Introduction
Acute megakaryoblastic leukemia (AMKL;
French-American-British M7) is a rare disease, occurring in 4 to
15% of children with acute myeloid leukemia (AML) worldwide
(1-4).
AMKL appears to be de novo in infants and in young children
without Down's syndrome (DS), and patients frequently present with
bone marrow fibrosis, hepatosplenomegaly and pancytopenia (5,6). The
diagnosis of AMKL is based on blast cell morphology, which is
suggestive of megakaryoblasts and the protein expression of
platelet-associated markers (CD41, CD42b or CD61) using
immunophenotyping (1,7).
The chromosomal translocation, t(1;22)(p13;q13)
occurs in 10-15% of pediatric non-DS-AMKL and is a specific
translocation in infants with AMKL. This translocation results in
the fusion of the RNA-binding motif protein-15 (RBM15) and
megakaryoblastic leukemia-1 (MKL1) genes (5,7-10).
The present case report describes a case of neonatal AMKL, with a
hitherto unreported 3-way translocation t(1;7;22)(p13;q21;q13).
Case report
A 31-year-old woman presented at the University
Hospitals Leuven (Belgium) at 36 weeks in her second pregnancy,
with a decrease in child movement for 5 days and the loss of brown
fluid per vaginam for 3 days. Until then, the pregnancy was
uncomplicated and no fetal abnormalities were observed.
An ultrasound examination of the fetus showed
hepatosplenomegaly and a cardiotocography revealed tachycardia and
small variations on the trace. An urgent cesarean section was
performed, and a baby girl was born at 36 weeks and 2 days of
pregnancy, with Apgar scores of 2, 6 and 7 after 1, 5 and 10 min,
respectively. The patient weighed 2,700 g, measured 48 cm in length
and had a head circumference of 33.5 cm. The abdomen was extremely
distended due to an enlarged liver and spleen, and was hard on
palpation.
Initial blood testing showed normochromic,
normocytic anemia (hemoglobin, 7.3 g/dl; reference range, 14.5-22.5
g/dl), with signs of active erythropoiesis, deep thrombopenia
(39x109/l; reference range, 150-450x109/l)
and leukocytosis of 23.4x109/l (reference range,
9.4-34x109/l) with 20% blasts, and absolute neutropenia
(2.3x109/l; reference range, 5-21x109/l). The
lactate dehydrogenase level was elevated to 1,403 U/l (reference
range, 135-250 U/l). The cells were then analyzed using
histopathology. The cells were fixed with absolute methanol for 10
min, stained with May Grünwald solution for 5 min, stained with
Giemsa solution for 5 min and then in buffer (pH 6.8) for 2 min,
all at room temperature. Images were captured using a Leica DM LED
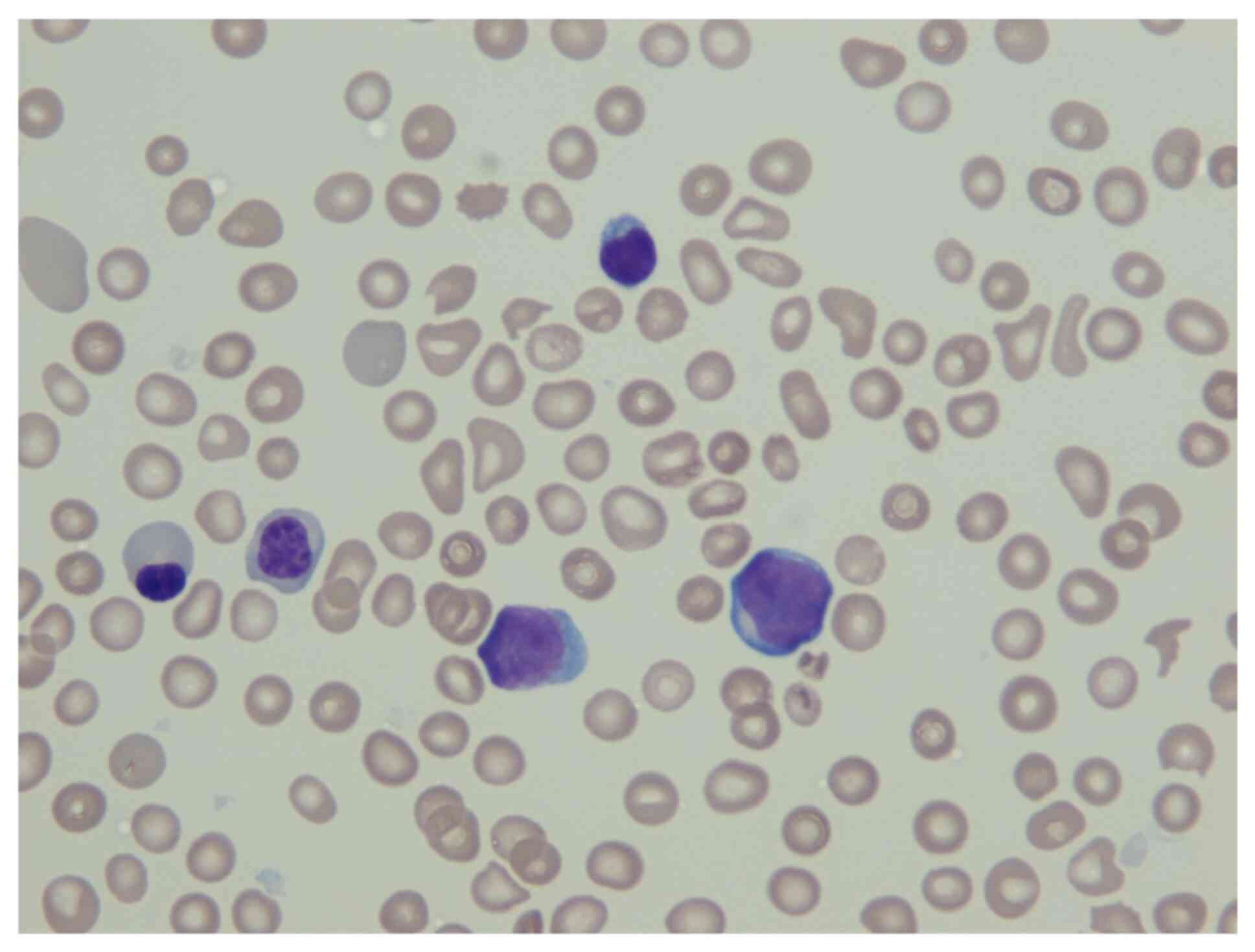
light microscope at x500 magnification. Fig. 1 illustrates the megakaryoblasts,
found in the peripheral blood smear, from May Grünwald staining,
together with two normoblasts with anisopoikilocytosis of the red
blood cells, a lymphocyte and irregular formed agranular platelets.
The megakaryoblasts were medium sized with a high nucleus/cytoplasm
ratio, a basophilic agranular cytoplasm and a round regular nucleus
with fine reticular chromatin.
Immunophenotyping of the peripheral blood was
performed using flow cytometry. For surface staining a 6-color
protocol was used: 100 µl peripheral blood was incubated for 10 min
with the following monoclonal antibodies: CD45-PerCP-Cy5.5 (20 µl;
1/2 diluted in Cell Wash; cat. no. 332784; BD Biosciences),
CD61-FITC (15 µl; undiluted; cat. no. 347407; BD Biosciences),
CD11b-PE (15 µl; undiluted; cat no. 333142; BD Biosciences),
CD13-PE (15 µl; undiluted; cat. no. 347406; BD Biosciences),
CD33-APC, (5 µl; undiluted; cat. no. 345800; BD Biosciences),
CD34-FITC (15 µl; undiluted; cat. no. 345801; BD Biosciences),
CD117-PE-Cy7 (5 µl; undiluted; cat. no. 339217; BD Biosciences),
anti-HLA-DR-APC-H7 (5 µl; undiluted; cat. no. 641411; BD
Biosciences), CD42b-PE (15 µl; undiluted; cat. no. IM1417U; Beckman
Coulter, Inc.) and CD36-FITC (15 µl; undiluted; cat. no. B49201;
Beckman Coulter, Inc.) at room temperature, then subsequently lysed
for 10 min using 2 ml FACS-Lysing solution (BD Biosciences). For
intracellular staining, a Fix and Perm reagent (ImTec Diagnostics
NV) was used and monoclonal antibodies against MPO-FITC (15 µl;
undiluted; cat. no. F0714; Dako; Agilent Technologies, Inc.) and
CD79a-PE (15 µl; undiluted; cat. no. 333152; BD Biosciences). After
staining, the samples were washed with 2 m Cell Wash (BD
Biosciences) and analyzed using a FacsCanto flow cytometer (BD
Biosciences) by collecting 100,000 events. For analysis, the
FacsDIVA software (version 6.1.2; BD Bioscience) was used. Blast
cells were gated based on their side-scatter and dim CD45
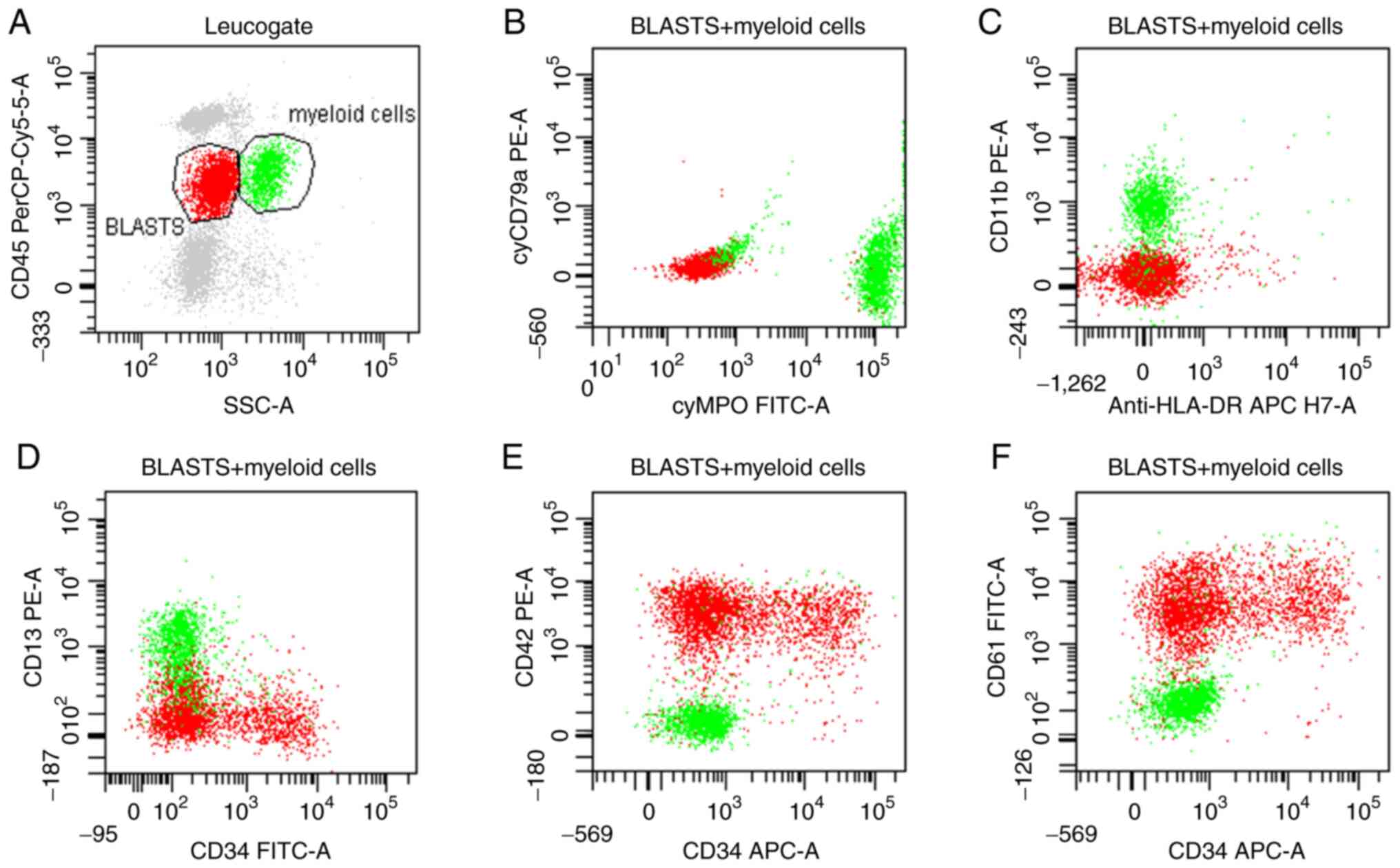
characteristics. Immunophenotyping of the peripheral blood showed a
population of 28% blasts, and were positive for CD34, CD61 and
CD42b and negative for cyMPO, CD13, CD117, CD33, CD36 and human
leukocyte antigen-DR (Fig. 2). As
the megakaryocytic markers, CD61 and CD42b were found to be
positive from the peripheral blood, this suggested the patient had
AMKL. Short-term culture of the peripheral blood, without mitogen,
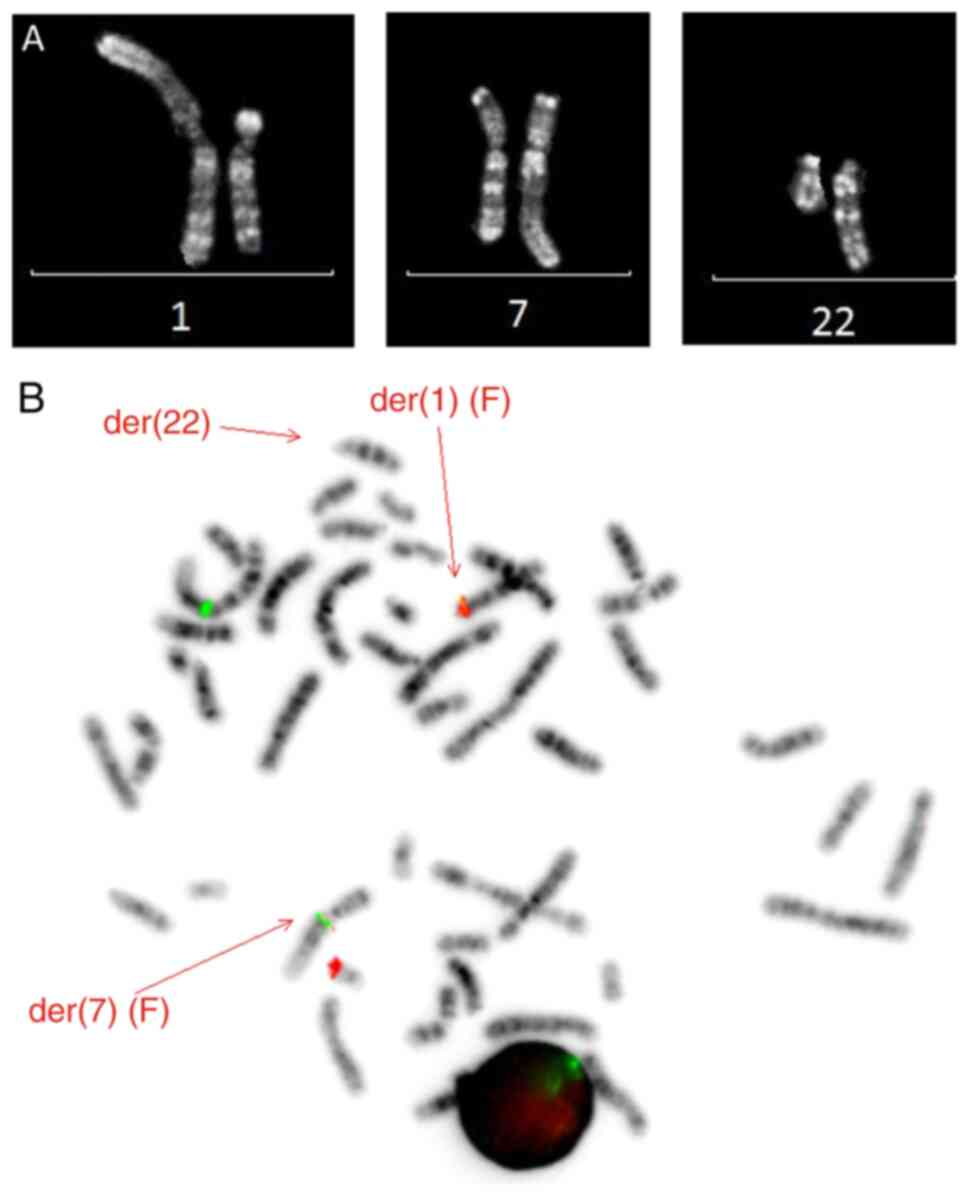
revealed a balanced translocation t(1;7;22)(p13;q21;q13), as the
sole aberration in all 13 analyzed metaphases (Fig. 3A). Fluorescent in situ
hybridization (FISH) was performed according to standard protocols
and manufacturer's procedures. The commercially available probe
RBM15-MKL Dual Fusion/Translocation FISH Probe (CytoTest) was used
on peripheral blood metaphases from the same culture as that
analyzed by conventional karyotyping. Images were captured using a
fluorescence microscope (magnification, x400) equipped with an
Axiophot 2 camera (Carl Zeiss AG) and a MetaSystems Isis imaging
system (MetaSystems). This showed one expected fusion signal on the
derivate 1 chromosome, while the second fusion was not located on
the derivative 22, but on the derivative 7. This demonstrated that
the observed three-way translocation was a variant of the recurrent
t(1;22)(p13;q13) (Fig. 3B).
Reverse transcription-quantitative PCR analysis of
the peripheral blood showed an overexpression of ecotropic viral
integration site 1 (EVI1) (data not shown). RT-PCR analysis was
performed as previously described by Gröschel et al
(11). RNA was extracted from the
leukemic blast cells using a Maxwell® RSC simplyRNA
Blood kit (Promega Corporation), according to the manufacturer's
protocol. cDNA was subsequently synthesized using a Superscript II
reverse transcriptase kit (Invitrogen; Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. EVI1 expression
was quantified against the housekeeping gene, ABL1
(ΔCp=Cp (MECOM)-Cp (ABL1); ratio, 2-ΔCp)
(11). EVI1 qPCR was performed
using the QuantStudio DX Real-Time PCR instrument (Applied
Biosystems; Thermo Fisher Scientific, Inc.) using 12.5 µl TaqMan
Fast Advanced Master Mix (Applied Biosystems; Thermo Fisher
Scientific, Inc.), 2.5 µl MECOM primer probe mix, 2.5 µl ABL1
primer probe mix, 7.5 µl water and 2.5 µl cDNA (maximum, 125 ng
RNA). The following primers (Integrated DNA Technologies, Inc.)
were used: MECOM EVI1 forward, 5'-AGTGCCCTGGAGATGAGTTG-3', and
reverse, 5'-TTTGAGGCTATCTGTGAAGTGC-3'; ABL-F-EAC forward,
5'-TGGAGATACACTCTAAGCATAACTAAAGGT-3', and reverse,
5'-GATGTAGTTGCTTGGGACCCA-3'. The following probes (IDT) were used:
ABL-P_EAC-HEX, HEX-CCATTTTTGGTTTGGGCTTCACACCATT-BHQ1, and EVI1_P2
FAM-CCCCAGTGAGGTATAAAGAGGAAGAATATA-BHQ1. The following
thermocycling conditions were used: 95˚C for 20 sec, 50˚C for 2
sec, at 95˚C for 1 sec and 60˚C for 20 sec (50 cycles). The SKOV3
cell line (3q26 amplified) was used as a calibrator for
quantification and as a positive control, while the HL60 cell line
and H2O were used as negative controls. Positivity was
defined as a sample with sigmoid amplification and a ratio
(normalized to SKOV3 ratio) >0.11.
On the first day, the patient developed an
intracranial hemorrhage, hypotension and renal failure. In
consultation with the parents, it was decided not to start
chemotherapy, as their child was critically ill. She died after 3
days.
Discussion
AMKL is a rare subtype of AML and is predominantly
found in infants (1,12). The chromosomal translocation
t(1;22)(p13;q13), which results in the RMB15/MKL1 fusion gene, is
specific to this subtype (5,8). Until
recently, the RMB15/MKL1 fusion gene was the only recurrent genetic
aberration detected in non-DS-AMKL; however, novel fusion genes
have been identified over the few years, such as CBFA2R3-GLIS2 and
NUP95-KDM5A (13).
The patient in the present case report presented
with hepatosplenomegaly, anemia and thrombopenia, which is
frequently observed in AMKL (5,6). Blood
analysis revealed the diagnosis of AMKL, with the typical findings
of megakaryocytes from a peripheral blood smear and was positive
for CD42b and CD61 from immunophenotyping. However, several cases
have been described where the diagnosis of AMKL was complicated due
to bone marrow fibrosis or extramedullary disease (14-17).
Karyotyping of the blast cells showed a three-way
variant of the known translocation t(1;22)(p13;q13) and FISH
analysis confirmed the RBM15-MKL1 fusion gene. To the best of our
knowledge, this is the first description of this translocation. A
search of the Mitelman database and the literature only revealed a
few variants of the translocation (1;22)(p13;q13), in addition to
the novel 3-way variant t(1;7;22)(p13;q21;q13), as aforementioned.
There were 2 3-way translocations described,
t(1;22;14)(p13;q13;q31) and t(1;22;4)(p13;q13;q35) (3,18), as
well as 3 additional 4-way translocations,
t(1;22;17;18)(p13;q13;q22;q12), t(1;6;6;22)(p13;p25;q13;q13) and
(t1;2;22;2)(p13;q21;q13;p23) (10,12).
Whether this rare variant carries the same prognosis, is currently
unclear and requires further research.
AMKL has a poor outcome, but with intensive
chemotherapy regimens, an improvement in survival time has been
achieved, with a reported 5-year overall survival rate of 70±6% in
the AML-BMF 04 trial vs. 45±8% in the AML-BMF 98 trial (9). The decision to renounce therapy in the
present case, was thoroughly discussed and was based on the
comorbidities the patient had.
Acknowledgements
The authors would like to thank Ms Monique Rubens
and Msc Geneviève Ameye (Department of Human Genetics, University
Hospitals Leuven, Belgium) for their assistance in the preparation
of the karyotyping images and FISH-analysis.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JM acquired the data from the experiments and the
patient, and wrote the original draft of the manuscript. LM, NB and
PV investigated the chromosomal aberrations. AU and SAJ contributed
to the interpretation of the results, and made substantial
contributions to conception and design. AU, LM, PV, NB and SAJ
critically reviewed and edited the draft version of the manuscript.
SAJ supervised the research. LM, NB and PV confirmed the
authenticity of all the raw data. All authors reviewed the results
and approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Written informed consent was obtained from the
parents of the patient for publication of the case report and any
accompanying images.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lion T, Haas OA, Harbott J, Bannier E,
Ritterbach J, Jankovic M, Fink FM, Stojimirovic A, Herrmann J,
Riehm HJ, et al: The translocation t(1;22)(p13;q13) is a nonrandom
marker specifically associated with acute megakaryocytic leukemia
in young children. Blood. 79:3325–3330. 1992.PubMed/NCBI
|
|
2
|
Athale UH, Razzouk BI, Raimondi SC, Long
X, Behm FG, Head DR, Srivastava DK, Rubnitz JE, Bowman L, Pui CH
and Ribeiro RC: Biology and outcome of childhood acute
megakaryoblastic leukemia: A single institution's experience.
Blood. 97:3727–3732. 2001.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Dastugue N, Lafage-Pochitaloff M, Pagès
MP, Radford I, Bastard C, Talmant P, Mozziconacci MJ, Léonard C,
Bilhou-Nabéra C, Cabrol C, et al: Cytogenetic profile of childhood
and adult megakaryoblastic leukemia (M7): A study of the Groupe
Français de Cytogénétique Hématologique (GFCH). Blood. 100:618–626.
2002.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Reinhardt D, Diekamp S, Langebrake C,
Ritter J, Stary J, Dworzak M, Schrauder A, Zimmermann M,
Fleischhack G, Ludwig WD, et al: Acute megakaryoblastic leukemia in
children and adolescents, excluding Down's syndrome: Improved
outcome with intensified induction treatment. Leukemia.
19:1495–1496. 2005.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Carroll A, Civin C, Schneider N, Dahl G,
Pappo A, Bowman P, Emami A, Gross S, Alvarado C, Phillips C, et al:
The t(1;22) (p13;q13) is nonrandom and restricted to infants with
acute megakaryoblastic leukemia: A pediatric oncology group study.
Blood. 78:748–752. 1991.PubMed/NCBI
|
|
6
|
Bernstein J, Dastugue N, Haas OA, Harbott
J, Heere NA, Huret JL, Landman-Parker J, Lebeau MM, Leonard C, Mann
G, et al: Nineteen cases of the t(1;22)(p13;q13) acute
megakaryblastic leukaemia of infants/children and a review of 39
cases: Report from a t(1;22) study group. Leukemia. 14:216–218.
2000.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Inaba H, Zhou Y, Abla O, Adachi S,
Auvrignon A, Beverloo HB, De Bont E, Chang TT, Creutzig U, Dworzak
M, et al: Heterogeneous cytogenetic subgroups and outcomes in
childhood acute megakaryoblastic leukemia: A retrospective
international study. Blood. 126:1575–1584. 2015.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Ma Z, Morris SW, Valentine V, Li M,
Herbrick JA, Cui X, Bouman D, Li Y, Mehta PK, Nizetic D, et al:
Fusion of two novel genes, RBM15 and MKL1, in the t(1;22)(p13;q13)
of acute megakaryoblastic leukemia. Nat Genet. 28:220–221.
2001.PubMed/NCBI View
Article : Google Scholar
|
|
9
|
Schweitzer J, Zimmermann M, Rasche M, Von
Neuhoff C, Creutzig U, Dworzak M, Reinhardt D and Klusmann JH:
Improved outcome of pediatric patients with acute megakaryoblastic
leukemia in the AML-BFM 04 trial. Ann Hematol. 94:1327–1336.
2015.PubMed/NCBI View Article : Google Scholar
|
|
10
|
de Rooij JD, Branstetter C, Ma J, Li Y,
Walsh MP, Cheng J, Obulkasim A, Dang J, Easton J, Verboon LJ, et
al: Pediatric non-down syndrome acute megakaryoblastic leukemia is
characterized by distinct genomic subsets with varying outcomes.
Nat Genet. 49:451–456. 2017.PubMed/NCBI View
Article : Google Scholar
|
|
11
|
Gröschel S, Lugthart S, Schlenk RF, Valk
PJ, Eiwen K, Goudswaard C, van Putten WJ, Kayser S, Verdonck LF,
Lübbert M, et al: High EVI1 expression predicts outcome in younger
adult patients with acute myeloid leukemia and is associated with
distinct cytogenetic abnormalities. J Clin Oncol. 28:2101–2107.
2010.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Torres L, Lisboa S, Vieira J, Cerveira N,
Santos J, Pinheiro M, Correia C, Bizarro S, Almeida M and Teixeira
MR: Acute megakaryoblastic leukemia with a four-way variant
translocation originating the RBM15-MKL1 fusion gene. Pediatr Blood
Cancer. 56:846–849. 2011.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Masetti R, Guidi V, Ronchini L, Bertuccio
NS, Locatelli F and Pession A: The changing scenario of non-down
syndrome acute megakaryoblastic leukemia in children. Crit Rev
Oncol Hematol. 138:132–138. 2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Margolskee E, Saab J, Geyer JT, Aledo A
and Mathew S: A novel variant t(1;22)
translocation-ins(22;1)(q13;p13p31)-in a child with acute
megakaryoblastic leukemia. Am J Case Rep. 18:422–426.
2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Gökçe M, Aytaç S, Ünal Ş, Altan İ, Gümrük
F and Çetin M: Acute megakaryoblastic leukemia with t(1;22)
mimicking neuroblastoma in an infant. Turk J Haematol. 32:64–67.
2015.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Kawasaki Y, Makimoto M, Nomura K, Hoshino
A, Hamashima T, Hiwatari M, Nakazawa A, Takita J, Yoshida T and
Kanegane H: Neonatal acute megakaryoblastic leukemia mimicking
congenital neuroblastoma. Clin Case Rep. 3:145–149. 2015.PubMed/NCBI View
Article : Google Scholar
|
|
17
|
Marques-Piubelli ML, Cordeiro MG,
Cristofani L, Barroso RS, Paes VR, Castelli JB and Rodrigues
Pereira Velloso ED: Acute megakaryoblastic leukemia with
t(1;22)(p13.3;q13.1); RBM15-MKL1 mimicking hepatoblastoma in an
infant: The role of karyotype in differential diagnosis. Pediatr
Blood Cancer. 67(e28111)2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Mercher T, Coniat MB, Monni R, Mauchauffe
M, Nguyen Khac F, Gressin L, Mugneret F, Leblanc T, Dastugue N,
Berger R and Bernard OA: Involvement of a human gene related to the
Drosophila spen gene in the recurrent t(1;22) translocation
of acute megakaryocytic leukemia. Proc Natl Acad Sci USA.
98:5776–5779. 2001.PubMed/NCBI View Article : Google Scholar
|