Introduction
Breast cancer is the most common malignancy in women
worldwide, including Thailand, accounting for 25% of all cancers in
women (1,2). Hormone receptor (HR)-negative breast
cancer or non-luminal breast cancer patients have a poorer outcome
than other subtypes and lack of hormonal therapy for long-term
control of the disease. Non-luminal breast cancer comprises triple
negative breast cancer (TNBC) and HER2 subtypes. Due to a lack of
target for therapy, systemic chemotherapy is the main treatment for
TNBC. In addition, approximately 20-50% of HER2-positive patients
showed resistance to Trastuzumab one year after treatment (3). Therefore, identification of novel
prognostic markers and alternative treatments is imperative for
both subtypes.
Androgen receptor (AR), a class I steroid receptor
is commonly expressed up to 70% in primary breast cancer and
approximately 50-75% in metastatic breast cancer (4,5). In
luminal subtype breast cancer, AR co-expression was associated with
better outcomes (6,7). AR expression was a significant
prognostic factor for disease-free survival (DFS), overall survival
(OS) and decreased risk of metastasis of non-luminal subtype breast
cancer in some studies (8-10).
By contrast, androgen can induce proliferation of
AR-positive/estrogen receptor (ER)-negative cells as commonly found
in molecular apocrine subtype which had AR expression of
approximately 50% (11-13).
Previous studies reported that RNA binding protein, Lin28A
(referred to as Lin28 in this study), which regulates let-7 miRNA,
stimulates HER2 expression and alters AR promoter activity
(14-16).
The upregulation of Lin28 in adults leads to carcinogenesis and
progressive cell proliferation in several malignancies, including
breast cancer (17-20).
However, the role of these proteins in non-luminal breast cancer
remains controversial. The aim of the present study was to evaluate
the clinical significance of AR and Lin28 in non-luminal subtype
breast cancer. It was found that AR and Lin28 are co-expressed.
Thus, Lin28 may be a novel marker for prognosis and targeted for
treatment in HER2 subtype breast cancer.
Patients and methods
Patients and data collection
In total, 284 patients were retrospectively
recruited at the Division of Head Neck and Breast Surgery,
Department of Surgery, Faculty of Medicine, Siriraj Hospital,
Mahidol University (Bangkok, Thailand) from January 2007 to January
2016. The patients with pathological stage I-III, invasive ductal
breast carcinomas, age at diagnosis equal to or more than 20 years,
and HR negative were included. The sample size was determined by
the formula for estimation of infinite population proportion using
parameters from a previous study (15). The proportions of Ki-67 status were
0.375 and 0.197 in groups 1 and 2. The ratios of proportion in both
groups were 2.590 (according to AR expression), α=0.05, 2-sided
test, and power 80%. This resulted in a sample size of 239. To
achieve statistically significant difference and the expected 10%
dropout of the patients, the total sample size was approximately
250 cases. The current study was approved by the ethics committee
of the Faculty of Medicine Siriraj Hospital, Mahidol University,
Bangkok, Thailand (COA no. Si733/2016). This study was performed in
a retrospective manner, therefore, no informed consent was obtained
from the patients.
The data collected from medical records comprised
age at diagnosis, pathological reports, surgical procedure,
adjuvant treatment (chemotherapy, hormonal therapy, targeted
therapy, and radiotherapy), and follow-up data. Repository formalin
fixed-paraffin embedded (FFPE) breast cancer and non-neoplasm
control tissues (prostate, tonsil, and testis) in excess of
standard pathological examination were obtained from the Department
of Pathology, Faculty of Medicine, Siriraj Hospital, Mahidol
University. The case record forms did not indicate any
identification that linked to individual patients.
Tissue microarray (TMA)
All H&E-stained slides and corresponding
paraffin blocks of each case including non-cancerous breast tissues
were collected and reviewed. The selected areas mapped on donor
paraffin blocks were punched by manual microarrayer with diameter 2
mm for 3 cores and placed into the applied recipient mold. Each
mold was melted at 60˚C for 6 min and re-embedded. Finally, each
slide contained triplicate of 17 cases, negative, and positive
tissue controls. The TMA blocks were sliced into 4 µm thickness.
The section ribbon was placed on the slide glass and air dried for
30 min.
Immunohistochemistry
Expression of AR and Lin28 was assessed by
immunohistochemistry (IHC) using mouse monoclonal anti-human AR
(AR441, dilution 1:300, Dako) and mouse monoclonal anti-human
Lin28A (55CT58.12.1, dilution 1:75, Sigma-Aldrich). AR staining was
performed by semi-autostainer (Agilent Technologies, Dako;
Autostainer Link 48). Deparaffinization, rehydration, and antigen
retrieval were performed by target retrieval solution high pH (pH
9.0) at 95˚C with PT Link (Dako PT link). Lin28 staining was
performed by manual procedure. PT Link (Dako PT link) with target
retrieval solution high pH (pH 9.0) was used. The sections were
incubated overnight at 4˚C with primary antibody. Subsequently, the
sections were warmed up at room temperature (25˚C) and rinsed twice
with PBS. Peroxidase-blocking solution (Dako Peroxidase Blocking
Code SM801) was used for endogenous blocking for approximately 5
min and then rinsed twice with PBS for 10 min. The sections were
incubated with HRP-conjugated secondary antibody (Envision FLEX-HRP
Code SM802) for 20 min. The visualization step was performed with
Envision FLEX DAB and Chromogen (Envision FLEX DAB and Chromogen
Code DM827) for 12 min and then rinsed with tap water for 5 min.
The sections were counterstained with hematoxylin. Finally, the
sections were dehydrated with alcohol series (95 and 100% alcohol
and acetone, respectively) and cleared with xylene.
The protein expression level was calculated by a
mean score of 3 cores. The AR-positive status was determined by an
established cut-off value of >20% of nucleus staining. Lin28
status was evaluated by cytoplasmic staining and scored according
to the Modified Allred Scoring system including staining intensity
and percentage of positive cells. The sum of staining and
percentage was classified as: 0-2, negative and 3-8, positive.
Scoring was performed by two experienced breast pathologists who
did not know the clinical data of patients.
Dual in situ hybridization (DISH)
For HER2 equivocal cases (IHC score 2+), HER2
amplification status was assessed by dual color in situ
hybridization (DISH). The process was performed by using a
cocktail-specific probe for HER2 and chromosome 17 (Chr 17) on a
single slide. The HER2 copies were detected using the HER2
DNP-labeled probe and visualized via ultraView SISH detection kit
[Ventana ultraView SISH dinitrophenyl (DNP), Ventana Medical
System, USA]. Centromeres of chromosome 17 were assigned by Chr17
DIG-labeled probe and visualized by ultraView Red ISH detection kit
[Ventana ultraViewRed ISH digoxigenin (DIG), Ventana Medical
System]. DISH staining was performed by auto-staining system
(BenchMark XT automated slide stainer, Ventana). The black signal
(HER2) to red signal (Chr 17) ratio was manually counted by light
microscope at a magnification, x20 for 20 cells and calculated. The
ratios of equal or more than 2.0 were considered as HER2
amplification.
Statistical analysis
Associations between protein expression and
clinicopathological parameters were analyzed using a Chi-square
test. Binary logistic regression was performed for multivariate
analysis using backward conditional method. Survival analysis was
performed by Log-rank test and survival curves were estimated by
Kaplan-Meier method. The DFS time was calculated from the date of
surgery to the date of cancer reccurrence, metastasis or death. The
OS time was calculated from the date of surgery to the date of
death. The Cox proportional hazards model was applied for
prediction of survival rate. Multivariate analysis was performed by
Cox regression to evaluate the effect of independent prognostic
factors on DFS and OS. The SPSS software version 21 was used for
statistical analysis. P<0.05 was considered to indicate a
statistically significant difference.
Results
Patient characteristics
A total of 284 patients were eligible and recruited
in this study. Patient characteristics are presented in Table I. HER2 equivocal cases by IHC were
further assessed for HER2 amplification by DISH (Fig. 1). The mean age at diagnosis was 55.39
years (±11.36 years). There were 131 HER2 subtype breast cancer
patients and 24 patients receiving HER2-targeted therapy. TNBC
subtype was 153 cases. Two hundred and two patients (71.1%) were
post-menopause. The mean tumor size was 20.9 mm (±10.5 mm). A tumor
size >20 mm was found in 190 cases (66.9%). Approximately half
of the patients were in stage II at diagnosis (143 cases, 50.4%).
There was no grade I tumor while the majority of the patients had
grade III tumor (67.3%). All the patients received chemotherapy
according to clinical practice guidelines and completed the course
of treatment.
 | Table IDemographics data of non-luminal
subtype patients. |
Table I
Demographics data of non-luminal
subtype patients.
| Parameters | Number, n=284
(%) |
|---|
| Age at diagnosis | 55.39 (±11.36) |
| Age at diagnosis [n
(%)] |
|
≤50
years | 87 (30.6) |
|
>50
years | 197 (69.4) |
| Tumor size [n
(%)] |
|
≤20 mm | 94 (33.1) |
|
>20-50
mm | 172 (60.6) |
|
>50
mm | 18 (6.3) |
| Histological grading
[n (%)] |
|
II | 93 (32.7) |
|
III | 191 (67.3) |
| Lymphovascular
invasion [n (%)] |
|
Absence | 201 (70.8) |
|
Presence | 83 (29.2) |
| Axillary nodal
metastasis [n (%)] |
|
No | 166 (58.5) |
|
Yes | 118 (41.5) |
| N stage [n (%)] |
|
N0 | 166 (58.5) |
|
N1 | 59 (20.8) |
|
N2 | 28 (9.9) |
|
N3 | 31 (10.9) |
| Staging [n (%)] |
|
I | 73 (25.7) |
|
II | 143 (50.4) |
|
III | 168 (23.9) |
| HER-2 status [n
(%)] |
|
Negative | 153 (53.9) |
|
Positive | 131 (46.1) |
| HER-2 targeted
therapy [n (%)]a |
|
No | 107 (81.7) |
|
Yes | 24 (18.3) |
AR and Lin28 expression
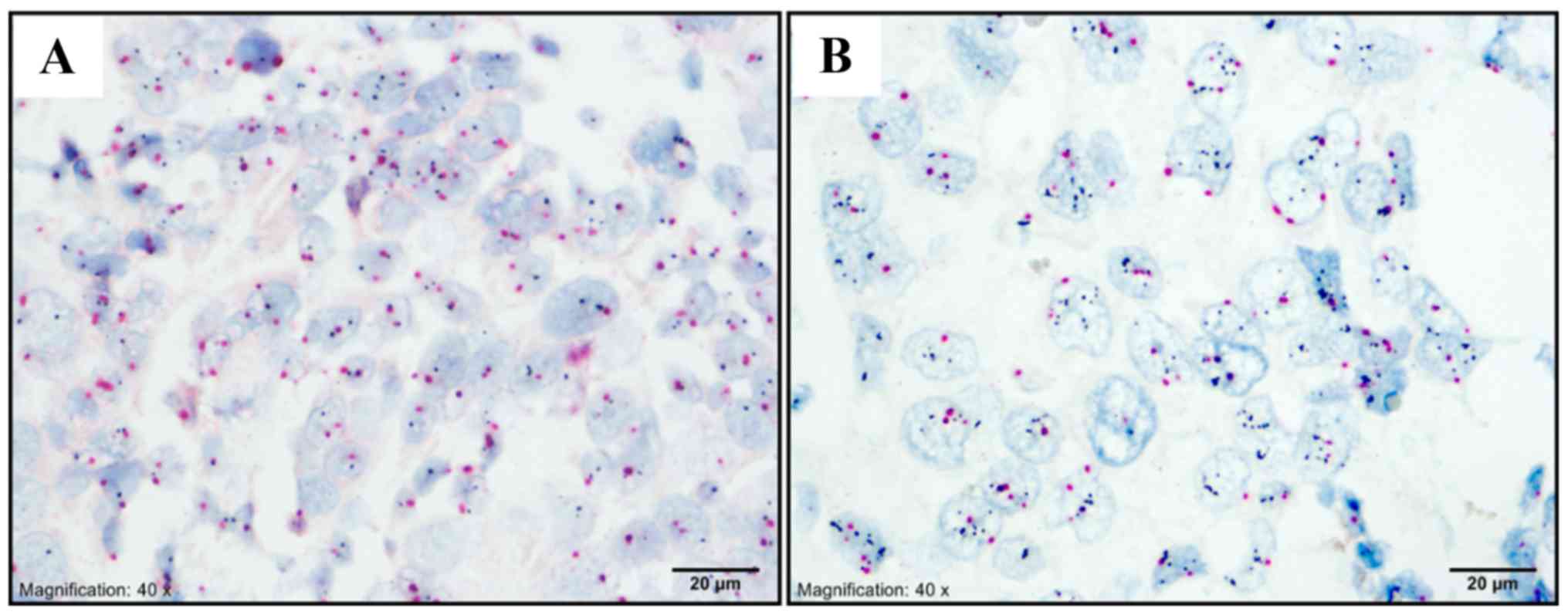
AR was expressed in 66 of 284 non-luminal tumors
(23.2%). AR expression was detected in 45 (68.2%) and 21 cases
(31.8%) in HER2 and TNBC subtype, respectively. Lin28 protein was
detected in 201 out of 284 patients (70.8%). From these, 164, 34,
and 3 patients had weak, moderate, and strong AR staining,
respectively. Ninety-two TNBC and 109 HER2 patients had Lin28
expression (Figs.
2-3).
Association between protein expression
and clinicopathological parameters
Among 284 patients, AR and Lin28 status was
significantly associated with HER2-positive status. In addition,
both proteins were co-expressed together in non-luminal breast
cancer (Tables
II-III). Multivariate analysis revealed that AR-positive status
was associated with the absence of axillary lymph node metastasis
(OR=0.428, 95% CI 0.224-0.817, P=0.010), HER2-positive (OR=2.948,
95% CI 1.555-5.587, P=0.001), and Lin28-positive status (OR=15.756,
95% CI 3.707-66.979, P<0.001). Lin28 expression was associated
with HER2-positive (OR=2.562, 95% CI 1.426-4.603, P=0.002), and AR
positive status (OR=15.437, 95% CI 3.649-65.312, P<0.001).
Survival analysis
The median follow-up time was 43 months (1-139
months). There were 51 events that occurred during follow up
including 3 loco-regional recurrences, 12 metastases, and 36
deaths. Two hundred and thirty-three patients were alive without
disease. Univariate analysis via log-rank test revealed that tumor
size, pathological staging, axillary lymph node metastasis, and
lymphovascular invasion (LVI) were associated with lower DFS
(P=0.042, P<0.001, P<0.001 and P<0.001, respectively).
Pathological staging, axillary lymph node metastasis, and LVI were
associated with lower OS (P<0.001, P<0.001 and P<0.001,
respectively). Multivariate analysis revealed that pathological
stage and LVI were the strong independent factors for DFS
(HR=2.769, 95% CI 1.383-5.544, P=004 and HR=2.748, 95% CI
1.391-5.428, P=004, respectively) and OS (HR=3.160, 95% CI
1.347-7.415, P=0.008 and HR=3.615, 95% CI 1.533-8.525, P=0.003,
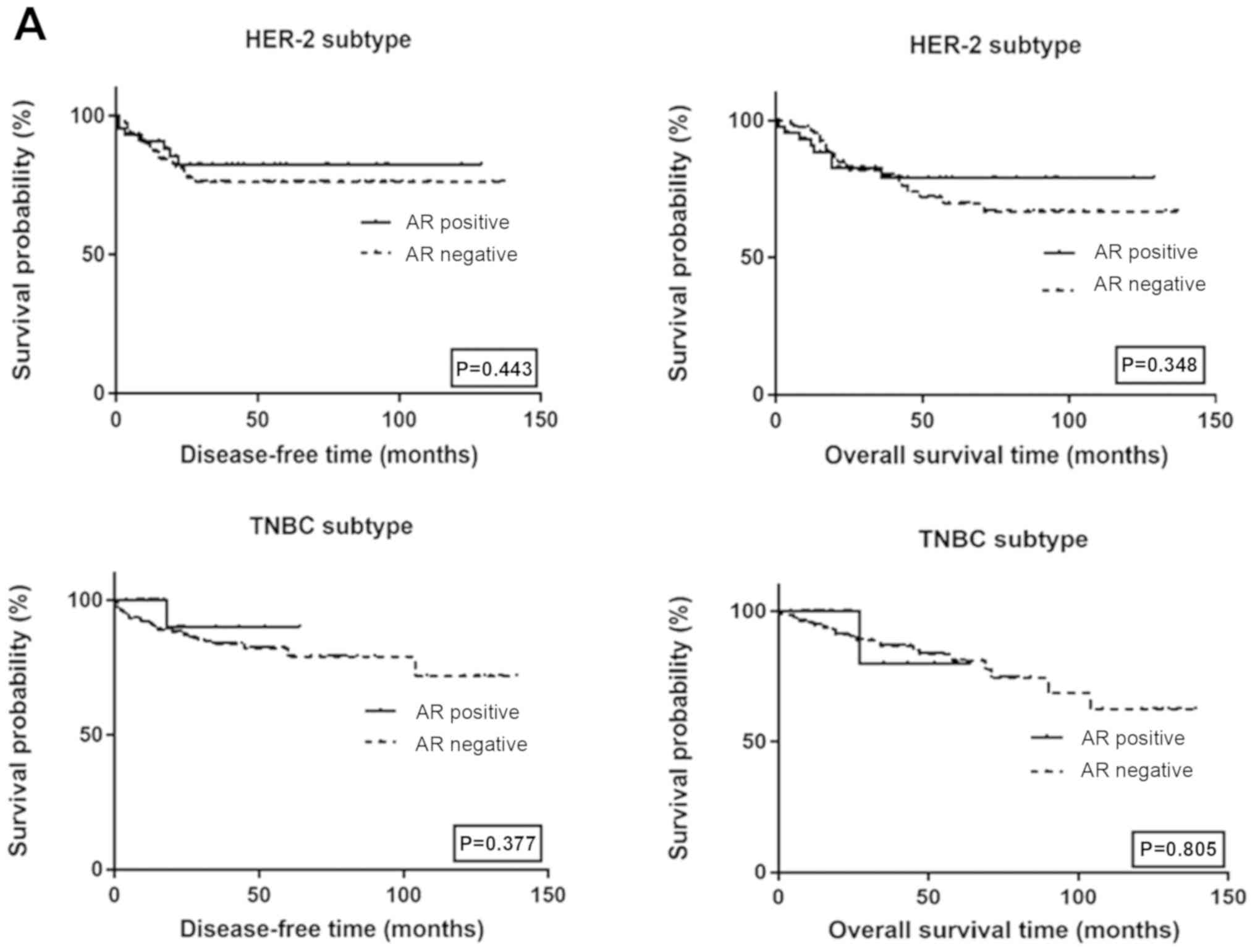
respectively). The survival curves among HER2 and TNBC subtypes by
AR and Lin28 status are shown in Fig.
4. There was no significant difference in survival among
different AR and Lin28 status.
Discussion
The present study demonstrated the associations
between the expression of AR and Lin28 in non-luminal breast
cancer. In HER2-overexpressed breast cancer, we also showed the
association between the expression of Lin28 and HER2.
A higher proportion of AR expression was observed in
HER2 subtype in the present study. Similar studies by Micello et
al (21) and Park et al
(22), showed that AR expression was
often detected in ER-negative/HER2-positive breast cancer. The
implications of HER2 and AR have been suggested in molecular basis.
HER2 is a transcriptional target of AR and able to activate ERK
activity (11,12). In vitro studies suggested that
androgen can induce proliferation in AR-positive/ER-negative cells
such as those commonly found in the molecular apocrine subtype
which exhibited AR co-expression of approximately 50% (3,13). He
et al reported that treatment with Enzalutamide, an AR
antagonist, reduced the ability of tumor growth via decreased cell
proliferation and increased cell death in HER2-positive breast
cancer, both in vitro and in vivo (3). AR-positive/ER-negative in HER2
overexpression or amplification in breast cancer has been reported
to be associated with unfavourable outcome when compared to those
with AR-negative (5,22-24).
However, in the present study, we did not find any significant
association between AR expression and unfavourable
clinicopathological parameters or worse survival outcomes.
One of the main AR transcriptional regulatory
cascades involves let-7 and Lin28. Lin28 is an RNA-binding protein
(RBP) that directly regulates let-7 miRNA. Aberration thereof could
lead to carcinogenesis and progressive cell proliferation in breast
cancer (25). In the HER2 subtype,
the association between AR expression and Lin28-positive status was
detected in the present study. Lin28 regulates the expression of AR
via c-myc, a proto-oncogene involved in cell proliferation
(16). The study by Feng et
al also demonstrated the relationship between Lin28 expression
and ER negative/HER2-positive in breast cancer cell (14). The Lin28 responsive element (LRE) is
200 nucleotides in length and is located in nearly 5' end of the
coding region of HER2 mRNA. The authors suggested that HER2 mRNA
contains a cis-acting element that is specifically recognized by
Lin28 within the coding region and activates translation in breast
cancer. An ‘A’ bulge flanked by two GC base pairs in the secondary
structure of HER2 mRNA served as the binding site for
Lin28(26). Shen et al,
reported that Lin28 and AR were co-expressed in
ER-negative/HER2-positive breast cancer tissues and cell lines,
suggesting a worse survival outcome (15,16).
In the present study, the positive association
between AR and Lin28 was noted. However, only one-third of positive
Lin28 breast cancer patients have positive AR expression. In
ER-negative/HER2-positive breast cancer, several signaling cascades
including the upregulation of Wnt and c-myc were involved in the
interaction between AR and HER2 (12,27).
Lin28 can activate the proliferation and growth of tumor cells via
Lin28/let7 pathway (20). Patients
with Lin28 expression tended to have a lower survival rate compared
to patients without Lin28 expression in HER2 subtype. This result
was in accordance with previous studies regarding the potential
role of Lin28 in decreased tumor suppressor miRNA and increased
oncoproteins in breast cancer (15,16,18,28-30).
In conclusion, this current results have
demonstrated the role of Lin28 in HER2-overexpressed breast cancer
and showed the potential prognostic factor. Thus, Lin28 may be a
novel marker for prognosis and future-targeted therapy for HER2
subtype breast cancer.
Acknowledgements
We would like to thank Miss Surat Phumphuang
(Division of Head Neck and Breast Surgery, Department of Surgery,
Faculty of Medicine, Siriraj Hospital, Mahidol University) for
helping with data collection and manuscript preparation.
Funding
This study was supported by Siriraj Graduate
Scholarship and Siriraj Grant for Research Development and Medical
Education, Faculty of Medicine Siriraj Hospital, Mahidol University
(grant no. R016133002).
Availability of data and materials
The datasets generated and/or used during the
present study are available from the corresponding author on
reasonable request.
Authors' contributions
DS, WP, KM, AK, TL and PO prepared the manuscript.
DS and PO treated the patients. NS and TC performed pathological
examination and data collection. WP, KM performed data collection
and the statistical analysis. PO provided the concept of the study
and finalized the manuscript. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
This study has been approved by the ethics committee
of the Faculty of Medicine, Siriraj Hospital, Mahidol University
(Bangkok, Thailand; COA no. Si733/2016).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sa-Nguanraksa D, Chuangsuwanich T,
Pongpruttipan T and O-Charoenrat P: High vascular endothelial
growth factor gene expression predicts poor outcome in patients
with non-luminal A breast cancer. Mol Clin Oncol. 3:1103–1108.
2015.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Sa-Nguanraksa D, Sasanakietkul T,
O-Charoenrat C, Kulprom A and O-Charoenrat P: Gail model
underestimates breast cancer risk in thai population. Asian Pac J
Cancer Prev. 20:2385–2389. 2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
He L, Du Z, Xiong X, Ma H, Zhu Z, Gao H,
Cao J, Li T, Li H, Yang K, et al: Targeting androgen receptor in
treating HER2 positive breast cancer. Sci Rep.
7(14584)2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Hickey TE, Robinson JL, Carroll JS and
Tilley WD: Minireview: The androgen receptor in breast tissues:
Growth inhibitor, tumor suppressor, oncogene? Mol Endocrinol.
26:1252–1267. 2012.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Rahim B and O'Regan R: AR signaling in
breast cancer. Cancers (Basel). 9(E21)2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Hu R, Dawood S, Holmes MD, Collins LC,
Schnitt SJ, Cole K, Marotti JD, Hankinson SE, Colditz GA and Tamimi
RM: Androgen receptor expression and breast cancer survival in
postmenopausal women. Clin Cancer Res. 17:1867–1874.
2011.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Peters AA, Buchanan G, Ricciardelli C,
Bianco-Miotto T, Centenera MM, Harris JM, Jindal S, Segara D, Jia
L, Moore NL, et al: Androgen receptor inhibits estrogen
receptor-alpha activity and is prognostic in breast cancer. Cancer
Res. 69:6131–6140. 2009.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Rampurwala M, Wisinski KB and O'Regan R:
Role of the androgen receptor in triple-negative breast cancer.
Clin Adv Hematol Oncol. 14:186–193. 2016.PubMed/NCBI
|
|
9
|
Jiang HS, Kuang XY, Sun WL, Xu Y, Zheng
YZ, Liu YR, Lang GT, Qiao F, Hu X and Shao ZM: Androgen receptor
expression predicts different clinical outcomes for breast cancer
patients stratified by hormone receptor status. Oncotarget.
7:41285–41293. 2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Sutton LM, Cao D, Sarode V, Molberg KH,
Torgbe K, Haley B and Peng Y: Decreased androgen receptor
expression is associated with distant metastases in patients with
androgen receptor-expressing triple-negative breast carcinoma. Am J
Clin Pathol. 138:511–516. 2012.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Naderi A and Hughes-Davies L: A
functionally significant cross-talk between androgen receptor and
ErbB2 pathways in estrogen receptor negative breast cancer.
Neoplasia. 10:542–548. 2008.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Chia KM, Liu J, Francis GD and Naderi A: A
feedback loop between androgen receptor and ERK signaling in
estrogen receptor-negative breast cancer. Neoplasia. 13:154–166.
2011.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Robinson JL, Macarthur S, Ross-Innes CS,
Tilley WD, Neal DE, Mills IG and Carroll JS: Androgen receptor
driven transcription in molecular apocrine breast cancer is
mediated by FoxA1. EMBO J. 30:3019–3027. 2011.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Feng C, Neumeister V, Ma W, Xu J, Lu L,
Bordeaux J, Maihle NJ, Rimm DL and Huang Y: Lin28 regulates HER2
and promotes malignancy through multiple mechanisms. Cell Cycle.
11:2486–2494. 2012.PubMed/NCBI View
Article : Google Scholar
|
|
15
|
Shen H, Yang Y, Zhao L, Yuan J and Niu Y:
Lin28A and androgen receptor expression in ER-/Her2+ breast cancer.
Breast Cancer Res Treat. 156:135–147. 2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Shen H, Zhao L, Feng X, Xu C, Li C and Niu
Y: Lin28A activates androgen receptor via regulation of c-myc and
promotes malignancy of ER-/Her2+ breast cancer. Oncotarget.
7:60407–60418. 2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Viswanathan SR, Powers JT, Einhorn W,
Hoshida Y, Ng TL, Toffanin S, O'Sullivan M, Lu J, Phillips LA,
Lockhart VL, et al: Lin28 promotes transformation and is associated
with advanced human malignancies. Nat Genet. 41:843–848.
2009.PubMed/NCBI View
Article : Google Scholar
|
|
18
|
Viswanathan SR and Daley GQ: Lin28: A
microRNA regulator with a macro role. Cell. 140:445–449.
2010.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Yang J, Bennett BD, Luo S, Inoue K, Grimm
SA, Schroth GP, Bushel PR, Kinyamu HK and Archer TK: LIN28A
modulates splicing and gene expression programs in breast cancer
cells. Mol Cell Biol. 35:3225–3243. 2015.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Balzeau J, Menezes MR, Cao S and Hagan JP:
The LIN28/let-7 pathway in cancer. Front Genet.
8(31)2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Micello D, Marando A, Sahnane N, Riva C,
Capella C and Sessa F: Androgen receptor is frequently expressed in
HER2-positive, ER/PR-negative breast cancers. Virchows Arch.
457:467–476. 2010.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Park S, Koo J, Park HS, Kim JH, Choi SY,
Lee JH, Park BW and Lee KS: Expression of androgen receptors in
primary breast cancer. Ann Oncol. 21:488–492. 2010.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Ni M, Chen Y, Lim E, Wimberly H, Bailey
ST, Imai Y, Rimm DL, Liu XS and Brown M: Targeting androgen
receptor in estrogen receptor-negative breast cancer. Cancer Cell.
20:119–131. 2011.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Pietri E, Conteduca V, Andreis D, Massa I,
Melegari E, Sarti S, Cecconetto L, Schirone A, Bravaccini S, Serra
P, et al: Androgen receptor signaling pathways as a target for
breast cancer treatment. Endocr Relat Cancer. 23:R485–498.
2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Nguyen LH and Zhu H: Lin28 and let-7 in
cell metabolism and cancer. Transl Pediatr. 4:4–11. 2015.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Lei XX, Xu J, Ma W, Qiao C, Newman MA,
Hammond SM and Huang Y: Determinants of mRNA recognition and
translation regulation by Lin28. Nucleic Acids Res. 40:3574–3584.
2012.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Ni M, Chen Y, Fei T, Li D, Lim E, Liu XS
and Brown M: Amplitude modulation of androgen signaling by c-MYC.
Genes Dev. 27:734–748. 2013.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Mayr F and Heinemann U: Mechanisms of
Lin28-mediated miRNA and mRNA regulation-a structural and
functional perspective. Int J Mol Sci. 14:16532–16553.
2013.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Tsialikas J and Romer-Seibert J: LIN28:
Roles and regulation in development and beyond. Development.
142(2397)2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Thammaiah CK and Jayaram S: Role of let-7
family microRNA in breast cancer. Noncoding RNA Res. 1:77–82.
2016.PubMed/NCBI View Article : Google Scholar
|