Cholangiocarcinoma (CCA) is a bile duct cancer with
a high mortality rate. A significant proportion of the CCA cases
and resultant mortalities worldwide are reported in the
northeastern region of Thailand, where the major risk factor is
infection by the liver fluke Opisthorchis viverrini through
the consumption of improperly cooked cyprinoid fish that contain
the parasite (1). There are
currently no specific biomarkers for early detection of early-stage
CCA, and patients are usually diagnosed when the disease has
already progressed to the advanced stage, resulting in a poor
prognosis. Liver resection is the standard therapy, but is not
suitable for all cases. The 5-year survival rate following liver
resection is <20%, depending on the aggressiveness, metastatic
propensity and invasiveness of the tumors. Furthermore, CCA is
resistant to chemotherapy and radiation (2). Investigation of the molecular
mechanisms underlying the pathogenesis and progression of CCA has
been an ongoing research focus, and several signaling molecules and
pathways have been demonstrated to be involved in the pathogenesis
and progression of CCA (3).
The Notch signaling pathway has been proposed as a
conservative pathway that plays a key role in cell differentiation,
proliferation and apoptosis (4).
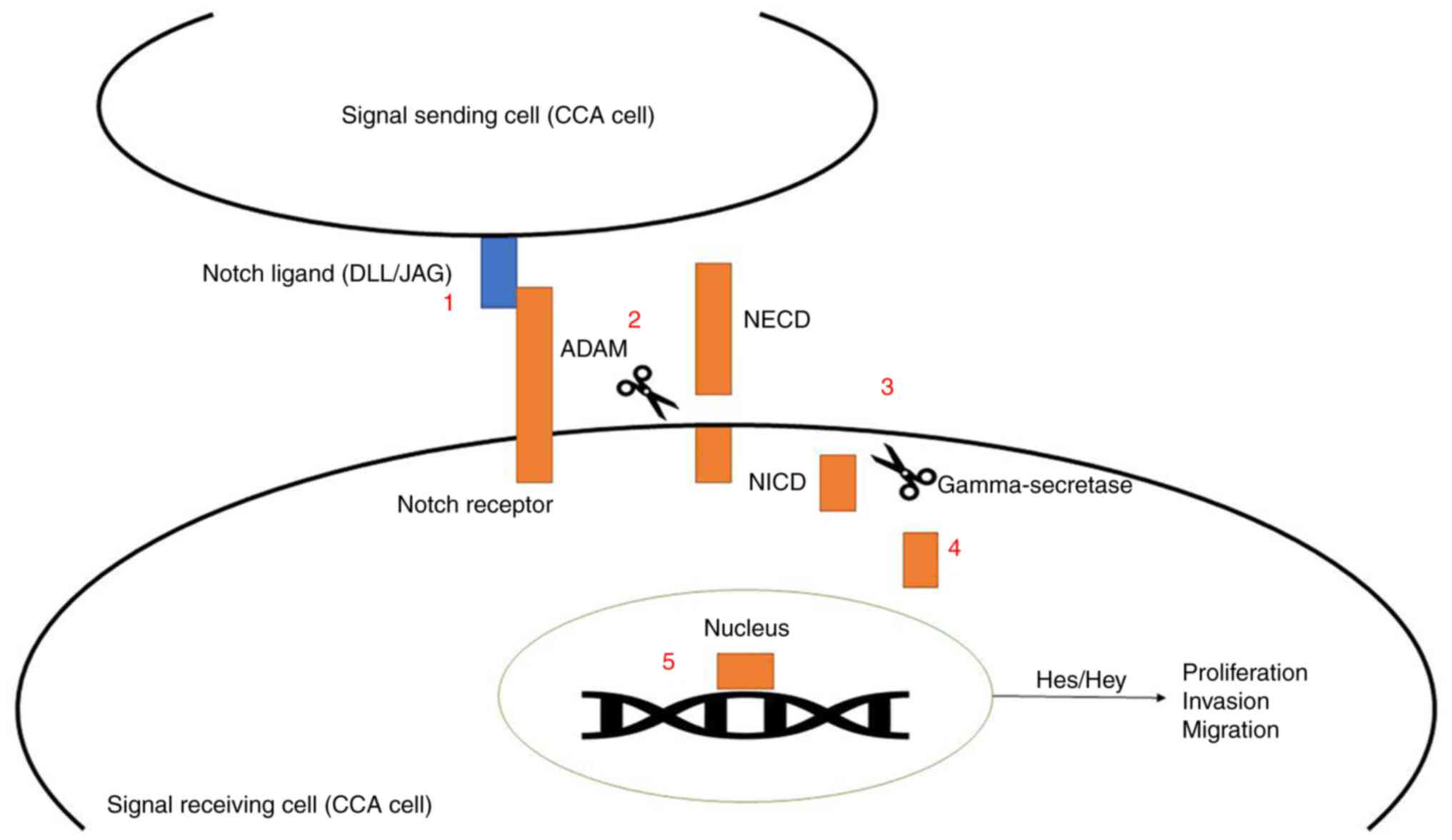
This signaling pathway is associated with several receptors and
ligands. The four identified receptors are Notch1, Notch2, Notch3
and Notch4. The two families of ligands involved are the Delta-like
family (DLL1, DLL3 and DLL4), and the Jagged family (Jagged1 and
Jagged2). The activation of Notch signaling relies on two
proteolytic enzymes in the a disintegrin and metalloprotease (ADAM)
families and γ-secretase enzyme, which cleaves Notch receptors into
two domains, i.e., Notch extracellular domain (NECD) and Notch
intracellular domain (NICD). Following cleavage, NICD translocates
to the nucleus and binds to transcription factors to promote
expression of target genes, such as members of the hairy and
enhancer of split (Hes) and Hes-related to YRPW motif (Hey)
families (5).
Overexpression of Notch signaling genes is
associated with the proliferation of certain cancers, including
ovarian and breast cancers and glioma (6-8).
However, overexpression of Notch signaling genes can also result in
cancer cell apoptosis, such as in cases of liver cancer, small cell
lung cancer and melanoma (9-11).
The Notch signaling pathway in CCA cells is summarized in Fig. 1. The objectives of the present
systematic review were to analyze the association of Notch
signaling with the pathogenesis and progression of CCA, and uncover
potential molecular targets for CCA control.
The present systematic review was performed by
combining the search results from three databases, i.e., PubMed,
ScienceDirect and Scopus. The search terms applied were
‘cholangiocarcinoma’ AND ‘Notch signaling’. All articles were
retrieved and downloaded to the EndNote X9 database (Thomson
Reuters Company, Canada) for further analysis. They were initially
screened by titles and abstracts to exclude irrelevant articles
(those not involving CCA or the Notch signaling pathway). Full-text
articles included after the initial screening were further
evaluated by applying the predefined eligibility criteria. The
inclusion criteria were as follows: i) Articles published between
January 2004 and March 2020; ii) articles available as full-text
articles in English; and iii) articles with in vitro/in vivo/ex
vivo studies related to the Notch signaling pathway in CCA
alone or CCA and hepatocellular carcinoma (HCC). The exclusion
criteria were as follows: i) Articles related to other diseases or
types of cancer; ii) articles related to pathways other than Notch
signaling; iii) duplicated articles; or iv) review articles,
letters to the editor, editorials, systematic analyses or
meta-analyses.
Two reviewers extracted data independently and
disparities were resolved by discussion and suggestions from the
third reviewer. The information extracted for analysis included:
First author's name and year of publication, objective(s) of the
study, type of Notch receptor investigated, type of study (in
vitro, in vivo and ex vivo), type of cell lines or
animals used, laboratory techniques used, and key results and
conclusions.
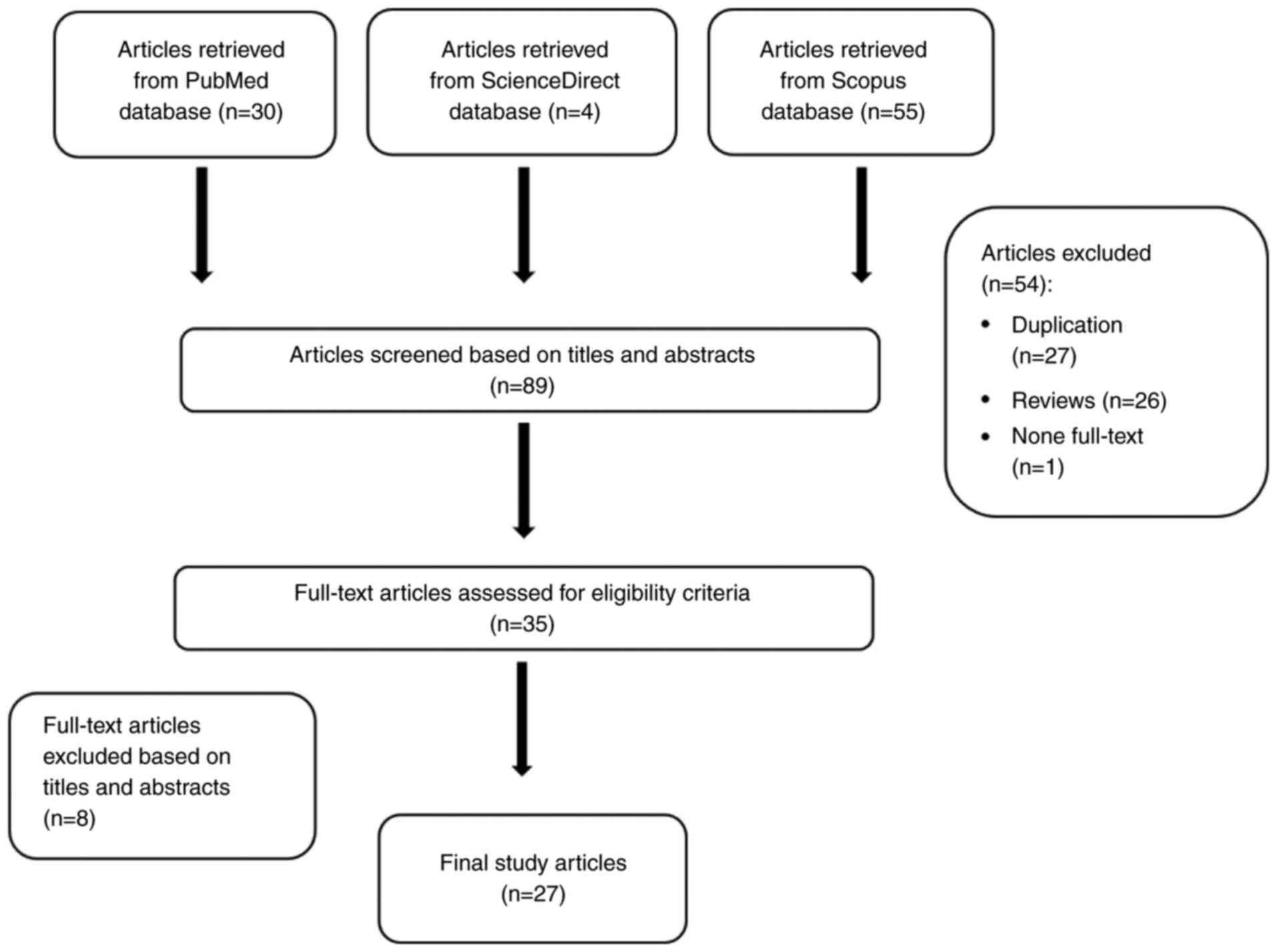
A total of 89 articles from PubMed, ScienceDirect
and Scopus databases were downloaded to the EndNote database. A
total of 54 articles were excluded, and further analysis of the
titles and abstracts of the remaining 36 articles led to the
exclusion of 8 articles (5 articles unrelated to CCA, and 3
articles unrelated to the Notch signaling pathway in CCA). Finally,
27 articles were included in the analysis. The flow diagram of the
study selection process is presented in Fig. 2, and the study summary is provided
in Tables I and II. The associations of Notch1 and Notch2
signaling with CCA development and progression were investigated in
6 articles each, while those of Notch3 and Notch4 signaling were
investigated in 3 and 1 article(s), respectively. The
investigations involved in vitro (n=8), in vivo
(n=12) and ex vivo (n=8) studies. The effects of modulators
of Notch signaling as potential chemotherapeutic targets for CCA
were investigated in 10, 5, 3 and 3 articles for Notch1, Notch2,
Notch3 and Notch4 signaling pathways, respectively. The
investigated modulators included cinobufagin,
verteporfin-photodynamic therapy (PDT), γ-secretase inhibitor
(GSI), γ-secretase inhibitor IX, endocannabinoids, corilagin, short
hairpin (sh)RNA, and small-molecule inhibitors (SMIs) of
aspartate-β-hydroxylase, anti-Notch1,2,3 and Jagged1, microRNA
(miRNA)-34a, PIK3-catalytic subunit alpha (PIK3CA), PIK75,
verteporfin, FLI-06, microfibrillar-associated protein 5 (MFAP5),
ALW-II-41-27, ephrin A1, lymphotoxin β receptor (LTβR), xanthohumol
and γ-secretase inhibitor
N-[N-(3,5-difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl
ester (DAPT) (Table II).
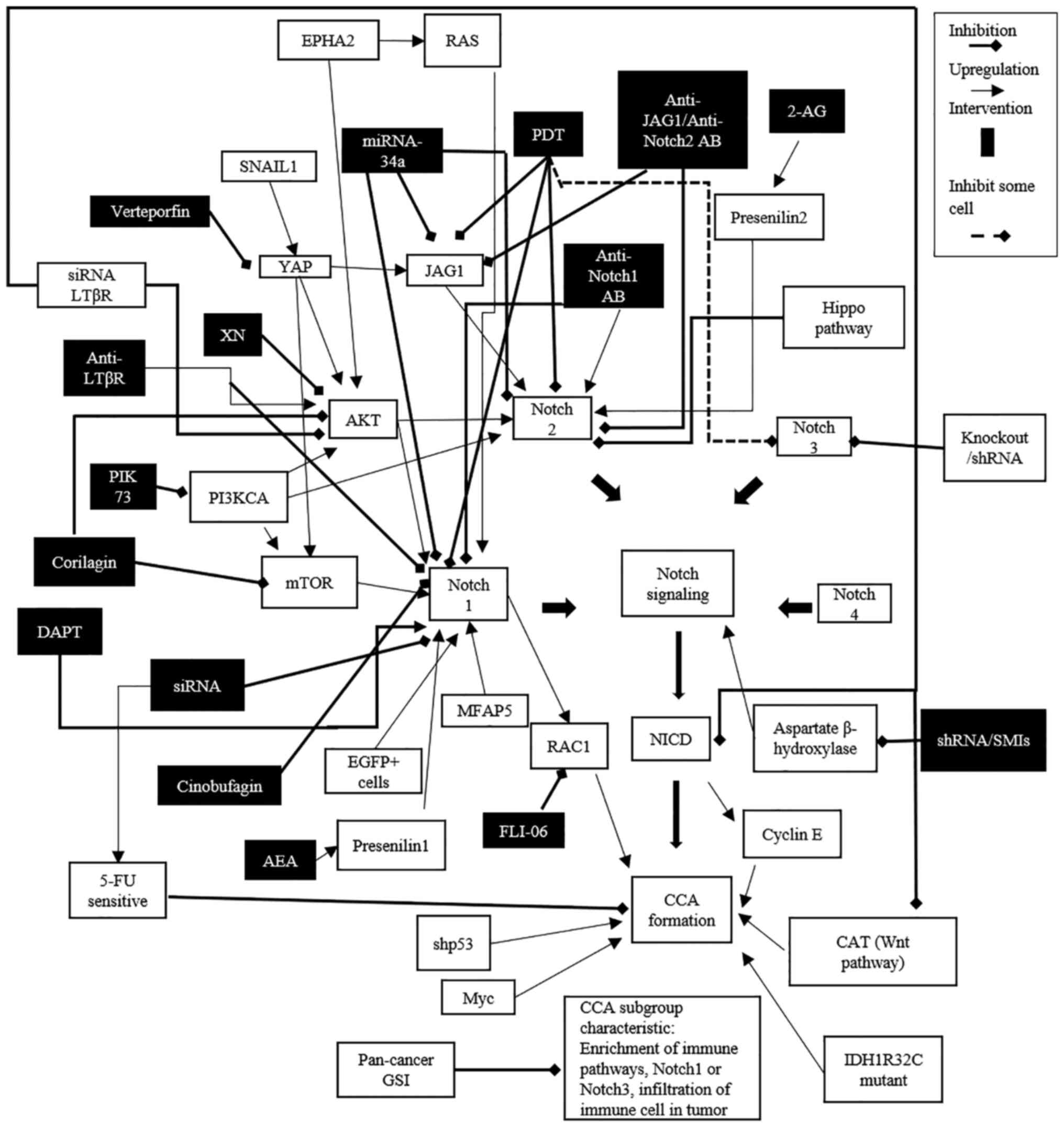
A summary of the currently available information on
the association between Notch signaling and the pathogenesis and
progression of CCA, including modulators (inhibitors or
stimulators) of the signaling pathway as potential candidates for
CCA chemotherapeutics, is presented in Fig. 3.
Upregulation of Notch signaling has been proposed as
the mechanism associated with the transformation of mature
hepatocytes into CCA cells (12-14).
Nevertheless, activation of Notch signaling in hepatic progenitor
cells, but not the transformation of hepatocytes, is proposed as
the mechanism underlying CCA development (15). Increased expression of Notch1 has
been linked to CCA development and progression (14,16-24).
Notch1 has also been associated with cyclin E, the coordinate
regulator protein in the G1 phase of the cell cycle; additionally,
cyclin E can induce DNA damage (15). Increased NICD1 expression has also
been associated with CCA development and progression through
upregulation of cyclin E-associated DNA damage (15,25).
Furthermore, Notch1 and Notch2 signaling have been reported to play
a critical role in CCA formation (12-14,24,26-29),
in which Jagged1 is the specific ligand. Upregulation of PIK3CA,
AKT, and Jagged1 directly activates Notch 2 signaling and induces
CCA development. Overexpression of Jagged1 enhances Notch2
signaling (26), while
anti-Jagged1 treatment suppresses Notch2 signaling (27,28,30,31).
At this time, the information on Notch3 and its role in CCA
development and progression arelimited (24,32).
Notch4 signaling has also been reported to promote the development
of intrahepatic CCA (ICC) and is associated with a poor survival
rate (24).
The activation of upstream Notch signaling
molecules, including Yes-associated protein (YAP), AKT, mTOR, SNAIL
and PIK3CA, are key processes that stimulate CCA formation through
the transformation of mature hepatocytes into CCA cells (12). AKT is the main upstream Notch
signaling molecule, which upregulates Notch1 and Notch2 (12-14,26,27,29,33).
mTOR is another upstream activator of the Notch1 receptor (12,16).
YAP is an upstream signaling molecule for both AKT and mTOR
(12), and co-expression of YAP
with AKT induces CCA development through activating Notch signaling
via the Notch2 receptor and Jagged1 ligand (12,14,28,34),
or with co-expression of NICD and shp53 (shRNA of p53) (35). Co-expression of AKT and Ras (the
protein product of the oncogene KRAS2), on the other hand, induces
tumorigenesis (21,27,29).
MFAP5, enhanced green fluorescent protein-positive cells, mTOR and
AKT, activate Notch signaling by increasing Notch1 expression,
thereby enhancing CCA cell proliferation (12,16,17).
Modulation of mutant genes, such as p53 (inactivation), isocitrate
dehydrogenase1 (activation), or other pathways, such as the Wnt
(β-catenin) pathway (activation), and the Myc pathway (activation)
with co-expression of Notch signaling (activation) has been
reported to induce CCA development (14,25,33,36).
Upregulation of Notch1 expression may also be caused by additional
factors, such as a high expression level of presenilin 1(37). Aspartate β-hydroxylase (ASPH)
enhances activation of Notch signaling and stimulates CCA cell
proliferation and migration. A high expression level of Notch1 can
activate Ras-related C3 botulinum toxin substrate 1, which promotes
CCA cell invasion and migration. ephrin type-A receptor 2 enhances
the expression level of Notch1 and promotes CCA growth through the
activation of AKT/RAS and by promoting lymphatic metastasis in ICC
(21).
The role of Notch signaling in cancer pathogenesis
and progression has also been demonstrated in several other types
of cancer, including embryonal carcinoma (38), glioblastoma (39), melanoma (40), ependymoma (41), breast cancer (42), HCC (34), ovarian cancer (43), endometrial carcinoma (44), esophageal squamous cell carcinoma,
gonadotroph pituitary adenomas (42), rhabdomyosarcoma (45), colon cancer (46), gastric cancer (47), gastrointestinal stromal tumors
(48), anaplastic thyroid cancer
(49), medullary thyroid cancer
(50), pancreatic cancer (51), glioblastoma multiforme (52) and neuroendocrine neoplasms
(53). The signaling molecules and
pathways involved vary according to the type of cancer. Notch3, in
addition to Notch1, appears to play an important role in breast
cancer development and progression through its activation of
cartilage oligomeric matrix protein expression (54).
Downregulation of Notch1 signaling by several
interventions has been demonstrated to be a promising strategy for
inhibition of CCA growth. These interventions include
administration of cinobufagin (a traditional Chinese medicine
extracted from parotid and skin glands of Chinese Toad) (21), xanthohumol (55), verteporfin-PDT (30), DAPT (23), PIK75 (PIK3CA-specific inhibitor),
verteporfin, anti-Notch1 antibody (27), miRNA-34a (31) and small interfering (si)RNA LTβR
(22). The downregulation of
Notch1 siRNA expression reduces Notch1 levels, which results in
inhibition of Notch signaling, suppression of cell proliferation,
and promotion of apoptosis (20,22,27,30,31,55).
Verteporfin-PDT downregulates the mRNA expression of Notch1, Notch2
and Jagged1(30); additionally,
verteporfin can reduce YAP levels, decrease cell proliferation and
induce apoptosis (14). Inhibition
of MFAP5 using the γ-secretase inhibitor FLI-06 also suppressed
Notch1 expression in CCA (20).
Inhibition of Notch2 signaling using anti-Notch2 or anti-Jagged1
antibodies also suppressed Notch2 signaling (27) and miRNA-34a expression (31). Direct inhibition of Notch2 using
miRNA-34a, PDT, anti-Notch2 or anti-Jagged1, and the Hippo pathway
cascade decreases Notch2 levels, promotes apoptosis and inhibits
cell proliferation. However, Notch1 and Notch2 signaling have been
demonstrated to interact antagonistically with each other (27,36).
Antagonists of Notch1 signaling can enhance Notch2 signaling, while
Notch2 depletion can increase the levels of various components of
the Notch1 signaling pathway, such as the endocannabinoids
anandamide (AEA) and 2-arachidonylglycerol (2-AG), or anti-Notch1
and anti-Notch2 antibodies (27,34).
AEA and 2-AG have been shown to exert different effects on Notch
signaling. AEA, which has antiproliferative activity, upregulates
Notch1 signaling via increasing the level of presenilin1, a
catalytic subunit of γ-secretase. On the other hand, 2-AG, which
has growth-promoting activity, upregulates Notch2 signaling via
increasing the expression of presenilin 2, another catalytic
subunit of γ-secretase. 2-AG activates Notch2 and enhances CCA cell
proliferation (36). There have
only been a limited number of studies related to Notch3 and its
role in CCA, although is has been shown that using gene knockout or
shRNA and SMIs to decrease Notch3 levels suppresses CCA growth
(32). Both shRNA and SMIs inhibit
ASPH and Notch signaling to suppress CCA cell proliferation and
migration (56).
The potential of the aforementioned interventions
for the control of other types of cancer has also been
demonstrated. These include cinobufagin for osteosarcoma (57), xanthohumol for hepatocarcinoma
(58), FLI-06 for tongue cancer
(59), GSI for glioblastoma cancer
stem cells (60), T-cell for acute
lymphoblastic leukemia (61),
osteosarcoma (62) and
triple-negative breast cancer (63), and DAPT for glioma (64), colorectal cancer (65), cervical cancer (66), gastric cancer (67), head/neck squamous cell carcinoma
(68), osteosarcoma (69), choriocarcinoma (70) and ovarian cancer (71). In addition, miRNA-34a has also been
reported to inhibit the progression of pancreatic cancer and
medulloblastoma (72,73), and siRNA interference has been
reported to inhibit cell proliferation in glioblastoma multiforme
(74).
In summary, overexpression/upregulation of the
expression of Notch ligands (e.g., Jagged1) and Notch receptors
(Notch1, Notch2, Notch3 and Notch4), as well as upregulation of the
expression of upstream Notch signaling molecules, promotes CCA
development and progression. Therefore, downregulation of Notch1
signaling through several interventions is a promising strategy for
inhibition of CCA development and progression. However, further
studies focusing on the application of these modulators of Notch
signaling in a clinical setting must be performed in the
future.
The authors thank Mr. Ethan Vindvamara (American
molecular biologist; Graduate Program in Bioclinical Sciences,
Chulabhorn International College of Medicine, Thammasat University,
Pathumthani, Thailand) for English editing of the manuscript.
Funding: The study was supported by the Research Team Promotion
Grant, National Research Council of Thailand (grant no. NRCT
820/2563), Thammasat University (Center of Excellence in
Pharmacology and Molecular Biology of Malaria and
Cholangiocarcinoma) and Thammasat University Research Fund
(contract no. TUFT 65/2564).
Data sharing is not applicable to this article, as
no datasets were generated or analyzed during the current
study.
PV, WC and KN conceived the study and analyzed and
interpreted the data. PN performed the background research,
collected the data and wrote the manuscript. WC and KN critically
revised the manuscript for important intellectual content. Data
authentication is not applicable. All the authors have read and
approved the final manuscript.
Not applicable.
Not applicable.
All the authors declare that they have no competing
interests.
|
1
|
Khan SA, Tavolari S and Brandi G:
Cholangiocarcinoma: Epidemiology and risk factors. Liver Int. 39
(Suppl 1):S19–S31. 2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Mao Y, Huang X, Shuang Z, Lin G, Wang J,
Duan F, Chen J and Li S: PARP inhibitor olaparib sensitizes
cholangiocarcinoma cells to radiation. Cancer Med. 7:1285–1296.
2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Papoutsoglou P, Louis C and Coulouarn C:
Transforming growth factor-beta (TGFbeta) signaling pathway in
cholangiocarcinoma. Cells. 8(960)2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Pancewicz-Wojtkiewicz J: Epidermal growth
factor receptor and notch signaling in non-small-cell lung cancer.
Cancer Med. 5:3572–3578. 2016.PubMed/NCBI View
Article : Google Scholar
|
|
5
|
Kandasamy K, Mohan SS, Raju R,
Keerthikumar S, Kumar GS, Venugopal AK, Telikicherla D, Navarro JD,
Mathivanan S, Pecquet C, et al: NetPath: A public resource of
curated signal transduction pathways. Genome Biol.
11(R3)2010.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Hopfer O, Zwahlen D, Fey MF and Aebi S:
The Notch pathway in ovarian carcinomas and adenomas. Br J Cancer.
93:709–718. 2005.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Stockhausen MT, Kristoffersen K and
Poulsen HS: The functional role of Notch signaling in human
gliomas. Neuro Oncol. 12:199–211. 2010.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Kontomanolis EN, Kalagasidou S, Pouliliou
S, Anthoulaki X, Georgiou N, Papamanolis V and Fasoulakis ZN: The
notch pathway in breast cancer progression. ScientificWorldJournal.
2018(2415489)2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Huang Q, Li J, Zheng J and Wei A: The
carcinogenic role of the notch signaling pathway in the development
of hepatocellular carcinoma. J Cancer. 10:1570–1579.
2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Sriuranpong V, Borges MW, Ravi RK, Arnold
DR, Nelkin BD, Baylin SB and Ball DW: Notch signaling induces cell
cycle arrest in small cell lung cancer cells. Cancer Res.
61:3200–3205. 2001.PubMed/NCBI
|
|
11
|
Shao H, Cai L, Grichnik JM, Livingstone
AS, Velazquez OC and Liu ZJ: Activation of Notch1 signaling in
stromal fibroblasts inhibits melanoma growth by upregulating
WISP-1. Oncogene. 30:4316–4326. 2011.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Yamamoto M, Xin B, Watanabe K, Ooshio T,
Fujii K, Chen X, Okada Y, Abe H, Taguchi Y, Miyokawa N, et al:
Oncogenic determination of a broad spectrum of phenotypes of
hepatocyte-derived mouse liver tumors. Am J Pathol. 187:2711–2725.
2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Wang J, Dong M, Xu Z, Song X, Zhang S,
Qiao Y, Che L, Gordan J, Hu K, Liu Y, et al: Notch2 controls
hepatocyte-derived cholangiocarcinoma formation in mice. Oncogene.
37:3229–3242. 2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Li X, Tao J, Cigliano A, Sini M, Calderaro
J, Azoulay D, Wang C, Liu Y, Jiang L, Evert K, et al: Co-activation
of PIK3CA and Yap promotes development of hepatocellular and
cholangiocellular tumors in mouse and human liver. Oncotarget.
6:10102–10115. 2015.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Zender S, Nickeleit I, Wuestefeld T,
Sörensen I, Dauch D, Bozko P, El-Khatib M, Geffers R, Bektas H,
Manns MP, et al: A critical role for notch signaling in the
formation of cholangiocellular carcinomas. Cancer Cell. 23:784–795.
2013.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Ishii T, Yasuchika K, Suemori H, Nakatsuji
N, Ikai I and Uemoto S: Alpha-fetoprotein producing cells act as
cancer progenitor cells in human cholangiocarcinoma. Cancer Lett.
294:25–34. 2010.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Zhou Q, Wang Y, Peng B, Liang L and Li J:
The roles of Notch1 expression in the migration of intrahepatic
cholangiocarcinoma. BMC Cancer. 13(244)2013.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wu WR, Zhang R, Shi XD, Zhu MS, Xu LB,
Zeng H and Liu C: Notch1 is overexpressed in human intrahepatic
cholangiocarcinoma and is associated with its proliferation,
invasiveness and sensitivity to 5-fluorouracil in vitro. Oncol Rep.
31:2515–2524. 2014.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Gu Y, Xiao L, Ming Y, Zheng Z and Li W:
Corilagin suppresses cholangiocarcinoma progression through Notch
signaling pathway in vitro and in vivo. Int J Oncol. 48:1868–1876.
2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Li JH, Zhu XX, Li FX, Huang CS, Huang XT,
Wang JQ, Gao ZX, Li SJ, Xu QC, Zhao W and Yin XY: MFAP5 facilitates
the aggressiveness of intrahepatic Cholangiocarcinoma by activating
the Notch1 signaling pathway. J Exp Clin Cancer Res.
38(476)2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Ren J, Wang S, Jin L, Ma F, Zhou D and Cai
Q: Cinobufagin inhibits tumor growth by inducing apoptosis through
Notch signaling pathways in human cholangiocarcinoma. Transl Cancer
Res. 8:2461–2469. 2019.
|
|
22
|
Sheng Y, Wei J, Zhang Y, Gao X, Wang Z,
Yang J, Yan S, Zhu Y, Zhang Z, Xu D, et al: Mutated EPHA2 is a
target for combating lymphatic metastasis in intrahepatic
cholangiocarcinoma. Int J Cancer. 144:2440–2452. 2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
O'Rourke CJ, Matter MS, Nepal C,
Caetano-Oliveira R, Ton PT, Factor VM and Andersen JB:
Identification of a pan-gamma-secretase inhibitor response
signature for notch-driven cholangiocarcinoma. Hepatology.
71:196–213. 2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Wu WR, Shi XD, Zhang R, Zhu MS, Xu LB, Yu
XH, Zeng H, Wang J and Liu C: Clinicopathological significance of
aberrant Notch receptors in intrahepatic cholangiocarcinoma. Int J
Clin Exp Pathol. 7:3272–3279. 2014.PubMed/NCBI
|
|
25
|
Ding N, Che L, Li XL, Liu Y, Jiang LJ, Fan
B, Tao JY, Chen X and Ji JF: Oncogenic potential of IDH1R132C
mutant in cholangiocarcinoma development in mice. World J
Gastroenterol. 22:2071–2080. 2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Huntzicker EG, Hötzel K, Choy L, Che L,
Ross J, Pau G, Pau G, Sharma N, Siebel CW, Chen X and French DM:
Differential effects of targeting Notch receptors in a mouse model
of liver cancer. Hepatology. 61:942–952. 2015.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Che L, Fan B, Pilo MG, Xu Z, Liu Y,
Cigliano A, Cossu A, Palmieri G, Pascale RM, Porcu A, et al: Jagged
1 is a major Notch ligand along cholangiocarcinoma development in
mice and humans. Oncogenesis. 5(e274)2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Zhang S, Wang J, Wang H, Fan L, Fan B,
Zeng B, Tao J, Li X, Che L, Cigliano A, et al: Hippo cascade
controls lineage commitment of liver tumors in mice and humans. Am
J Pathol. 188:995–1006. 2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Xu M, Wang J, Xu Z, Li R, Wang P, Shang R,
Cigliano A, Ribback S, Solinas A, Pes GM, et al: SNAIL promotes the
cholangiocellular phenotype, but not epithelial-mesenchymal
transition, in a murine hepatocellular carcinoma model. Cancer Res.
79:5563–5574. 2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Cerec V, Andreola F and Pereira S: Effect
of verteporfin-PDT on the Notch signaling pathway in
cholangiocarcinoma (CCA) cell lines: SPIE; 7380, Photodynamic
Therapy: Back to the Future, 73806J 2009.
|
|
31
|
Kwon H, Song K, Han C, Zhang J, Lu L, Chen
W and Wu T: Epigenetic Silencing of miRNA-34a in human
cholangiocarcinoma via EZH2 and DNA methylation: Impact on
regulation of notch pathway. Am J Pathol. 187:2288–2299.
2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Guest RV, Boulter L, Dwyer BJ, Kendall TJ,
Man TY, Minnis-Lyons SE, Lu WY, Robson AJ, Gonzalez SF, Raven A, et
al: Notch3 drives development and progression of
cholangiocarcinoma. Proc Natl Acad Sci USA. 113:12250–12255.
2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Scarzello AJ, Jiang Q, Back T, Dang H,
Hodge D, Hanson C, Subleski J, Weiss JM, Stauffer JK,
Chaisaingmongkol J, et al: LTβR signalling preferentially
accelerates oncogenic AKT-initiated liver tumours. Gut.
65:1765–1775. 2016.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Tschaharganeh DF, Xue W, Calvisi DF, Evert
M, Michurina TV, Dow LE, Banito A, Katz SF, Kastenhuber ER,
Weissmueller S, et al: p53-dependent Nestin regulation links tumor
suppression to cellular plasticity in liver cancer. Cell.
158:579–592. 2014.PubMed/NCBI View Article : Google Scholar
|
|
35
|
El Khatib M, Bozko P, Palagani V, Malek
NP, Wilkens L and Plentz RR: Activation of Notch signaling is
required for cholangiocarcinoma progression and is enhanced by
inactivation of p53 in vivo. PLoS One. 8(e77433)2013.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Frampton G, Coufal M, Li H, Ramirez J and
DeMorrow S: Opposing actions of endocannabinoids on
cholangiocarcinoma growth is via the differential activation of
Notch signaling. Exp Cell Res. 316:1465–1478. 2010.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Fox V, Gokhale PJ, Walsh JR, Matin M,
Jones M and Andrews PW: Cell-cell signaling through NOTCH regulates
human embryonic stem cell proliferation. Stem Cells. 26:715–723.
2008.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Chen J, Kesari S, Rooney C, Strack PR,
Chen J, Shen H, Wu L and Griffin JD: Inhibition of notch signaling
blocks growth of glioblastoma cell lines and tumor neurospheres.
Genes Cancer. 1:822–835. 2010.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Balint K, Xiao M, Pinnix CC, Soma A, Veres
I, Juhasz I, Brown EJ, Capobianco AJ, Herlyn M and Liu ZJ:
Activation of Notch1 signaling is required for
beta-catenin-mediated human primary melanoma progression. J Clin
Invest. 115:3166–3176. 2005.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Puget S, Grill J, Valent A, Bieche I,
Dantas-Barbosa C, Kauffmann A, Dessen P, Lacroix L, Geoerger B, Job
B, et al: Candidate genes on chromosome 9q33-34 involved in the
progression of childhood ependymomas. J Clin Oncol. 27:1884–1892.
2009.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Chen J, Chang H, Peng X, Gu Y, Yi L, Zhang
Q, Zhu J and Mi M: 3,6-dihydroxyflavone suppresses the
epithelial-mesenchymal transition in breast cancer cells by
inhibiting the Notch signaling pathway. Sci Rep.
6(28858)2016.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Wang X, Wu B and Zhong Z: Downregulation
of YAP inhibits proliferation, invasion and increases cisplatin
sensitivity in human hepatocellular carcinoma cells. Oncol Lett.
16:585–593. 2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Wei X, Jia Y, Lou H, Ma J, Huang Q, Meng
Y, Sun C, Yang Z, Li X, Xu S, et al: Targeting YAP suppresses
ovarian cancer progression through regulation of the PI3K/Akt/mTOR
pathway. Oncol Rep. 42:2768–2776. 2019.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Götte M, Greve B, Kelsch R, Müller-Uthoff
H, Weiss K, Kharabi Masouleh B, Sibrowski W, Kiesel L and Buchweitz
O: The adult stem cell marker Musashi-1 modulates endometrial
carcinoma cell cycle progression and apoptosis via Notch-1 and
p21WAF1/CIP1. Int J Cancer. 129:2042–2049. 2011.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Cheng C, Cui H, Zhang L, Jia Z, Song B,
Wang F, Li Y, Liu J, Kong P, Shi R, et al: Genomic analyses reveal
FAM84B and the NOTCH pathway are associated with the progression of
esophageal squamous cell carcinoma. Gigascience.
5(1)2016.PubMed/NCBI View Article : Google Scholar
|
|
46
|
De Salvo M, Raimondi L, Vella S, Adesso L,
Ciarapica R, Verginelli F, Pannuti A, Citti A, Boldrini R, Milano
GM, et al: Hyper-activation of Notch3 amplifies the proliferative
potential of rhabdomyosarcoma cells. PLoS One.
9(e96238)2014.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Singh S, Arcaroli J, Chen Y, Thompson DC,
Messersmith W, Jimeno A and Vasiliou V: ALDH1B1 is crucial for
colon tumorigenesis by modulating Wnt/β-catenin, notch and PI3K/Akt
signaling pathways. PLoS One. 10(e0121648)2015.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Zhang XS, Hu YH, Gao HY, Lan XW and Xue
YW: Downregulation of Notch1 inhibits the invasion and metastasis
of human gastric cancer cells SGC7901 and MKN74 in vitro through
PTEN activation and dephosphorylation of Akt and FAK. Mol Med Rep.
16:2318–2324. 2017.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Dumont AG, Yang Y, Reynoso D, Katz D,
Trent JC and Hughes DP: Anti-tumor effects of the Notch pathway in
gastrointestinal stromal tumors. Carcinogenesis. 33:1674–1683.
2012.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Yu XM, Phan T, Patel PN, Jaskula-Sztul R
and Chen H: Chrysin activates Notch1 signaling and suppresses tumor
growth of anaplastic thyroid carcinoma in vitro and in vivo.
Cancer. 119:774–781. 2013.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Jaskula-Sztul R, Pisarnturakit P,
Landowski M, Chen H and Kunnimalaiyaan M: Expression of the active
Notch1 decreases MTC tumor growth in vivo. J Surg Res. 171:23–27.
2011.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Dong X, Lin Q, Aihara A, Li Y, Huang CK,
Chung W, Tang Q, Chen X, Carlson R, Nadolny C, et al: Aspartate
β-Hydroxylase expression promotes a malignant pancreatic cellular
phenotype. Oncotarget. 6:1231–1248. 2015.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Sturla LM, Tong M, Hebda N, Gao J, Thomas
JM, Olsen M and de la Monte SM: Aspartate-β-hydroxylase (ASPH): A
potential therapeutic target in human malignant gliomas. Heliyon.
2(e00203)2016.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Adler JT, Hottinger DG, Kunnimalaiyaan M
and Chen H: Histone deacetylase inhibitors upregulate Notch-1 and
inhibit growth in pheochromocytoma cells. Surgery. 144:956–961;
discussion 961-2. 2008.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Walden D, Kunnimalaiyaan S, Sokolowski K,
Clark TG and Kunnimalaiyaan M: Antiproliferative and apoptotic
effects of xanthohumol in cholangiocarcinoma. Oncotarget.
8:88069–88078. 2017.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Huang CK, Iwagami Y, Aihara A, Chung W, de
la Monte S, Thomas JM, Olsen M, Carlson R, Yu T, Dong X and Wands
J: Anti-Tumor effects of second generation β-Hydroxylase inhibitors
on cholangiocarcinoma development and progression. PLoS One.
11(e0150336)2016.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Cao Y, Yu L, Dai G, Zhang S, Zhang Z, Gao
T and Guo W: Cinobufagin induces apoptosis of osteosarcoma cells
through inactivation of Notch signaling. Eur J Pharmacol.
794:77–84. 2017.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Sun Z, Zhou C, Liu F, Zhang W, Chen J, Pan
Y, Ma L, Liu Q, Du Y, Yang J and Wang Q: Inhibition of breast
cancer cell survival by Xanthohumol via modulation of the Notch
signaling pathway in vivo and in vitro. Oncol Lett. 15:908–916.
2018.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Gan RH, Lin LS, Xie J, Huang L, Ding LC,
Su BH, Peng XE, Zheng DL and Lu YG: FLI-06 intercepts notch
signaling and suppresses the proliferation and self-renewal of
tongue cancer cells. Onco Targets Ther. 12:7663–7674.
2019.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Dai L, He J, Liu Y, Byun J, Vivekanandan
A, Pennathur S, Fan X and Lubman DM: Dose-dependent proteomic
analysis of glioblastoma cancer stem cells upon treatment with
γ-secretase inhibitor. Proteomics. 11:4529–4540. 2011.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Tammam J, Ware C, Efferson C, O'Neil J,
Rao S, Qu X, Gorenstein J, Angagaw M, Kim H, Kenific C, et al:
Down-regulation of the Notch pathway mediated by a gamma-secretase
inhibitor induces anti-tumour effects in mouse models of T-cell
leukaemia. Br J Pharmacol. 158:1183–1195. 2009.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Tanaka M, Setoguchi T, Hirotsu M, Gao H,
Sasaki H, Matsunoshita Y and Komiya S: Inhibition of Notch pathway
prevents osteosarcoma growth by cell cycle regulation. Br J Cancer.
100:1957–1965. 2009.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Li ZL, Chen C, Yang Y, Wang C, Yang T,
Yang X and Liu SC: Gamma secretase inhibitor enhances sensitivity
to doxorubicin in MDA-MB-231 cells. Int J Clin Exp Pathol.
8:4378–4387. 2015.PubMed/NCBI
|
|
64
|
Liu X, Xu QR, Xie WF and Wang MD: DAPT
suppresses the proliferation of human glioma cell line SHG-44.
Asian Pac J Trop Med. 7:552–556. 2014.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Li H, Li D and Meng N: Effects of RUNX3
mediated Notch signaling pathway on biological characteristics of
colorectal cancer cells. Int J Oncol. 50:2059–2068. 2017.PubMed/NCBI View Article : Google Scholar
|
|
66
|
He G, Mu T, Yuan Y, Yang W, Zhang Y, Chen
Q, Bian M, Pan Y, Xiang Q, Chen Z and Sun A: Effects of notch
signaling pathway in cervical cancer by curcumin mediated
photodynamic therapy and its possible mechanisms in vitro and in
vivo. J Cancer. 10:4114–4122. 2019.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Peng X, Zhou J, Li B, Zhang T, Zuo Y and
Gu X: Notch1 and PI3K/Akt signaling blockers DAPT and LY294002
coordinately inhibit metastasis of gastric cancer through mutual
enhancement. Cancer Chemother Pharmacol. 85:309–320.
2020.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Zhao ZL, Zhang L, Huang CF, Ma SR, Bu LL,
Liu JF, Yu GT, Liu B, Gutkind JS, Kulkarni AB, et al: NOTCH1
inhibition enhances the efficacy of conventional chemotherapeutic
agents by targeting head neck cancer stem cell. Sci Rep.
6(24704)2016.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Qin J, Wang R, Zhao C, Wen J, Dong H, Wang
S, Li Y, Zhao Y, Li J, Yang Y, et al: Notch signaling regulates
osteosarcoma proliferation and migration through Erk
phosphorylation. Tissue Cell. 59:51–61. 2019.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Tian Q, Xue Y, Zheng W, Sun R, Ji W, Wang
X and An R: Overexpression of hypoxia-inducible factor 1α induces
migration and invasion through Notch signaling. Int J Oncol.
47:728–738. 2015.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Jiang LY, Zhang XL, Du P and Zheng JH:
γ-Secretase Inhibitor, DAPT inhibits self-renewal and stemness
maintenance of ovarian cancer stem-like cells in vitro. Chin J
Cancer Res. 23:140–146. 2011.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Ji Q, Hao X, Zhang M, Tang W, Yang M, Li
L, Xiang D, Desano JT, Bommer GT, Fan D, et al: MicroRNA miR-34
inhibits human pancreatic cancer tumor-initiating cells. PLoS One.
4(e6816)2009.PubMed/NCBI View Article : Google Scholar
|
|
73
|
de Antonellis P, Medaglia C, Cusanelli E,
Andolfo I, Liguori L, De Vita G, Carotenuto M, Bello A, Formiggini
F, Galeone A, et al: MiR-34a targeting of Notch ligand delta-like 1
impairs CD15+/CD133+ tumor-propagating cells and supports neural
differentiation in medulloblastoma. PLoS One.
6(e24584)2011.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Wang J, Wang C, Meng Q, Li S, Sun X, Bo Y
and Yao W: siRNA targeting Notch-1 decreases glioma stem cell
proliferation and tumor growth. Mol Biol Rep. 39:2497–2503.
2012.PubMed/NCBI View Article : Google Scholar
|