1. Introduction
Numerous studies have shown that microbes have the
ability to drive the development of cancer (1). A wide variety of microorganisms
including viruses (2), and bacteria
(3) have been robustly associated
with higher risk of cancer. Microbes such as Helicobacter
pylori (H. pylori) have been shown to significantly
increase the risk of developing gastric cancer and duodenal cancer
(4); cytomegalovirus (CMV)
associated with subtypes of colorectal cancer (CRC) (5) and inflammatory breast cancer (IBC)
(6); human papilloma virus (HPV)
infection is associated with many cervical cancers, penile cancers,
anogenital, and head and neck cancers (7); human T-cell leukemia virus-1 (HTLV-1)
is the oncogenic cause of Adult T cell leukemia (8); human herpes virus 8 (HHV8) infection is
associated with subtypes of Castleman disease as well as sarcomas
(9,10); and Epstein Barr virus (EBV) is
associated with lymphomas derived from B-cells, NK-cells, T-cells,
and carcinomas derived from epithelial cells (11,12).
Though many cancers are known to be driven by or associated with
microbial infections, it should be noted that only a fraction of
these infections actually leads to cancer. Only 3% of H.
pylori infections result in gastric cancer (13), approximately 1.5% of EBV infections
are associated with cancer worldwide (11), ~5% of HTLV-1 infected patients
develop HTLV-1 associated myelopathy/tropical spastic paraparesis
(HAM/TSP) (14). Furthermore, it is
interesting that microbes that have been linked with high risk of
cancer have a high level of endemic infection. EBV is prevalent in
~90% of the world's population (15), CMV have frequency of infection
ranging from 50 to 100% in the general adult population worldwide
(16), and H. pylori
colonizes approximately 50% of world population (4). Thus, it is evident that other factors,
in addition to infection by a microorganism of a particular
species, must be significant in modifying risks associated with
cancer. Factors such as patient genetic background (14), environmental factors (17), status of the immune system (18), and microbial genotype (19) have been known to play important roles
in determining cancer risk factors in the presence of microbial
infection. Therefore, it is difficult to predict the risk of cancer
imposed by microorganisms solely based on their occurrence in an
individual. In this review we focus on the importance of microbial
genotypes in determining cancer risk factors (Fig. 1; Table
I), and discuss the importance of incorporation of this
information into clinical diagnosis and prognosis.
 | Table IAssociation of microbial genotype
with increased risk of cancer. |
Table I
Association of microbial genotype
with increased risk of cancer.
| A, H.
pylori |
|---|
| Author, year | Cancer type | Associated
genotype | Strain
identification method | Increase in
risk | (Refs.) |
|---|
| Wroblewski et
al, 2010, Basso et al, 2008 | Gastric cancer and
premalignant lesions | CagA+ | Presence of
CagA gene and variation in its EPIYA motif | OR=7.37, 95% CI:
1.98-27.48 for one EPIYA OR=32.5, 95% CI: 8.41-125.58 for 2 or more
EPIYA for gastric cancer | (4,21) |
| Basso et al,
2008, Foegeding et al, 2016 | Gastric cancer and
peptic ulcer | Genotype s1, m1,
and i1 | Variation in
VacA gene | OR=5.02, 95% CI:
2.10-11.98, P<0.001 for Gastric cancer OR=2.58, 95% CI:
1.19-5.61, P<0.05 for peptic ulcer | (21,22) |
| Gerhard et
al, 1999 | Duodenal ulcer and
gastric cancer | babA2+ | Presence of
babA2 gene | babA2 genotype
present significantly more P=0.0002 for DU P=0.033 for gastric
adenocarcinoma (Chi-squared test) | (23) |
| Yamaoka et
al, 2002 | Duodenal ulcer | OipA+ | Presence of
OipA gene | OR (adjusted)=5,
95% CI: 2.1-11.9 | (24) |
| Hussein, 2010 | Duodenal ulcer and
gastric cancer in some population | DupA+ | Presence of
DupA gene | OR=1.4, 95% CI:
1.1-1.7, P=0.001 | (25) |
| Franco et
al, 2009 | Gastric cancer | FlaA mutant | Point mutation in
Fla gene | Decreased risk | (26) |
| B, CMV |
| Author, year | Cancer type | Associated
genotype | Strain
identification method | Increase in
risk | (Refs.) |
| Chen et al,
2015 | Colorectal
cancer | Genotype B | Variation in
UL144 gene | HR=5.79, 95% CI:
1.30-25.81, P=0.021 | (5) |
| Mohamed et
al, 2014 | Inflammatory breast
cancer (IBC) | gB-1+gB-3 | Variation in
UL55 (gB) gene | Significantly
higher occurrence in IBC compared to non-IBC (P=0.029, Fisher's
exact test) | (6) |
| Mohamed et
al, 2014 | Inflammatory breast
cancer (IBC) | gN-1+gN-3b | Variation in
UL73 (gN) gene | Significantly
higher occurrence in IBC compared to non-IBC (P=0.034, Fisher's
exact test) | (6) |
| Mohamed et
al, 2014 | Inflammatory breast
cancer (IBC) | gN3+gN-4b/c | Variation in
UL73 (gN) gene | Significantly
higher occurrence in IBC compared to non-IBC (P=0.033, Fisher's
exact test) | (6) |
| C, EBV |
| Author, year | Cancer type | Associated
genotype | Strain
identification method | Increase in
risk | (Refs.) |
| Pai et al,
2007 | Nasopharyngeal
carcinoma | Cao CTAR2 | Variation in in the
C-terminal activating region of LMP1 gene | HR=2.01,
P=0.026 | (27) |
| D, HHV8 |
| Author, year | Cancer type | Associated
genotype | Strain
identification method | Increase in
risk | (Refs.) |
| Tozetto-Mendoza
et al, 2016 | AIDS-associated
Kaposi's | Genotype B | Variation in ORF K1
sarcoma | Presence of only
genotype B was significant P=0.0182 (Fisher's exact test) | (29) |
| E, HPV |
| Author, year | Cancer type | Associated
genotype | Strain
identification method | Increase in
risk | (Refs.) |
| Powell et
al, 2011 | Cervical
cancer | HPV-16 | Variations in
L1 gene | OR (adjusted for
age)=2770, 95% CI: 1050-7320 | (31) |
| Powell et
al, 2011 | | HPV-18 | Variations in
L1 gene | OR (adjusted for
age)=950, 95% CI: 330-2740 | (31) |
| D'Souza et
al, 2007 | Oropharyngeal
cancer | HPV-16 | Variations in
L1gene | OR=14.6, 95% CI:
6.3-36.6 | (32) |
| D'Souza et
al, 2007 | Anal cancer | HPV-16 | Variations in
L1 gene | OR=2.58, 95% CI:
1.31-5.08, P=0.006 | (32) |
| Bruno et al,
2007 | | HPV-31 | Variations in
L1 gene | OR=4.74, 95% CI:
2.00-11.22, P=0.0004 | (35) |
| F, HCV |
| Author, year | Cancer type | Associated
genotype | Strain
identification method | Increase in
risk | (Refs.) |
| Bruno et al,
2007 | Hepatocellular
carcinoma | Genotype 1b | Variation in 5'UTR
region | HR=3.02,
CI:1.40-6.53 compared to genotype 2a/c | (35) |
| G, HBV |
| Author, year | Cancer type | Associated
genotype | Strain
identification method | Increase in
risk | (Refs.) |
| Yu et al,
2005 Yeh et al, 2004 | Hepatocellular
carcinoma | Genotype C | Variations in
coding regions HBs and HBx | OR (adjusted)=5.11,
95% CI: 3.2-8.81 compared to other genotypes | (36,37) |
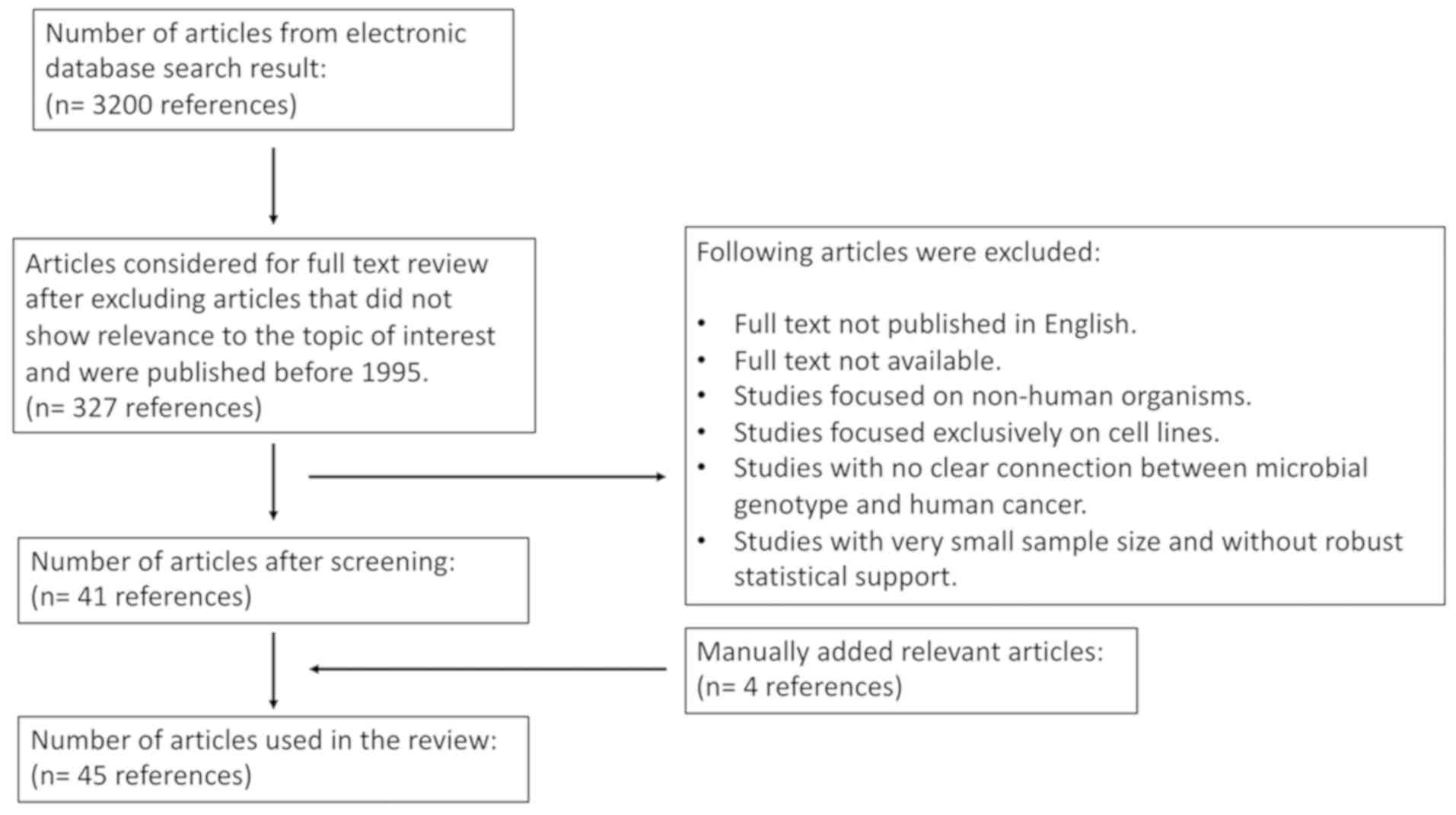
2. Literature search
Articles with information associated with microbial
genotype contributing to the cancer risk were researched in
electronic databases. Thirty-two hundred articles resulted from our
initial search. From this list, articles that showed
relevance to our topic of interest based on their title and
published between 1995 and May of 2019 were selected. This resulted
in 327 articles. Further, articles that were not published in
English, did not have full text available, focused on non-human
organisms and cell lines, did not have clear connection between
microbial genotype and human cancer, and contained very small
sample size without robust statistical support were excluded from
our study. Four articles that contained information of interest and
were not in our list were added manually. Our final list consisted
of 45 articles that were used in our study.
3. H. pylori genotypes and
cancer risks
Numerous studies have shown that among many
genotypes of microbes, some variants are associated with a higher
risk of cancer than others. In H. pylori, the presence of
the Cag pathogenicity island (Cag PAI), a 40 kb insertion with
27-31 genes flanked by 31 bp direct repeats, is associated with a
high risk of gastric cancer (GC) (4). More specifically, a gene called
Cytotoxin-associated gene A (CagA) within the Cag PAI is
associated with increased risk of gastric cancer and premalignant
lesions (20). CagA contains
an EPIYA motif, and variation in this motif is associated with
differential phosphorylation of CagA which is shown to greatly
influence the occurrence of H. pylori mediated GC (4). In fact, the odds of developing GC in
the presence of one EPIYA segment was increased to seven times
compared to CagA negative patients (OR=7.37, 95% CI: 1.98-27.48)
and the presence of two or more EPIYA segments increased the odds
to thirty-two-fold higher (OR=32.5, 95% CI: 8.41-125.58) (21). In addition, all H. pylori
possess another gene that encodes for a toxin called VacA and
variation in its three regions: Signal region (s), middle region
(m), and intermediate region (i) have been associated with
differential risk of GC induced by H. pylori. Specifically,
individuals infected with Genotype s1, m1 and i1 have a higher risk
of GC (OR=5.02, 95% CI: 2.10-11.98, P<0.001) and peptic ulcers
(OR=2.58, 95% CI: 1.19-5.61, P<0.05) compared to individuals
infected with Genotype s2, m2 and i2 (21,22). The
presence of intact Blood group antigen binding adhesin
(babA2) is another factor that has been linked with
increased pathogenicity of H. pylori. The prevalence of
babA2 positive H. pylori strains were found to be
significantly higher in duodenal ulcers (DU) (P=0.0002, chi squared
test) and gastric adenocarcinomas (P=0.033, Chi-squared test);
interestingly, the risk factor for babA2 increases even more
when it is found together with CagA and VacA s1
strains (23). Outer inflammatory
protein (OipA) is another factor that is known to increase the
pathogenicity of H. pylori. OipA is either found in
an intact form or an out of frame form; the presence of intact
OipA has been reported to increase the risk of DU and was
the most robust discrimination factor for separating outcomes of DU
and gastritis (OR=5, 95% CI: 2.1-11.9) (24). Interestingly it has been also
reported that the intact form OipA is often linked to more
pathogenic determinants of H. pylori such as vacAs1, vacAm1,
babA2 and most strongly with CagA positive genotypes. The
authors speculate that there is selection for the linkage observed
between cagA and in frame OipA, which is supported by
the observation that bacteria harboring cagA colonize poorly
when oipA is mutated. Such selective forces may be
responsible for sustaining more virulent forms of H. pylori.
The presence of Duodenal ulcer promoting gene a (dupA) gene
in the genome of H. pylori has been also associated with
increased risk of DU worldwide (OR=1.4, 95% CI: 1.1-1.7, P=0.001)
and in addition has also been shown to have increased risk of
gastric ulcer and GC in various parts of the world, although
dupA seems to promote DU and GC only in certain population
worldwide (4,25). Furthermore, an H. pylori
strain containing a point mutation in a gene encoding a flagella
subunit called Flagellin A (FlaA) was shown to
decrease the virulence of H. pylori, which the authors
speculate might be due to its decreased motility as a result of the
mutation (26).
4. Viral genotypes and cancer risks
Epstein Barr virus
Nasopharyngeal carcinoma (NPC) has a high local
control probability after treatment by primary radiotherapy.
Despite the prevalence of successful treatment, ~19-29% of patients
develop distant metastases with controlled locoregional disease,
and variations in the LMP1 gene in EBV have been linked to
distant metastasis of NPC (27). A
study involving 249 Taiwanese patients was conducted to test if a
variation in LMP1, called Cao C-terminal activation region 2
(Cao CTAR2), was associated with the distant metastasis in patients
with NPC treated with radiation or chemotherapy. It was observed
that patients with tumor harboring the Cao-CTAR2 variant had twice
the risk of developing distant metastasis (HR=2.01, P=0.026) and a
lower overall survival probability (P=0.047) compared to patients
without Cao-CTAR2. The authors also discovered that Cao-CTAR2 had
prognostic significance that was independent of patient clinical
characteristics and conveyed that this variant could be potentially
used as an additional marker for selecting patients that might
require more intensive treatment (27). In Hodgkin's Lymphoma derived cell
line, expressing Cao-CTAR2 variant of LAMP1 showed weak
expression of various cytokines, notably IFN-γ, and alteration in
cell cycle. IFN-γ has been shown to have anti-viral response
against EBV, and its lowered expression might be the way through
which Cao-CTAR2 infected cells escape the immune anti-viral
surveillance and promote cancer (28).
Human herpes virus 8
HHV8 triggered AIDS-associated Kaposi's sarcoma
(AIDS-KS) is the most severe and resistant form of KS tumor. One
recent study aimed to investigate the association between HHV8
variability and development of AIDS-KS used fragments of ORF K1 for
genotyping HHV8 virus (29). ORF K1
contains two highly variable regions, VR1 and VR2, that allow the
identification of major genotypes of HHV8 and encode for a
transmembrane molecule crucial for the HHV8 lifecycle and
synergistically works with HIV-1 Tat to promote tumors. This study
compared the variability in the ORF K1 of HHV8 in Brazilian
individuals with and without KS. Among genotypes A, B, C and F, it
was discovered that only genotype B showed predominance compared to
other genotypes (P = 0.0182, Fisher's exact test) suggesting this
genotype may be associated with better prognosis of KS tumor
(29).
Human papillomavirus
HPV is linked to several cancers worldwide of which
its association with cervical cancer is particularly noteworthy.
Infection with HPV is linked to cervical cancers worldwide, and
among all the described genotypes, 70% of all cervical cancers are
linked to HPV-16 and 18 infection, although several other HPV types
are also categorized under high risk (30). In a case control study of 264
invasive cervical cancer patients and 8,428 controls in Wales, it
was discovered that there was a substantial high risk of cervical
cancer in patients infected with HPV-16 [OR (adjusted for
age)=2770; 95% CI, 1050-7320] and HPV-18 [OR (adjusted for
age)=950; 95% CI, 330-2740]. In addition to HPV types 16 and 18
several other HPV types (31,33,35,39,45,52,56,58,59,66) were also
linked to an increased risk for cervical cancer [OR (adjusted for
age) between 20.2 to 386] (31). In
addition, HPV is also associated with various noncervical cancers,
and strikingly HPV-16 and 18 are again shown to be linked with 90%
of these HPV related non-cervical cancers (7). In a case
control study, it was found that HPV-16 infection increased the
odds of oropharyngeal cancer by 14-fold (OR=14.6, 95% CI: 6.3-36.6)
(32). Another study that was
performed to determine whether HPV genotypes were associated with
anal pathology in HIV-infected males with a history of
anal-receptive intercourse; it was discovered that HPV types16
(38%), 18 (19%), 45 (22%), and 52 (19%) were the most common types
found in patients, and there was an increased odds of prevalence of
high-grade intra-anal disease (which is an anal cancer precursor)
when infected with HPV-16 (OR=2.58, 95% CI: 1.31-5.08, P=0.006) and
HPV-31 (OR=4.74, 95% CI: 2.00-11.22, P=0.0004) (33). This evidence demonstrates the
importance of discriminating HPV types in order to determine the
extent of associated risk for development of various cancers.
Hepatitis C and B viruses
Hepatocellular carcinoma (HCC) is the most common
type of liver cancer and also the most frequent cause of deaths in
patients with compensated cirrhosis; its prevalence has been linked
to infection with hepatitis C virus (HCV) (34). In a prospective study of 163
consecutive HCV-infected patients with cirrhosis, two genotypes
were observed to be predominant; 64% of the patients were infected
with genotype 1b and 32% of the patients were infected with
genotype 2a/c. After a median follow-up of 10.7 years
(range=0.6-17.22 years), it was observed that the cumulative
incidence of HCC was significantly higher (P=0.0001) and mortality
after HCV infection was significantly greater (P<0.00001) in
patients infected with genotype 1b compared to genotype 2a/c. In
fact patients infected with genotype 1b HCV carried a greater than
three-fold risk factor of developing HCC compared to patients
infected with genotype 2a/c (HR=3.02, 95% CI: 1.4-6.53)
demonstrating that patients infected with genotype 1b have
substantial high risk of developing HCC compared to patients
infected with genotype 2a/c (35).
In HCV, variations in the 5' untranslated region (UTR) region,
instead of coding regions, was used to determine the viral
genotypes (35).
Hepatitis B virus (HBV) infection with circulating
HBV surface antigenemia is associated with a 20-fold increased risk
of development of HCC relative to that in the absence of
antigenemia. In a study conducted in Taiwanese men over 30 years of
age, it was observed that HBV genotype C was associated with an
increased risk of HCC compared with other HBV genotypes (adjusted
OR=5.11, 95% CI: 3.2-8.81) suggesting that patients infected with
HBV genotype C were five times more likely to develop HCC compared
to infection with other genotypes. Furthermore, both viral load and
the presence of genotype C showed an additive effect and patients
infected with this genotype with high viral load had up to
26.6-fold risk of developing HCC compared to patients infected with
other genotypes and low viral load, showing that HBV mediated HCC
is dependent on the viral genotype and also the viral load
(36). Genotyping and quantification
of HBV in this study was done utilizing the variations in the
coding regions HBs and HBx in a single reaction using real-time PCR
and melting curve analysis (37).
Human cytomegalovirus
In a study investigating genetic polymorphisms in
human cytomegalovirus (HCMV) and its correlation with clinical
outcomes in patients with colorectal cancer (CRC) the HCMV Genotype
B was associated with shorter disease-free survival (DFS) and
presence of this genotype was independently able to predict the
recurrence of tumor in patients (HR=5.79, 95% CI: 1.30-25.81,
P=0.021) (5). Furthermore, the gene
UL144 was most frequently expressed in samples with the
genotype B infection compared to infection with other genotypes.
Because UL144 has been shown to have role in the host immune
response, the authors speculate that UL144 may play an
immunomodulatory role in the tumor microenvironment of CRC
(5). In addition to a specific
genotype of HCMV contributing towards a higher risk of CRC, the
presence of mixed HCMV genotypes has been described to have a
higher risk of developing inflammatory breast cancer (IBC). IBC is
known to be a highly metastatic, aggressive and fatal form of
breast cancer. In a study aimed to establish incidence and type of
HCMV genotypes present in carcinoma tissues between HCMV infected
IBC and non IBC patients, Intermediate Early (IE) gene was used to
detect the presence of HCMV DNA and variation in genes UL55
(gB) and UL73 (gN) were used to determine the viral
genotypes (6). Among the various
genotypes and combination of genotypes tested in this study, IBC
patients had a significantly higher prevalence of mixed genotypes:
gB-1+gB-3 (P=0.029, Fisher's exact test), gN-1+gN-3b (P=0.034,
Fisher's exact test), and gN-3+gN-4b/c (P=0.033, Fisher's exact
test) compared to non-IBC patients. Additionally, the presence of
mixed HCMV genotypes in IBC patients significantly correlated with
lymphovascular invasion and formation of dermal lymphatic emboli
which was not observed in patients without IBC (6). Together, these findings revealed that
the prevalence of mixed HCMV genotypes might be an important factor
in driving the progression of IBC.
5. Interaction between human and microbe
genetic factors
In addition to the microbial genotype, the human
genotype also plays an important role in determining cancer risk
factors (4,14). Interestingly there have been several
reports of a synergistic interaction between host and microbial
genetic factors leading to differential risk of developing cancer.
Patients harboring susceptible variants of interleukin-1β (IL-1B)
infected with more virulent strain of H. pylori were shown
to have up to 87-fold higher risk of developing GC compared to
baseline (Table II) (38). Another independent study showed that
the prevalence of individuals with specific variations in IL-1B and
IL-1 receptor antagonist (IL-1RN) when infected with strains
positive for CagA and harboring virulent versions of
VacA showed an increased risk of developing severe
histological alterations that precede gastric cancer including
intestinal metaplasia, atrophic gastritis, severe lymphocytic
infiltration, and severe granulocytic infiltration compared to
individuals only harboring susceptible IL-1B and IL-1RN alleles
(Table II) (39). Together these studies show that there
is an interaction between human and microbial genotype that could
result in an even more heightened cancer risk, and the ability to
detect these variations would provide crucial information to more
precisely predict cancer risks associated with bacterial
infections.
 | Table IIDifferences in cancer risks as a
result of interaction between microbe and host genetic factors. |
Table II
Differences in cancer risks as a
result of interaction between microbe and host genetic factors.
| A, H.
pylori |
|---|
| Author, year | Cancer type | Microbe
genopype | Human genotype | Odds ratio | (Refs.) |
|---|
| Figueiredo et
al, 2002 | Gastric cancer | VacA s1 | -- | OR=17, 95% CI:
7.8-38 | (38) |
| Gastric cancer | VacA m1 | -- | OR=6.7, 95% CI:
3.6-12 | |
| Gastric cancer | CagA | -- | OR=15, 95% CI:
7.4-29 | |
| Gastric cancer | - | IL-1B-511*T | OR=3.3, 95% CI:
1.3-8.2 | |
| Gastric cancer | VacA s1 | IL-1B-511*T | OR=87, 95% CI:
11-679 | |
| Gastric cancer | VacA m1 | IL-1B-511*T | OR=7.4, 95% CI:
3.2-17 | |
| Gastric cancer | CagA | IL-1B-511*T | OR=25, 95% CI:
8.2-77 | |
| Gastric cancer | VacA s1 | IL-1RN*2 | OR=32, 95% CI:
7.8-134 | |
| Gastric cancer | VacA m1 | IL-1RN*2 | OR=8.8, 95% CI:
2.2-35 | |
| Gastric cancer | CagA | IL-1RN*2 | OR=23, 95% CI:
7-7.2 | |
| Rad et al,
2003 | Severe lymphocytic
infiltration | - | IL-1B-511*T
IL-1RN*2 | OR=2.6, 95% CI:
1.3-5.7 | (39) |
| Severe granulocytic
infiltration | - | IL-1B-511*T
IL-1RN*2 | OR=2.7, 95% CI:
1.3-5.7 | |
| Intestinal
metaplasia | - | IL-1B-511*T
IL-1RN*2 | OR=2.3, 95% CI:
1.2-4.6 | |
| Atrophic
gastritis | - | IL-1B-511*T
IL-1RN*2 | OR=1.7, 95% CI:
0.8-3.4 | |
| Severe lymphocytic
infiltration | CagA VacA s1 | IL-1B-511*T
IL-1RN*2 | OR=24.8, 95% CI:
5.2-117.3 | |
| Severe granulocytic
infiltration | CagA VacA s1 | IL-1B-511*T
IL-1RN*2 | OR=9.5, 95% CI:
2.8-32.1 | |
| Intestinal
metaplasia | CagA VacA s1 | IL-1B-511*T 2
IL-1RN* | OR=6.0, 95% CI:
2.4-15.5 | |
| Atrophic
gastritis | CagA VacA s1 | IL-1B-511*T
IL-1RN*2 | OR=2.4, 95% CI:
0.93-6.2 | |
| B, HPV |
| Author, year | Cancer type | Microbe
genopype | Human genotype | Odds ratio | (Refs.) |
| Zehbe et al,
2003 | Cervical
cancer | HPV-16 E6 L83V | HLA-B*44 | OR=3.5, 95% CI:
1.1-11.1 | (40) |
| Cervical
cancer | HPV-16 E6 L83V | HLA-B*51 | OR=4.2, 95% CI:
1.19-14.69 | |
| Cervical
cancer | HPV-16 E6 L83V | HLA-B*57 | OR=4.67, 95% CI:
1.2-18.6 | |
| Wu et al,
2007 | Cervical
cancer | HPV-16 As | DQB1*060101 | OR=4.47, 95% CI:
2.16-9.27 | (42) |
| Cervical
cancer | HPV-16 As |
DRB1*150101-DQB1*0602 | OR=0.31, 95% CI:
0.09-1.08 | |
| Cervical
cancer | HPV-16 As |
DRB1*070101-DQB1*0201 | OR=0.16, 95% CI:
0.02-1.26 | |
| Cervical
cancer | HPV-16 E6
prototype-positive | DQB1*060101 | OR=5.95, 95% CI:
2.16-9.27 | |
| Cervical
cancer | HPV-16 E6
prototype-positive | DQB1*030201 | OR=10.87, 95% CI:
3.16-37.48 | |
| Cervical
cancer | HPV-16 E6
prototype-positive | DPB1*1301 | OR=7.40, 95% CI:
2.18-25.17 | |
| Cervical
cancer | HPV-16 E6
prototype-positive | DRB1-DQB1 | No significant
effect | |
As previously described, HPV is associated with the
majority of cervical cancers with HPV 16 being the predominant
factor in most cervical cancers. Interestingly, several studies
have provided support that interaction between various human
leukocyte antigen (HLA) alleles and HPV 16 subtypes may contribute
towards differential risk factor for cervical cancer. A study in
Swedish women showed that polymorphisms in HLA class I antigens
HLA-B*44, HLA-B*51, or HLA-B*57 had approximately 4-5-fold increase
in risk for cervical cancer in presence of a L83V variant in E6
gene of HPV-16 (Table II) (40,41).
Interestingly it was discovered that HLA-B*15 allele was completely
absent in Swedish women with cervical cancer which was also
observed in studies conducted in a German and Italian cohort.
Although this study consisted of a rather small sample size (n=27),
the authors speculate that the HLA-B*15 allele may contribute to an
effective immune response against HPV-16 infection (40). Another study in Chinese women showed
that polymorphisms in HLA class II alleles had differential risk
for cervical cancer in presence of two HPV 16 variants. There was a
higher risk factor associated with DQB1*060101 allele, and a lower
risk factor associated with DRB1*150101-DQB1*0602 and
DRB1*070101-DQB1*0201 alleles in individuals infected with HPV-16
As variant (Table II) (42). Additionally, women with HLA types
DQB1*060101, DQB1*030201, and DPB1*1301 had a higher risk of
cervical cancer, while HLA type DRB1-DQB1 showed no significant
risk when infected with HPV-16 E6 prototype-positive variant
(Table II) (42). Together these studies demonstrate
that certain combinations of HLA types and HPV 16 types may be
crucial factors in determining oncogenicity, further supporting the
importance of understanding human-microbe genetic interactions for
improved cancer diagnosis and prognosis.
6. Conclusion and prospects
There is overwhelming evidence that microbial
genotype information is crucial for determining cancer risks in
patients with microbial infection. This observation provides, at a
minimum, a partial explanation for the disparity between the high
prevalence of infection with some malignancy-associated pathogens
in human populations and the fact that only some of those infected
develop malignancies. Several clinical assays that are currently in
use detect microbes only at the species level, which makes it often
difficult to accurately predict the associated risks. Incorporation
of the genotype information into clinical assays would provide a
further step in assessing risk factors associated with specific
strains and has the potential to greatly enhance cancer diagnosis
and prognosis. Such information could also greatly enhance
treatment decisions. For example, patients infected with high risk
genotypes at diagnosis could be considered for different and more
intensive therapy than patients infected with intermediate or low
risk genotypes. In addition, individuals infected with high risk
genotypes could be targeted with antimicrobial treatment in order
to prevent the development of malignancy.
With the advent of novel sequencing technologies
that are able to generate massive amount of sequence data in
parallel, and using ever improving state of the art bioinformatic
pipelines, we have the power to detect not only the variations in
the human genome but also robustly detect variations in the
microbial genomes within an individual (43). Leveraging information described in
the literature, combined with the cutting-edge sequencing
technology and its analysis, data from informative genomic sites
can be used to greatly improve prediction of cancer risks
associated with microbial infections potentially also aiding in
cancer treatment. In fact, various research groups have already
started implementing this technology to detect virulent strains of
microbes present in patient samples. An NGS based analysis was used
to detect variation in CagA and VacA genes in H.
pylori and discriminate between variants associated with GC and
MALT lymphoma (44). Further,
metagenomics approaches have been also used to characterize highly
virulent bacterial strains (45).
Moving forward, studies conducted using approaches such as shotgun
metagenomics could further provide with better microbial genomic
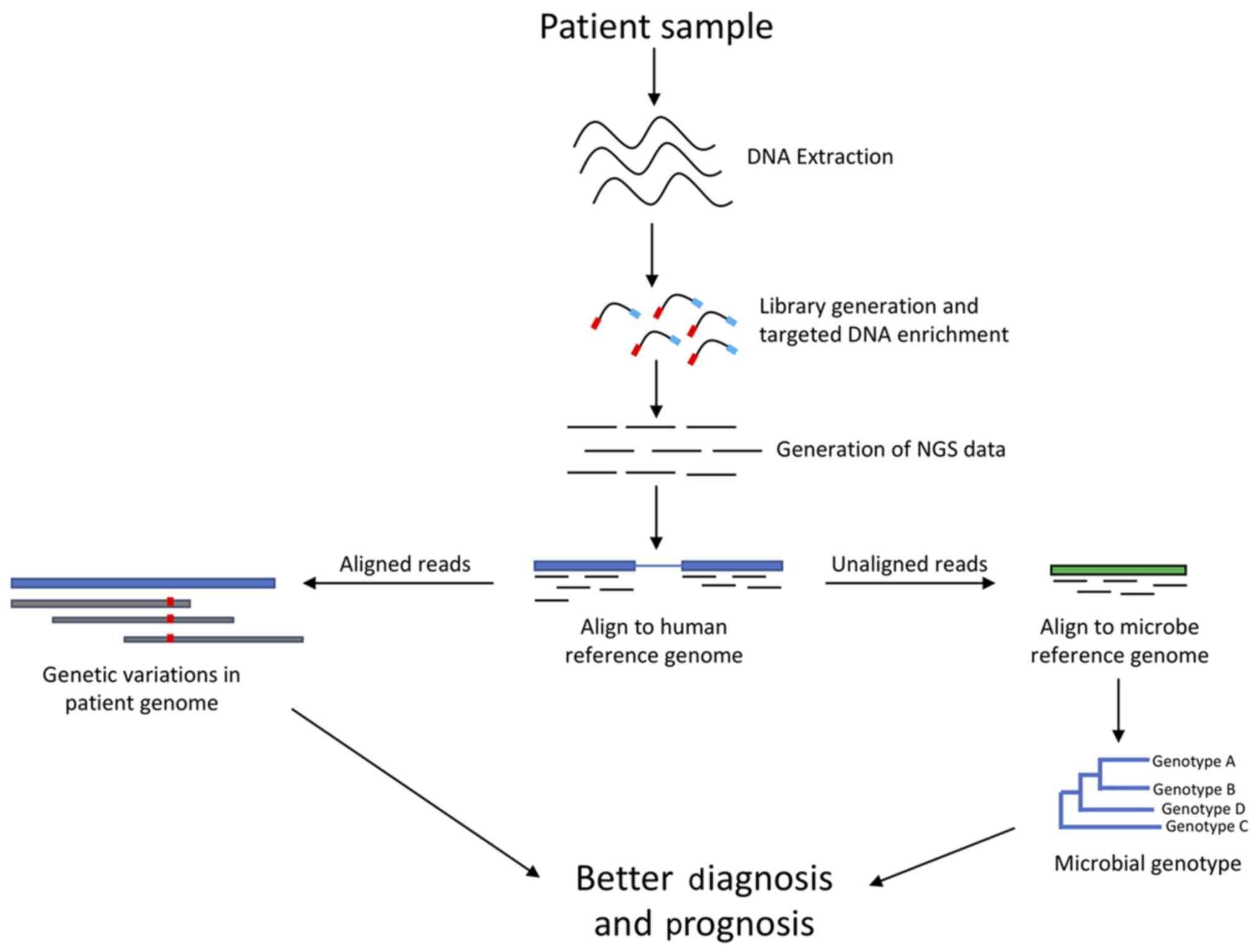
markers associated with high risk of cancer. Our lab has recently
developed a targeted NGS panel called that targets not only
important human genomic sites but also regions in the microbial
genome (Fig. 2). Thus, this panel is
not only able to provide information on genetic variations in
patients, but also can be used for detection of variations present
in targeted microbial sequences. Such panels can be employed into
clinical assays to target informative sites in the microbes of
interest and retrieve genotype information without having to
perform whole genome sequencing, which is rather expensive and time
consuming. Additionally, interactions between microbe and human
genetic factors has also been shown to influence cancer risks in
patients, where specific genotype combinations between humans and
microbes can lead to further heightened or differential risk of
cancer (Table II). Targeted human
and microbial panels could be utilized in clinical assays to
generate invaluable data in a single workflow to detect the
prevalence of these high-risk human-microbe genotype combinations
in patients.
Microbial intervention leading to cancer has been
very well established, however not all infections lead to cancer
development. Microbial genotype is one of the crucial factors that
dictates the risk factors associated with these infections. With
the power of sequencing technologies and our understanding of the
human and microbial genomes, we can now begin to fine tune clinical
assays that could provide a much better understanding of cancer
risk associated with microbial infections. Such assays could also
potentially lead to interventions to prevent the development of
malignancy and direct their therapy when present.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
DJ and RSO designed the current study, wrote the
manuscript and analyzed the data. AB, DJ, SD, KS, and NB edited the
manuscript and analyzed the data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Dzutsev A, Badger JH, Perez-Chanona E, Roy
S, Salcedo R, Smith CK and Trinchieri G: Microbes and cancer. Annu
Rev Immunol. 35:199–228. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Hudnall SD (ed): Viruses and Human Cancer.
Springer-Verlag, New York, NY, 2014.
|
|
3
|
Mager DL: Bacteria and cancer: Cause,
coincidence or cure? A review. J Transl Med. 4(14)2006.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Wroblewski LE, Peek RM Jr and Wilson KT:
Helicobacter pylori and gastric cancer: Factors that
modulate disease risk. Clin Microbiol Rev. 23:713–739.
2010.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Chen HP, Jiang JK, Chan CH, Teo WH, Yang
CY, Chen YC, Chou TY, Lin CH and Chan YJ: Genetic polymorphisms of
the human cytomegalovirus UL144 gene in colorectal cancer and its
association with clinical outcome. J Gen Virol. 96:3613–3623.
2015.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Mohamed HT, El-Shinawi M, Nouh MA, Bashtar
AR, Elsayed ET, Schneider RJ and Mohamed MM: Inflammatory breast
cancer: High incidence of detection of mixed human cytomegalovirus
genotypes associated with disease pathogenesis. Front Oncol.
4(246)2014.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Lowy DR and Schiller JT: Reducing
HPV-associated cancer globally. Cancer Prev Res (Phila). 5:18–23.
2012.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Poiesz BJ, Ruscetti FW, Gazdar AF, Bunn
PA, Minna JD and Gallo RC: Detection and isolation of type C
retrovirus particles from fresh and cultured lymphocytes of a
patient with cutaneous T-cell lymphoma. Proc Natl Acad Sci USA.
77:7415–7419. 1980.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Chadburn A, Said J, Gratzinger D, Chan JK,
de Jong D, Jaffe ES, Natkunam Y and Goodlad JR: HHV8/KSHV-positive
lymphoproliferative disorders and the spectrum of plasmablastic and
plasma cell neoplasms: 2015 SH/EAHP Workshop Report-Part 3. Am J
Clin Pathol. 147:171–187. 2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Nagy A, Bhaduri A, Shahmarvand N,
Shahryari J, Zehnder JL, Warnke RA, Mughal T, Ali S and Ohgami RS:
Next-generation sequencing of idiopathic multicentric and
unicentric Castleman disease and follicular dendritic cell
sarcomas. Blood Adv. 2:481–491. 2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Farrell PJ: Epstein-barr virus and cancer.
Annu Rev Pathol. 14:29–53. 2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Hoffmann JC, Chisholm KM, Cherry A, Chen
J, Arber DA, Natkunam Y, Warnke RA and Ohgami RS: An analysis of
MYC and EBV in diffuse large B-cell lymphomas associated with
angioimmunoblastic T-cell lymphoma and peripheral T-cell lymphoma
not otherwise specified. Hum Pathol. 48:9–17. 2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Uemura N, Okamoto S, Yamamoto S, Matsumura
N, Yamaguchi S, Yamakido M, Taniyama K, Sasaki N and Schlemper RJ:
Helicobacter pylori infection and the development of gastric
cancer. N Engl J Med. 345:784–789. 2001.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Talledo M, López G, Huyghe JR, Verdonck K,
González E, Clark D, Vanham G, Gotuzzo E, Van Camp G and Van Laer
L: Possible implication of NFKB1A and NKG2D genes in susceptibility
to HTLV-1-associated myelopathy/tropical spastic paraparesis in
Peruvian patients infected with HTLV-1. J Med Virol. 84:319–326.
2012.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Edwards RH, Sitki-Green D, Moore DT and
Raab-Traub N: Potential selection of LMP1 variants in
nasopharyngeal carcinoma. J Virol. 78:868–881. 2004.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Michaelis M, Doerr HW and Cinatl J: The
story of human cytomegalovirus and cancer: Increasing evidence and
open questions. Neoplasia. 11:1–9. 2015.
|
|
17
|
Parsa N: Environmental factors inducing
human cancers. Iran J Public Health. 41:1–9. 2012.PubMed/NCBI
|
|
18
|
Muenst S, Läubli H, Soysal SD, Zippelius
A, Tzankov A and Hoeller S: The immune system and cancer evasion
strategies: Therapeutic concepts. J Intern Med. 279:541–562.
2016.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Chen JN, Ding YG, Feng ZY, Li HG, He D, Du
H, Wu B and Shao CK: Association of distinctive Epstein-Barr virus
variants with gastric carcinoma in Guangzhou, southern China. J Med
Virol. 82:658–667. 2010.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Hatakeyama M: Helicobacter pylori
CagA and gastric cancer: A paradigm for hit-and-run carcinogenesis.
Cell Host Microbe. 15:306–316. 2014.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Basso D, Zambon CF, Letley DP, Stranges A,
Marchet A, Rhead JL, Schiavon S, Guariso G, Ceroti M, Nitti D, et
al: Clinical relevance of Helicobacter pylori cagA and vacA
gene polymorphisms. Gastroenterology. 135:91–99. 2008.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Foegeding NJ, Caston RR, McClain MS, Ohi
MD and Cover TL: An overview of Helicobacter pylori VacA
toxin biology. Toxins (Basel). 8(E173)2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Gerhard M, Lehn N, Neumayer N, Borén T,
Rad R, Schepp W, Miehlke S, Classen M and Prinz C: Clinical
relevance of the Helicobacter pylori gene for blood-group
antigen-binding adhesin. Proc Natl Acad Sci USA. 96:12778–12783.
1999.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Yamaoka Y, Kikuchi S, el-Zimaity HMT,
Gutierrez O, Osato MS and Graham DY: Importance of Helicobacter
pylori oipA in clinical presentation, gastric inflammation, and
mucosal interleukin 8 production. Gastroenterology. 123:414–424.
2002.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Hussein NR: The association of dupA and
Helicobacter pylori-related gastroduodenal diseases. Eur J
Clin Microbiol Infect Dis. 29:817–821. 2010.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Franco AT, Friedman DB, Nagy TA,
Romero-Gallo J, Krishna U, Kendall A, Israel DA, Tegtmeyer N,
Washington MK and Peek RM Jr: Delineation of a carcinogenic
Helicobacter pylori proteome. Mol Cell Proteomics.
8:1947–1958. 2009.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Pai PC, Tseng CK, Chuang CC, Wei KC, Hao
SP, Hsueh C, Chang KP and Tsang NM: Polymorphism of C-terminal
activation region 2 of Epstein-Barr virus latent membrane protein 1
in predicting distant failure and post-metastatic survival in
patients with nasopharyngeal carcinoma. Head Neck. 29:109–119.
2007.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Sueur C, Lupo J, Mas P, Morand P and Boyer
V: Difference in cytokine production and cell cycle progression
induced by Epstein-Barr virus Lmp1 deletion variants in Kmh2, a
Hodgkin lymphoma cell line. Virol J. 11(94)2014.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Tozetto-Mendoza TR, Ibrahim KY, Tateno AF,
Oliveira CM, Sumita LM, Sanchez MC, Luna EJ, Pierrotti LC, Drexler
JF, Braz-Silva PH, et al: Genotypic distribution of HHV-8 in AIDS
individuals without and with Kaposi sarcoma: Is genotype B
associated with better prognosis of AIDS-KS? Medicine (Baltimore).
95(e5291)2016.PubMed/NCBI View Article : Google Scholar
|
|
30
|
de Martel C, Plummer M, Vignat J and
Franceschi S: Worldwide burden of cancer attributable to HPV by
site, country and HPV type. Int J Cancer. 141:664–670.
2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Powell NG, Hibbitts SJ, Boyde AM, Newcombe
RG, Tristram AJ and Fiander AN: The risk of cervical cancer
associated with specific types of human papillomavirus: A
case-control study in a UK population. Int J Cancer. 128:1676–1682.
2011.PubMed/NCBI View Article : Google Scholar
|
|
32
|
D'Souza G, Kreimer AR, Viscidi R, Pawlita
M, Fakhry C, Koch WM, Westra WH and Gillison ML: Case-control study
of human papillomavirus and oropharyngeal cancer. N Engl J Med.
356:1944–1956. 2007.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Salit IE, Tinmouth J, Chong S, Raboud J,
Diong C, Su D, Sano M, Lytwyn A, Chapman W and Mahony J: Screening
for HIV-associated anal cancer: Correlation of HPV genotypes, p16,
and E6 transcripts with anal pathology. Cancer Epidemiol Biomarkers
Prev. 18:1986–1992. 2009.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Benvegnù L, Gios M, Boccato S and Alberti
A: Natural history of compensated viral cirrhosis: A prospective
study on the incidence and hierarchy of major complications. Gut.
53:744–749. 2004.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Bruno S, Crosignani A, Maisonneuve P,
Rossi S, Silini E and Mondelli MU: Hepatitis C virus genotype 1b as
a major risk factor associated with hepatocellular carcinoma in
patients with cirrhosis: A seventeen-year prospective cohort study.
Hepatology. 46:1350–1356. 2007.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Yu MW, Yeh SH, Chen PJ, Liaw YF, Lin CL,
Liu CJ, Shih WL, Kao JH, Chen DS and Chen CJ: Hepatitis B virus
genotype and DNA level and hepatocellular carcinoma: A prospective
study in men. J Natl Cancer Inst. 97:265–272. 2005.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Yeh SH, Tsai CY, Kao JH, Liu CJ, Kuo TJ,
Lin MW, Huang WL, Lu SF, Jih J, Chen DS and Chen PJ: Quantification
and genotyping of hepatitis B virus in a single reaction by
real-time PCR and melting curve analysis. J Hepatol. 41:659–666.
2004.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Figueiredo C, Machado JC, Pharoah P,
Seruca R, Sousa S, Carvalho R, Capelinha AF, Quint W, Caldas C, van
Doorn LJ, et al: Helicobacter pylori and interleukin 1
genotyping: An opportunity to identify high-risk individuals for
gastric carcinoma. J Natl Cancer Inst. 94:1680–1687.
2002.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Rad R, Prinz C, Neu B, Neuhofer M, Zeitner
M, Voland P, Becker I, Schepp W and Gerhard M: Synergistic effect
of Helicobacter pylori virulence factors and interleukin-1
polymorphisms for the development of severe histological changes in
the gastric mucosa. J Infect Dis. 188:272–281. 2003.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Zehbe I, Mytilineos J, Wikström I,
Henriksen R, Edler L and Tommasino M: Association between human
papillomavirus 16 E6 variants and human leukocyte antigen class I
polymorphism in cervical cancer of Swedish women. Hum Immunol.
64:538–542. 2003.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Mirabello L, Clarke MA, Nelson CW, Dean M,
Wentzensen N, Yeager M, Cullen M and Boland JF: NCI HPV Workshop.
Schiffman M and Burk RD: The intersection of HPV epidemiology,
genomics and mechanistic studies of HPV-mediated carcinogenesis.
Viruses. 10(E80)2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Wu Y, Liu B, Lin W, Xu Y, Li L, Zhang Y,
Chen S and Xu A: HPV-16 E6 variants and HLA class II polymorphism
among Chinese women with cervical cancer. J Med Virol. 79:439–446.
2007.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Sridhar K, Singh A, Butzmann A, Jangam D
and Ohgami RS: Molecular genetic testing methodologies in
hematopoietic diseases: Current and future methods. Int J Lab
Hematol. 41 (Suppl 1):S102–S116. 2019.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Hashinaga M, Suzuki R, Akada J, Matsumoto
T, Kido Y, Okimoto T, Kodama M, Murakami K and Yamaoka Y:
Differences in amino acid frequency in CagA and VacA sequences of
Helicobacter pylori distinguish gastric cancer from gastric
MALT lymphoma. Gut Pathog. 8(54)2016.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Deurenberg RH, Bathoorn E, Chlebowicz MA,
Couto N, Ferdous M, García-Cobos S, Kooistra-Smid AM, Raangs EC,
Rosema S, Veloo AC, et al: Application of next generation
sequencing in clinical microbiology and infection prevention. J
Biotechnol. 243:16–24. 2017.PubMed/NCBI View Article : Google Scholar
|