Introduction
Globally, colorectal cancer (CRC) is the second most
common cancer in women and the third most common in men (1). Although the treatments are improved for
CRC, increasing the 5-year survival rate, the overall estimated
death rate is still 50–60% (2).
There is not a good method to diagnose for this cancer, because
although tumor markers greatly improve it, the invasive nature of
current procedures, as colonoscopy, limits their application.
Identification of useful non-invasive biomarkers in order to
facilitate the correct diagnosis and treatment is critical to
improve patient survival.
MicroRNAs (miRNAs) are a type of small RNA (18–22
nucleotides, nt) that mediates post-transcriptional gene silencing
by binding to mRNAs. The role of miRNA in carcinogenesis has been
increasingly recognized; miRNAs affect many oncogenes and tumor
suppressor genes. miRNA-induced deregulation in CRC has been well
documented and for this reason, those could be exploited as
biomarkers in CRC due to its high tissue specificity, stability and
the differences in the expression level between normal and tumor
tissues (3–6). The detection of miRNAs in serum samples
has raised the possibility that they could be used as non-invasive
biomarkers for different types of cancer (7–9).
In our study, we evaluated the miRNA profile in 16
samples of tumor tissue from patients with CRC by next-generation
sequencing (NGS) and compared the expression levels of 22 miRNAs
between CRC, hyperplastic polyps and adenoma patients with control
subjects to search differences that could be useful for a better
understanding of CRC carcinogenesis and could be potential
biomarkers.
Patients and methods
Subjects
We included 45 CRC cases (24 colon cancer and 21
rectum cancer), 11 advanced adenomas, 14 hyperplastic polyps and 48
controls from a multicenter hospital-based case-control study
conducted in Colombia. All cases were incident and confirmed by
histopathology, while controls were individuals without
gastrointestinal symptoms attending the outpatient services of
primary care units. Advanced adenomas were adenomas with size ≥1
cm, tubulovillous or villous adenomas or with high-grade dysplasia.
Subjects were unrelated and their age ranged between 30 and 76
years. Neither cases nor controls had a personal history of other
cancers and received neither chemotherapy nor radiotherapy. Trained
health professionals collected blood samples and administered
structured questionnaires on socio-economic characteristics and
other risk factors, once each participant gave written informed
consent. Tissues were collected during colonoscopy. This study was
approved by the Ethics Committee of the Instituto Nacional de
Cancerología, Bogotá, Colombia and by all the other Ethical Boards
from participant health institutions upon request.
miRNAs isolation and
quantification
Total RNA was extracted from 10 mg of tumor tissue
using Trizol (Thermo Fisher Scientific, Whaltan, USA) and after
that, miRCURY™ RNA Isolation Kit–Tissue (Exiqon, Copenhagen,
Denmark) was used for miRNA isolation. Serum miRNAs were extracted
from 200 µl using miRCURY RNA Isolation Kit-Biofluids (Exiqon,
Copenhagen, Denmark). Concentration and quality of the samples was
assessed using Agilent 2100 Bioanalyzer (Agilent Technologies,
Santa Clara, California, USA) and Qubit 3.0 Fluorometer (Thermo
Fisher Scientific, Whaltan, USA), following the manufacturer's
instructions. Isolated RNA was stored at −80°C until use.
Libraries construction and
sequencing
Libraries were constructed from 1 µg of extracted
miRNAs using Truseq Small RNA kit (Illumina, San Diego, USA),
following manufacturer's instructions. A pool of 16 libraries was
obtained with an adjusted concentration of 10 nM in a final volume
of 100 µl. The final concentration of the pooled samples was 2 nM.
The sequencing was performed in a MiSeq, using MiSeq Reagent Kit v2
(Illumina®, San Diego, USA), following the
manufacturer's instructions.
Bioinformatics analysis of detected
miRNAs
Bioinformatics analysis was made following the
pipeline from Hackenberg et al and the tool miRanalyzer
(10). The reads shorter than 17 nt
and longer than 26 nt were discard. Selected reads were aligned
against different databases, including the RefSeq to detect mRNA,
the miRBase database (11) to detect
mature miRNAs, and the GRCh37 human reference genome assembly to
predict possible new miRNAs. The miRanalyzer tool also does a
prediction over candidate miRNAs. We also took into account the
following high confidence criteria to select novel miRNA sequence
defined by miRbase (11).
Expression levels of selected miRNA in
serum and tumor tissue
The Universal cDNA synthesis kit II
(Exiqon®, Copenhagen, Denmark) was used for reverse
transcription (RT), according to the manufacturer's instructions.
The UniSp6 RNA Spike-in control was added during the cDNA
synthesis. For quantitative PCR, the obtained cDNA was diluted
1:100 in nuclease free water and then 5 µl of cDNA along with 5 µl
of Exilent SYBR Green master mix were transferred to a custom-made
Pick & Mix microRNA PCR panel that included primers for 22
target miRNAs (Exiqon®, Copenhagen, Denmark). These
miRNAs were selected based on the abundance of reads found in the
tumor tissue libraries and others by literature review of the most
abundant in serum from CRC patients. The hsa-miR were: 10a-5p,
10b-5p, 192-5p, 22-3p, 26a-5p, 148a-3p, 92a-3p, 143-3p, 486-5p,
141-3p, 27b-3p, 29a-3p, 221-3p, 200c-3p, 145-5p, 423-3p, 155-5p,
223-3p, 320a, 21-5p, 20a-5p and 221-3p. Amplification was performed
in duplicates in a Light Cycler 480 Real-Time PCR System (Roche,
Basel, Switzerland). Amplification curves were analyzed using the
Roche LC software, both for determination of Ct values and for
melting curve analysis. Normality of miRNA levels was assessed by
Shapiro-Wilks test. Correlations between serum and tissue levels
were made using Pearson test.
Data analysis and normalization of
RT-PCR
Amplification efficiency was calculated by using
Exiqon GenEx software specifically adapted to miRCURY LNA™
Universal RT microRNA PCR products, following the manufacturer's
instructions. First, we made a pre-processing and normalization of
our data in GenEx. Only miRNAs detected with Ct <37 were
included for analysis. During quality control steps, samples with a
>50% of missing data and miRNAs with <40% of valid data were
excluded. The software selected the hsa-miR-92a-3p as reference
gene for normalization. This normalization step correspond to the
first delta Ct, namely delta to the normalization factor, of the
2−ΔΔCt method (12).
After that, the serum Ct from polyps, adenomas and CRC patients
were converted to relative quantities, comparing to control group,
and by this step the data was expressed completely as
N=2−ΔΔCt method (12).
Expression data was converted to log2 scale for further analysis.
Comparisons between groups in serum samples were done by Welch's
ANOVA method adjusted by sex and differential expressed genes were
identified based on a Bonferroni corrected P-value of <0.002
(alpha of 0.05/22 tests). Finally, we used Pearson correlation
analysis of the miRNAs expression values found in serum and tumor
of CRC samples.
Bioinformatics analysis of target
genes for detected miRNAs and related biological pathways
In order to determine target genes of the identified
miRNAs, we used DIANA-TarBase v7.0 (13), that predict molecular targets of
miRNAs in coding sequences 3′UTR. Related biological pathways
associated with target genes and miRNAs were made using the Kyoto
Encyclopedia of Genes and Genomes (KEEG) (14).
Results
Libraries in tumor CRC
Expression pattern of known
miRNAs
Sixteen tumor samples were assessed by NGS, five
correspond to colon cancer and eleven to rectal cancer, 60% were
from males and the mean age was 59,1 years. 763 known mature miRNAs
were detected in the sixteen libraries by at least one alignment in
miRBase (11). The read counts of
the mature miRNAs from sixteen libraries were pooled. The known
mature miRNAs showed a wide range of expression values spanning
from 1 to 222455 read counts. 176 of 763 known miRNAs detected had
only one read count and 167 had more than 100 read counts. Nine
miRNAs had expression levels above 2% and it represents 70.4% of
the total read counts (hsa-miR: 10a-5p, 192-5p, 10b-5p, 22-3p,
26a-5p, 148a-3p, 181a-5p, 92a-3p and 143-5p) (Table IA).
 | Table I.Read count percentage of 22 miRNAs
selected to be analyzed by reverse transcription-PCR. |
Table I.
Read count percentage of 22 miRNAs
selected to be analyzed by reverse transcription-PCR.
| A, miRNAs selected
by abundance of reads found in the tumor tissue libraries |
|---|
|
|---|
| miRNA | Read counts, % |
|---|
| 10a-5p | 22.45 |
| 192-5p | 16.02 |
| 10b-5p | 9.63 |
| 22-3p | 4.80 |
| 26a-5p | 4.70 |
| 148a-3p | 4.69 |
| 181a-5p | 2.99 |
| 92a-3p | 2.98 |
| 143-3p | 2.11 |
|
| B, miRNAs
selected by literature review |
|
| miRNA | Read counts,
% |
|
| 486-5p | 1.93 |
| 141-3p | 1.50 |
| 27b-3p | 1.45 |
| 29a-3p | 0.33 |
| 221-3p | 0.29 |
| 200c-3p | 0.27 |
| 145-5p | 0.10 |
| 423-3p | 0.10 |
| 155-5p | 0.09 |
| 320a | 0.09 |
| 223-3p | 0.08 |
| 21-5p | 0.04 |
| 20a-5p | 0.02 |
Prediction and expression levels of
potential novel miRNAs
In total, eight potential novel miRNAs with fuzzy
Dicer pattern were identified in the libraries; no potential novel
miRNA was detected with a perfect Dicer pattern. Seven candidates
were present in four or more libraries. In silico analysis
of these sequences against miRBase, led to identify that each of
these candidates had a partial or total complementarity with mature
miRNAs (Table II).
 | Table II.Location, cluster sequence and
expression levels of miRNAs candidates found in the sixteen
libraries of colorectal cancer tissues. |
Table II.
Location, cluster sequence and
expression levels of miRNAs candidates found in the sixteen
libraries of colorectal cancer tissues.
|
|
| Full cluster
genomic coordinates (build GRCh37) |
|
|
|
| Alignments in
miRBase |
|---|
|
|
|
|
|
|
|
|
|
|---|
| Chr | Name | Chr start | Chr end | Strand | Read count | Read cluster
sequence | Size (base) | No. libraries | Complementary
to | Score |
|---|
| 1 | cand_1.1 | 220291187 | 1102596 | + | 230 |
CTGTCAATTCATAGGTCA | 18 | 1 | miR-192-5p | 90 |
| 3 | cand_3.1 | 49057565 | 1102596 | + | 127 |
ACGGGAGTGATCGTGTCATT | 20 | 8 | miR-425-5p | 100 |
| 5 | cand_5.1 | 148808467 | 148808597 | – | 13 |
GAGATGCAGCACTGCACC | 18 | 6 | miR-143-3p | 90 |
| 10 | cand_10.1 | 104196251 | 104196359 | – | 5,920 |
GaCCTATGGAATTCAGTTCTCAGa | 20–22 | 8 | miR-146b-5p | 105 |
|
| cand_10.2 | 105154025 | 105154149 | + | 20 |
CGACCGACGCCACGCCGAGT | 20 | 4 | miR-1307-3p | 100 |
|
| cand_10.3 | 105154041 | 105154143 | + | 41 |
CCGGTCGAGGTCCGGTCGA | 19 | 7 | miR-1307-5p | 95 |
| 17 | cand_17.1 | 46657191 | 46657319 | + | 51 |
CCCTAGATACGAATTTG | 17 | 9 | miR-10a-3p | 85 |
| X | cand_X.1 | 45605572 | 45605704 | + | 16 |
CCCAGCAGACAATGTAGCT | 19 | 4 | miR-221-3p | 95 |
Pathways related with most common
found miRNAs
We did a search of gene targets and pathways related
of the nine most common miRNAs found by sequencing in the
‘Colorectal cancer pathway’ (hsa05210) in TarBase 7.0/KEGG
(13). We found that all of these
miRNAs had gene targets involved in different pathways related with
CRC. The most common pathways involved are WNT, MAPK, PI3K/Akt,
TGF-β, DCC, p53 and microsatellite instability (MSI).
miRNAs levels in serum
From the most abundant miRNAs detected by sequencing
in tumor tissue, along with others thirteen differentially
expressed in serum from CRC patients according to literature
(Table IB), we selected 22 miRNAs to
be analysed by RT-PCR in the serum of patients including 45 CRC, 11
advanced adenomas, 14 hyperplastic polyps and 48 controls were
enrolled in this study. Table III
shows the distribution of age and gender according to phenotype.
Pre-processing data, using Exiqon GenEx software, excluded six
serum samples because they had >50% of missing data. Table I shows the percentage of total read
counts, by NGS in sixteen samples, of the thirteen miRNAs selected
according to literature.
 | Table III.Characteristics of patients and
controls enrolled in the present study. |
Table III.
Characteristics of patients and
controls enrolled in the present study.
| Variables | Control (n=48) | Polyps (n=14) | Adenoma (n=11) | CRC (n=45) |
|---|
| Age (mean) | 51.9 | 56.7 | 52.2 | 59.7 |
| Male | 58.3 | 35.7 | 45.5 | 53.3 |
| Female | 41.7 | 64.3 | 54.5 | 46.7 |
| Colon cancer | – | – | – | 53.3% |
| Rectum cancer | – | – | – | 46.7% |
From the twenty-two miRNAs selected to evaluate
differences in their levels in serum between groups, pre-processing
data excluded two miRNAs (miR-10a-5p and miR-221-5p) because they
had <40% of valid data. The data was expressed completely as
N=2−ΔΔCt method, miR-92-3p was selected by GenEx as
reference gene for normalization. Among the remaining 19 miRNAs, we
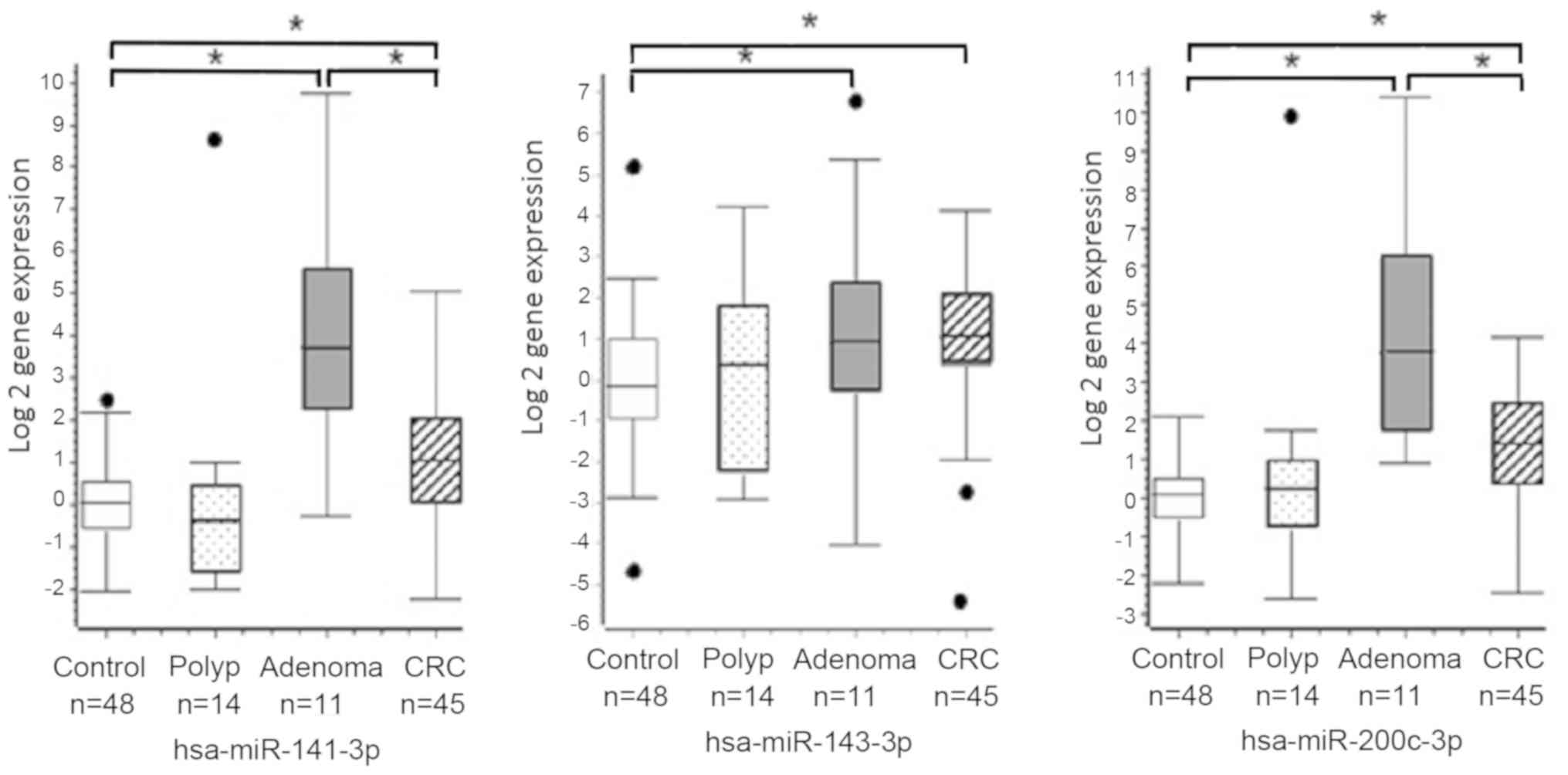
found significant higher serum expression levels of miR-143-3p,
miR-141-3p and miR-200c-3p in the CRC and adenoma groups compared
to controls by Mann-Whitney test with Bonferroni corrected P-value
(P<0.002; Fig. 1). In addition,
we also found significant higher levels of miR-141-3p and
miR-200c-3p in serum of adenoma patients compared to CRC group
(P<0.002).
Other miRNAs did not show statistical significant
differences between CRC patients and controls. Serum miRNA levels
between polyps patients and controls were very similar and their
behavior were the same. We also assessed levels of miRNAs in the
available twenty-two tumor tissues of CRC patients by RT-PCR. None
of the correlations in levels of miR-143-3p, miR-141-3p and
miR-200c, between tissue and serum samples from CRC patients
assessed Pearson test by were significant (P=0.225, P=0.867 and
P=0.652 respectively) (data not shown). Sixteen tumor samples used
for NGS were assessed by RT-PCR and their corresponding serum
samples too.
We used DIANA-miRPath v.3, to evidence the
involvement of these three miRNAs in the CRC pathway (hsa05210)
(15). This analysis led to identify
many gene targets related with different pathways, such as:
Apoptosis, PI3K-Akt, Wnt, MSI and TGF-β (Table IV).
 | Table IV.Gene targets and pathways regulated
by hsa-miR-143-3p, hsa-miR-141-3p and hsa-miR-200c-3p in the
colorectal cancer pathway (hsa-05210; from Diana Tool-miRPath v.3)
(12). |
Table IV.
Gene targets and pathways regulated
by hsa-miR-143-3p, hsa-miR-141-3p and hsa-miR-200c-3p in the
colorectal cancer pathway (hsa-05210; from Diana Tool-miRPath v.3)
(12).
|
| Gene targets |
|---|
|
|
|
|---|
| Pathway | hsa-miR-143-3p | hsa-miR-141-3p |
hsa-miR-200c-3p |
|---|
| Apoptosis | BCL2a | BCL2a, BAX | BCL2a |
| PI3K-Akt | KRASa, AKT1, MAPK1 | PIK3R1, RAC1,
MAPK9 | KRASa, RHOA, JUN |
| WNT | None | TCF7L1a, CCND1a, CTNNB1 | TCF7L1a, TCF7L2, CCND1a, APC |
| MSI | None | MSH2 | None |
| TGF-β | None | TGFB2 | SMAD2 |
Discussion
In the present work, deep sequencing and RT-qPCR
were used to analyze the expression levels of miRNAs in tumor
tissue and serum from patients with CRC. By deep sequencing, this
study detects 763 mature miRNAs in CRC tissues from sixteen
patients. Of the nine most expressed miRNA in our samples, three,
miR-10b-5p, −26a-5p and −92a-3p, have been reported that can act as
oncomiRs (6,16–28),
three, miR-192-5p, miR-148a-3p and miR-143-3p behave like
anti-oncomiRs (6,17,29–37).
With respect to miR-10a-5p, miR-22-3p and miR-181a-5p, their
tumorigenesis role is inconsistent. These miRNAs with dual roles in
carcinogenesis prove that many targets from many pathways can be
regulated by one miRNA and their effect on expression is very
complex at cellular and tisular levels.
One advantage of miRNA studies by deep sequencing is
that this technique allows the detection of novel miRNAs. Our
analysis found eight new miRNA candidates. All candidates showed
partial or total complementarity with mature miRNAs (scores 90–105)
based on miRBase analysis. These sequences with some grade or total
complementary could be produced by miRNAs bidirectional
transcription and processing (38,39). It
is possible that miRNA:miRNA duplex can be formed in the cell,
operating in competition with each other. Further experimental
studies are needed in order to assess the role of these miRNA
candidates in colorectal carcinogenesis, before register them into
public databases such as miRBase.
We found three miRNAs with significantly higher
expression in serum of CRC patients vs. controls (i.e. miR-143-3p,
miR-141-3p and miR-200c-3p) and two of them were more expressed in
patients with adenomas compared those with CRC (i.e. miR-141-3p and
miR-200c-3p). Interestingly, miR-141-3p and miR-200c-3p derive from
the same precursor, miR-8. Serum miR-141-3p and miR-200c-3p was
found over expressed in CRC patients compared to controls, as
previously reported (40–47). We found that patients with adenomas
had the highest serum levels of miR-141-3p and miR-200c-3p,
compared to all the others (i.e., controls, polyps and CRC groups).
These two miRNAs are good candidates for CRC screening and
prevention, as they could be measured through minimally invasive
procedures; nevertheless, further population-based studies are
needed for validation purposes.
The results seem to be contradictory. On the one
hand, lower levels of miRNAs have been found in CRC (18,48–53). On
the other hand, our findings are consistent with Luo et al
study (54) regarding higher levels
of miR-143-3p in CRC patients. The role of miRNAs in cancer is very
complex and depends of many particular factors and not alone cancer
type. Differences found in various studies in circulating levels of
miRNA can be related with ethnicity, gender and technical variance,
but there are other confounding lifestyle factors, such as smoking,
physical activity, etc., that are hardly verifiable and correctly
taken into consideration (9).
Like Waters et al (55), we did not find any correlation of
miRNA levels between serum and tissue of cancer patients. The
absence of this correlation could be attributed to the complex
nature of the circulating miRNAs sources. It has been found that
circulating tumor cells and exosomal release from tumor cells
contribute to circulating miRNAs (56,57).
Also, other factors such as the host immune response or
inflammation, could modulate miRNAs circulation levels and cause
these levels to be different to from those of the tissues.
Therefore, the levels of miRNA in circulation not only reflect what
happens in the tumor tissue, but also show what happens in the
whole human body.
Pathway analysis of the target genes of these miRNAs
uncovered a significant number of genes involved in many CRC
pathways, in accordance with reports highlighting that the hallmark
feature of CRC is the hyperactivation of the WNT pathway, usually
caused by mutations in the tumor suppressor gene APC (~75% of all
tumors) (58), mutations in CTNNB1
(β-catenin), or in other Wnt signaling activators (59–61).
In conclusion, this study found 763 miRNAs in tissue
from CRC and eight candidates to novel miRNAs. In serum, we found
that three miRNAs, miR-141-3p, miR-143-3p and miR-200c-3p, were
significantly higher in CRC vs. controls, and that two of them,
miR-141-3p and miR-200c, were also significantly lower in CRC vs.
adenomas. The measurement of miRNAs in the blood could complement
current screening methods for CRC and might provide new insights
into mechanisms of tumorigenesis and metastasis. However, the
differences between studies highlight the necessity to perform
further investigation.
Acknowledgements
The authors would like to thank Dr Ivan Lesende
(Universidad de la Coruña-Spain) for his technical support with the
library construction, Gustavo Hernández (Instituto Nacional de
Cancerología, Bogotá, Colombia) for collaboration with statistical
analysis and Unidad de Genética y Resistencia Antimicrobiana,
Centro Internacional de Genómica Microbiana for made
sequencing.
Funding
The present study was supported by Instituto
Nacional de Cancerología-E.S.E. de Colombia (DNP code 41030610-592)
y la Universidad Nacional de Colombia-Sede Bogotá (Hermes code
23561/QUIPU code 201010022022).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
HJA wrote the protocol, performed experiments and
contributed to the writing of the manuscript. MCS analyzed and
interpreted the data, and wrote the manuscript. XM conducted the
statistical analysis and contributed to the writing of the
manuscript. RR performed the bioinformatics analysis and
contributed to the writing of the manuscript. AHS contribute to the
design of the protocol, the analysis and interpretation of the
data, and the writing of the manuscript. MLS contributed to the
design of the protocol, performed the experiments and was a major
contributor in writing the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Instituto Nacional de Cancerología, Bogotá,
Colombia and by all the other Ethical Boards from participant
health institutions upon request. Each participant gave written
informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kumar R, Price TJ, Beeke C, Jain K, Patel
G, Padbury R, Young GP, Roder D, Townsend A, Bishnoi S and
Karapetis CS: Colorectal cancer survival: An analysis of patients
with metastatic disease synchronous and metachronous with the
primary tumor. Clin Colorectal Cancer. 13:87–93. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cekaite L, Eide PW, Lind GE, Skotheim RI
and Lothe RA: MicroRNAs as growth regulators, their function and
biomarker status in colorectal cancer. Oncotarget. 7:6476–6505.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chi Y and Zhou D: MicroRNAs in colorectal
carcinoma-from pathogenesis to therapy. J Exp Clin Cancer Res.
35:432016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yi R, Li Y, Wang FL, Miao G, Qi RM and
Zhao YY: MicroRNAs as diagnostic and prognostic biomarkers in
colorectal cancer. World J Gastrointest Oncol. 8:330–340. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Slattery ML, Herrick JS, Pellatt DF,
Stevens JR, Mullany LE, Wolff E, Hoffman MD, Samowitz WS and Wolff
RK: MicroRNA profiles in colorectal carcinomas, adenomas and normal
colonic mucosa: Variations in miRNA expression and disease
progression. Carcinogenesis. 37:245–261. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable blood-based
markers for cancer detection. Proc Natl Acad Sci USA.
105:10513–10518. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K,
Guo J, Zhang Y, Chen J, Guo X, et al: Characterization of microRNAs
in serum: A novel class of biomarkers for diagnosis of cancer and
other diseases. Cell Res. 18:997–1006. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tiberio P, Callari M, Angeloni V, Daidone
MG and Appierto V: Challenges in using circulating miRNAs as cancer
biomarkers. Biomed Res Int. 2015:7314792015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hackenberg M, Rodríguez-Ezpeleta N and
Aransay AM: miRanalyzer: An update on the detection and analysis of
microRNAs in high-throughput sequencing experiments. Nucleic Acids
Res 39 (Web Server issue). W132–W138. 2011. View Article : Google Scholar
|
|
11
|
Kozomara A and Griffiths-Jones S: miRBase:
Annotating high confidence microRNAs using deep sequencing data.
Nucleic Acids Res 42 (Database Issue). D68–D73. 2014. View Article : Google Scholar
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Vlachos IS, Paraskevopoulou MD, Karagkouni
D, Georgakilas G, Vergoulis T, Kanellos I, Anastasopoulos IL,
Maniou S, Karathanou K, Kalfakakou D, et al: DIANA-TarBase v7.0:
Indexing more than half a million experimentally supported
miRNA:mRNA interactions. Nucleic Acids Res 43 (Database Issue).
D153–D159. 2015. View Article : Google Scholar
|
|
14
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Vlachos IS, Zagganas K, Paraskevopoulou
MD, Georgakilas G, Karagkouni D, Vergoulis T, Dalamagas T and
Hatzigeorgiou AG: DIANA-miRPath v3.0: Deciphering microRNA function
with experimental support. Nucleic Acids Res. 43:W460–W466. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Schee K, Lorenz S, Worren MM, Günther CC,
Holden M, Hovig E, Fodstad O, Meza-Zepeda LA and Flatmark K: Deep
sequencing the MicroRNA transcriptome in colorectal cancer. PLoS
One. 8:e661652013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Della Vittoria Scarpati G, Calura E, Di
Marino M, Romualdi C, Beltrame L, Malapelle U, Troncone G, De
Stefano A, Pepe S, De Placido S, et al: Analysis of differential
miRNA expression in primary tumor and stroma of colorectal cancer
patients. Biomed Res Int. 2014:8409212014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Motoyama K, Inoue H, Takatsuno Y, Tanaka
F, Mimori K, Uetake H, Sugihara K and Mori M: Over- and
under-expressed microRNAs in human colorectal cancer. Int J Oncol.
34:1069–1075. 2009.PubMed/NCBI
|
|
19
|
Hur K, Toiyama Y, Schetter AJ, Okugawa Y,
Harris CC, Boland CR and Goel A: Identification of a
metastasis-specific MicroRNA signature in human colorectal cancer.
J Natl Cancer Inst. 107:dju4922015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nishida N, Yamashita S, Mimori K, Sudo T,
Tanaka F, Shibata K, Yamamoto H, Ishii H, Doki Y and Mori M:
MicroRNA-10b is a prognostic indicator in colorectal cancer and
confers resistance to the chemotherapeutic agent 5-fluorouracil in
colorectal cancer cells. Ann Surg Oncol. 19:3065–3071. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang Y, Li Z, Zhao X, Zuo X and Peng Z:
miR-10b promotes invasion by targeting HOXD10 in colorectal cancer.
Oncol Lett. 12:488–494. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Abdelmaksoud-Dammak R, Chamtouri N, Triki
M, Saadallah-Kallel A, Ayadi W, Charfi S, Khabir A, Ayadi L,
Sallemi-Boudawara T and Mokdad-Gargouri R: Overexpression of
miR-10b in colorectal cancer patients: Correlation with TWIST-1 and
E-cadherin expression. Tumour Biol. 39:10104283176959162017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Strubberg AM and Madison BB: MicroRNAs in
the etiology of colorectal cancer: Pathways and clinical
implications. Dis Model Mech. 10:197–214. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Qian X, Zhao P, Li W, Shi ZM, Wang L, Xu
Q, Wang M, Liu N, Liu LZ and Jiang BH: MicroRNA-26a promotes tumor
growth and angiogenesis in glioma by directly targeting prohibitin.
CNS Neurosci Ther. 19:804–812. 2013.PubMed/NCBI
|
|
25
|
Vishnubalaji R, Hamam R, Abdulla MH,
Mohammed MA, Kassem M, Al-Obeed O, Aldahmash A and Alajez NM:
Genome-wide mRNA and miRNA expression profiling reveal multiple
regulatory networks in colorectal cancer. Cell Death Dis.
6:e16142015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jinushi T, Shibayama Y, Kinoshita I,
Oizumi S, Jinushi M, Aota T, Takahashi T, Horita S, Dosaka-Akita H
and Iseki K: Low expression levels of microRNA-124-5p correlated
with poor prognosis in colorectal cancer via targeting of SMC4.
Cancer Med. 3:1544–1552. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pellatt DF, Stevens JR, Wolff RK, Mullany
LE, Herrick JS, Samowitz W and Slattery ML: Expression profiles of
miRNA subsets distinguish human colorectal carcinoma and normal
colonic mucosa. Clin Transl Gastroenterol. 7:e1522016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lv H, Zhang Z, Wang Y, Li C, Gong W and
Wang X: MicroRNA-92a promotes colorectal cancer cell growth and
migration by inhibiting KLF4. Oncol Res. 23:283–290. 2016.
View Article : Google Scholar
|
|
29
|
Chen Y, Song Y, Wang Z, Yue Z, Xu H, Xing
C and Liu Z: Altered expression of MiR-148a and MiR-152 in
gastrointestinal cancers and its clinical significance. J
Gastrointest Surg. 14:1170–1179. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Takahashi M, Cuatrecasas M, Balaguer F,
Hur K, Toiyama Y, Castells A, Boland CR and Goel A: The clinical
significance of MiR-148a as a predictive biomarker in patients with
advanced colorectal cancer. PLoS One. 7:e466842012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yang J, Ma D, Fesler A, Zhai H,
Leamniramit A, Li W, Wu S and Ju J: Expression analysis of microRNA
as prognostic biomarkers in colorectal cancer. Oncotarget.
8:52403–52412. 2016.PubMed/NCBI
|
|
32
|
Dong Y, Yu J and Ng SS: MicroRNA
dysregulation as a prognostic biomarker in colorectal cancer.
Cancer Manag Res. 6:405–422. 2014.PubMed/NCBI
|
|
33
|
Hibino Y, Sakamoto N, Naito Y, Goto K, Oo
HZ, Sentani K, Hinoi T, Ohdan H, Oue N and Yasui W: Significance of
miR-148a in colorectal neoplasia: Downregulation of miR-148a
contributes to the carcinogenesis and cell invasion of colorectal
cancer. Pathobiology. 82:233–241. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yu B, Liu X and Chang H: MicroRNA-143
inhibits colorectal cancer cell proliferation by targeting MMP7.
Minerva Med. 108:13–19. 2017.PubMed/NCBI
|
|
35
|
Hu Y, Ma Z, He Y, Liu W, Su Y and Tang Z:
PART-1 functions as a competitive endogenous RNA for promoting
tumor progression by sponging miR-143 in colorectal cancer. Biochem
Biophys Res Commun. 490:317–323. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Guo H, Chen Y, Hu X, Qian G, Ge S and
Zhang J: The regulation of Toll-like receptor 2 by miR-143
suppresses the invasion and migration of a subset of human
colorectal carcinoma cells. Mol Cancer. 12:772013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sun G, Cheng YW, Lai L, Huang TC, Wang J,
Wu X, Wang Y, Huang Y, Wang J, Zhang K, et al: Signature miRNAs in
colorectal cancers were revealed using a bias reduction small RNA
deep sequencing protocol. Oncotarget. 7:3857–3872. 2016.PubMed/NCBI
|
|
38
|
Tyler DM, Okamura K, Chung WJ, Hagen JW,
Berezikov E, Hannon GJ and Lai EC: Functionally distinct regulatory
RNAs generated by bidirectional transcription and processing of
microRNA loci. Genes Dev. 22:26–36. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Landgraf P, Rusu M, Sheridan R, Sewer A,
Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M,
et al: A mammalian microRNA expression atlas based on small RNA
library sequencing. Cell. 129:1401–1414. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang JY, Wang CL, Wang XM and Liu FJ:
Comprehensive analysis of microRNA/mRNA signature in colon
adenocarcinoma. Eur Rev Med Pharmacol Sci. 21:2114–2129.
2017.PubMed/NCBI
|
|
41
|
Sun Y, Liu Y, Cogdell D, Calin GA, Sun B,
Kopetz S, Hamilton SR and Zhang W: Examining plasma microRNA
markers for colorectal cancer at different stages. Oncotarget.
7:11434–11449. 2016.PubMed/NCBI
|
|
42
|
Ding L, Yu LL, Han N and Zhang BT: miR-141
promotes colon cancer cell proliferation by inhibiting MAP2K4.
Oncol Lett. 13:1665–1671. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Xi Y, Formentini A, Chien M, Weir DB,
Russo JJ and Ju J, Kornmann M and Ju J: Prognostic values of
microRNAs in colorectal cancer. Biomark Insights. 2:113–121.
2006.PubMed/NCBI
|
|
44
|
Chen J, Wang W, Zhang Y, Hu T and Chen Y:
The roles of miR-200c in colon cancer and associated molecular
mechanisms. Tumour Biol. 35:6475–6483. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Toiyama Y, Hur K, Tanaka K, Inoue Y,
Kusunoki M, Boland CR and Goel A: Serum miR-200c is a novel
prognostic and metastasis-predictive biomarker in patients with
colorectal cancer. Ann Surg. 259:735–743. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hur K, Toiyama Y, Takahashi M, Balaguer F,
Nagasaka T, Koike J, Hemmi H, Koi M, Boland CR and Goel A:
MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT)
in human colorectal cancer metastasis. Gut. 62:1315–1326. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhang GJ, Zhou T, Liu ZL, Tian HP and Xia
SS: Plasma miR-200c and miR-18a as potential biomarkers for the
detection of colorectal carcinoma. Mol Clin Oncol. 1:379–384. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Michael MZ, O'Connor SM, van Holst
Pellekaan NG, Young GP and James RJ: Reduced accumulation of
specific microRNAs in colorectal neoplasia. Mol Cancer Res.
1:882–891. 2003.PubMed/NCBI
|
|
49
|
Slaby O, Svoboda M, Fabian P, Smerdova T,
Knoflickova D, Bednarikova M, Nenutil R and Vyzula R: Altered
expression of miR-21, miR-31, miR-143 and miR-145 is related to
clinicopathologic features of colorectal cancer. Oncology.
72:397–402. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Jiang X, Wang W, Yang Y, Du L, Yang X,
Wang L, Zheng G, Duan W, Wang R, Zhang X, et al: Identification of
circulating microRNA signatures as potential noninvasive biomarkers
for prediction and prognosis of lymph node metastasis in gastric
cancer. Oncotarget. 8:65132–65142. 2017.PubMed/NCBI
|
|
51
|
Li D, Hu J, Song H, Xu H, Wu C, Zhao B,
Xie D, Wu T, Zhao J and Fang L: miR-143-3p targeting LIM domain
kinase 1 suppresses the progression of triple-negative breast
cancer cells. Am J Transl Res. 9:2276–2285. 2017.PubMed/NCBI
|
|
52
|
He Z, Yi J, Liu X, Chen J, Han S, Jin L,
Chen L and Song H: MiR-143-3p functions as a tumor suppressor by
regulating cell proliferation, invasion and epithelial-mesenchymal
transition by targeting QKI-5 in esophageal squamous cell
carcinoma. Mol Cancer. 15:512016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Li C, Yin Y, Liu X, Xi X, Xue W and Qu Y:
Non-small cell lung cancer associated microRNA expression
signature: Integrated bioinformatics analysis, validation and
clinical significance. Oncotarget. 8:24564–24578. 2017.PubMed/NCBI
|
|
54
|
Luo X, Stock C, Burwinkel B and Brenner H:
Identification and evaluation of plasma microRNAs for early
detection of colorectal cancer. PLoS One. 8:e628802013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Waters PS, McDermott AM, Wall D, Heneghan
HM, Miller N, Newell J, Kerin MJ and Dwyer RM: Relationship between
circulating and tissue microRNAs in a murine model of breast
cancer. PLoS One. 7:e504592012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Valadi H, Ekström K, Bossios A, Sjöstrand
M, Lee JJ and Lötvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Cancer Genome Atlas Network: Comprehensive
molecular characterization of human colon and rectal cancer.
Nature. 487:330–337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Liu W, Dong X, Mai M, Seelan RS, Taniguchi
K, Krishnadath KK, Halling KC, Cunningham JM, Boardman LA, Qian C,
et al: Mutations in AXIN2 cause colorectal cancer with defective
mismatch repair by activating beta-catenin/TCF signalling. Nat
Genet. 26:146–147. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
60
|
Suzuki H, Watkins DN, Jair KW, Schuebel
KE, Markowitz SD, Chen WD, Pretlow TP, Yang B, Akiyama Y, Van
Engeland M, et al: Epigenetic inactivation of SFRP genes allows
constitutive WNT signaling in colorectal cancer. Nat Genet.
36:417–422. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
61
|
Koo BK, Spit M, Jordens I, Low TY, Stange
DE, van de Wetering M, van Es JH, Mohammed S, Heck AJ, Maurice MM
and Clevers H: Tumour suppressor RNF43 is a stem-cell E3 ligase
that induces endocytosis of Wnt receptors. Nature. 488:665–669.
2012. View Article : Google Scholar : PubMed/NCBI
|