fuchs CS:
Cyclin D1 A870G polymorphism and the risk of colorectal cancer and
adenoma. Br J Cancer. 94:928–934. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang WS, Tang R, Lin PY, Changchien CR,
Chen JS, Chiang JM, Yeh CY, Wang JY and Hsieh LL: Impact of the
cyclin D1 A870G polymorphism on susceptibility to sporadic
colorectal cancer in Taiwan. Dis Colon Rectum. 49:602–608. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Probst-Hensch NM, Sun CL, Van Den Berg D,
Ceschi M, Koh WP and Yu MC: The effect of the cyclin D1 (CCND1)
A870G polymorphism on colorectal cancer risk is modified by
glutathione-S-transferase polymorphisms and isothiocyanate intake
in the Singapore Chinese health study. Carcinogenesis.
27:2475–2482. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Krüger S, Engel C, Bier A, Mangold E,
Pagenstecher C, Doeberitz Mv, Holinski-Feder E, Moeslein G, Keller
G, Kunstmann E, et al: Absence of association between cyclin D1
(CCND1) G870A polymorphism and age of onset in hereditary
nonpolyposis colorectal cancer. Cancer Lett. 236:191–197. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Grünhage F, Jungck M, Lamberti C, Berg C,
Becker U, Schulte-Witte H, Plassmann D, Rahner N, Aretz S,
Friedrichs N, et al: Association of familial colorectal cancer with
variants in the E-cadherin (CDH1) and cyclin D1 (CCND1) genes. Int
J Colorectal Dis. 23:147–154. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tan XL, Nieters A, Kropp S, Hoffmeister M,
Brenner H and Chang-Claude J: The association of cyclin D1 G870A
and E-cadherin C-160A polymorphisms with the risk of colorectal
cancer in a case control study and meta-analysis. Int J Cancer.
122:2573–2580. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Forones NM, de Lima JM, de Souza LG and da
Silva ID: Cyclin D1 A870G polymorphism in Brazilian colorectal
cancer patients. J Gastrointest Cancer. 39:118–123. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu B, Zhang Y, Jin M, Ni Q, Liang X, Ma
X, Yao K, Li Q and Chen K: Association of selected polymorphisms of
CCND1, p21 and caspase8 with colorectal cancer risk. Mol Carcinog.
49:75–84. 2010.PubMed/NCBI
|
|
36
|
Kanaan Z, Eichenberger MR, Young M,
Colliver D, Crawford N, Cobbs GA, Hein DW and Galandiuk S: An
alternative cyclin-D1 splice site is not linked to inflammatory
bowel disease-associated neoplasia. Int J Biol Markers. 25:27–31.
2010.PubMed/NCBI
|
|
37
|
Yaylim-Eraltan I, Arikan S, Yildiz Y,
Cacina C, Ergen HA, Tuna G, Görmüs U, Zeybek U and Isbir T: The
influence of cyclin D1 A870G polymorphism on colorectal cancer risk
and prognosis in a Turkish population. Anticancer Res.
30:2875–2880. 2010.PubMed/NCBI
|
|
38
|
Jelonek K, Gdowicz-Klosok A, Pietrowska M,
Borkowska M, Korfanty J, Rzeszowska-Wolny J and Widlak P:
Association between single-nucleotide polymorphisms of selected
genes involved in the response to DNA damage and risk of colon,
head and neck and breast cancers in a Polish population. J Appl
Genet. 51:343–352. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Talseth BA, Ashton KA, Meldrum C, Suchy J,
Kurzawski G, Lubinski J and Scott RJ: Aurora-A and Cyclin D1
polymorphisms and the age of onset of colorectal cancer in
hereditary nonpolyposis colorectal cancer. Int J Cancer.
122:1273–1277. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sameer AS, Parray FQ, Dar MA, Nissar S,
Banday MZ, Rasool S, Gulzar GM, Chowdri NA and Siddiqi MA: Cyclin
D1 G870A polymorphism and risk of colorectal cancer: A case control
study. Mol Med Rep. 7:811–815. 2013.PubMed/NCBI
|
|
41
|
Govatati S, Singamsetty GK, Nallabelli N,
Malempati S, Rao PS, Madamchetty VK, Govatati S, Kanapuram R,
Narayana N, Bhanoori M, et al: Contribution of cyclin D1 (CCND1)
and E-cadherin (CDH1) alterations to colorectal cancer
susceptibility: A case-control study. Tumour Biol. 35:12059–12067.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hall M and Peters G: Genetic alterations
of cyclins, cyclin-dependent kinases and Cdk inhibitors in human
cancer. Adv Cancer Res. 68:67–108. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Solomon DA, Wang Y, Fox SR, Lambeck TC,
Giesting S, Lan Z, Senderowicz AM, Conti CJ and Knudsen ES: Cyclin
D1 splice variants. Differential effects on localization, RB
phosphorylation and cellular transformation. J Biol Chem.
278:30339–30347. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Onay UV, Aaltonen K, Briollais L, Knight
JA, Pabalan N, Kilpivaara O, Andrulis IL, Blomqvist C, Nevanlinna H
and Ozcelik H: Combined effect of CCND1 and COMT polymorphisms and
increased breast cancer risk. BMC Cancer. 8:62008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Mandal RK and Mittal RD: Are cell cycle
and apoptosis genes associated with prostate cancer risk in North
Indian population? Urol Oncol. 30:555–561. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Casson AG, Zheng Z, Evans SC, Geldenhuys
L, van Zanten SV, Veugelers PJ, Porter GA and Guernsey DL: Cyclin
D1 polymorphism (G870A) and risk for esophageal adenocarcinoma.
Cancer. 104:730–739. 2005. View Article : Google Scholar : PubMed/NCBI
|