Introduction
Myelodysplastic/myeloproliferative neoplasm with
neutrophilia (MDS/MPN-N) exhibits a higher acute leukemia
transformation rate (up to 40%) compared with other
myelodysplastic/myeloproliferative neoplasms, including chronic
myelomonocytic leukemia (CMML), myelodysplastic/myeloproliferative
neoplasm with SF3B1 mutation and thrombocytosis and
myelodysplastic/myeloproliferative neoplasm, not otherwise
specified (1). Therefore, the
diagnosis of MDS/MPN-N and the differentiation from other
myelodysplastic/myeloproliferative neoplasms remains pivotal,
although it requires a complex interplay of hematopathological and
molecular genetic assessment. The increasing availability of
targeted sequencing and the first applications of whole genome
sequencing in routine use are expanding the diagnostic
armamentarium of MDS/MPN-N. The diagnostic criteria of the World
Health Organization (WHO) for MDS/MPN-N include leukocytosis (a
white blood cell count >13x109/l), left-shifted and
dysplastic granulopoiesis and a blast count <20% (1,2).
Case report
Patient's history and hematological
investigations
A patient was referred to the Hospital
Wels-Grieskirchen (Wels, Austria) in March 2022 with a leukocyte
count of 105 109/l. As reactive leukocytosis usually
does not exceed 100 109/l leukocytes, which is even true
for patients with sepsis, this result indicated the presence of a
myeloid neoplasm (3). Initial
laboratory assessment showed normal to slightly increased levels of
C-reactive protein and procalcitonin, which excluded an infectious
etiology for the notable elevation in leukocyte counts; however,
lactate dehydrogenase levels were notably increased, indicative of
increased cell turnover (4). In
further laboratory assessments, there was no evidence of autoimmune
disease. Supplementary abdominal sonography revealed a normal size
proportion of the liver and a slightly enlarged spleen (14 cm),
which was confirmed by computed tomography. Moreover, there was no
evidence of lymphadenopathy. Therefore, secondary causes of the
elevated leukocyte count were not considered (5,6).
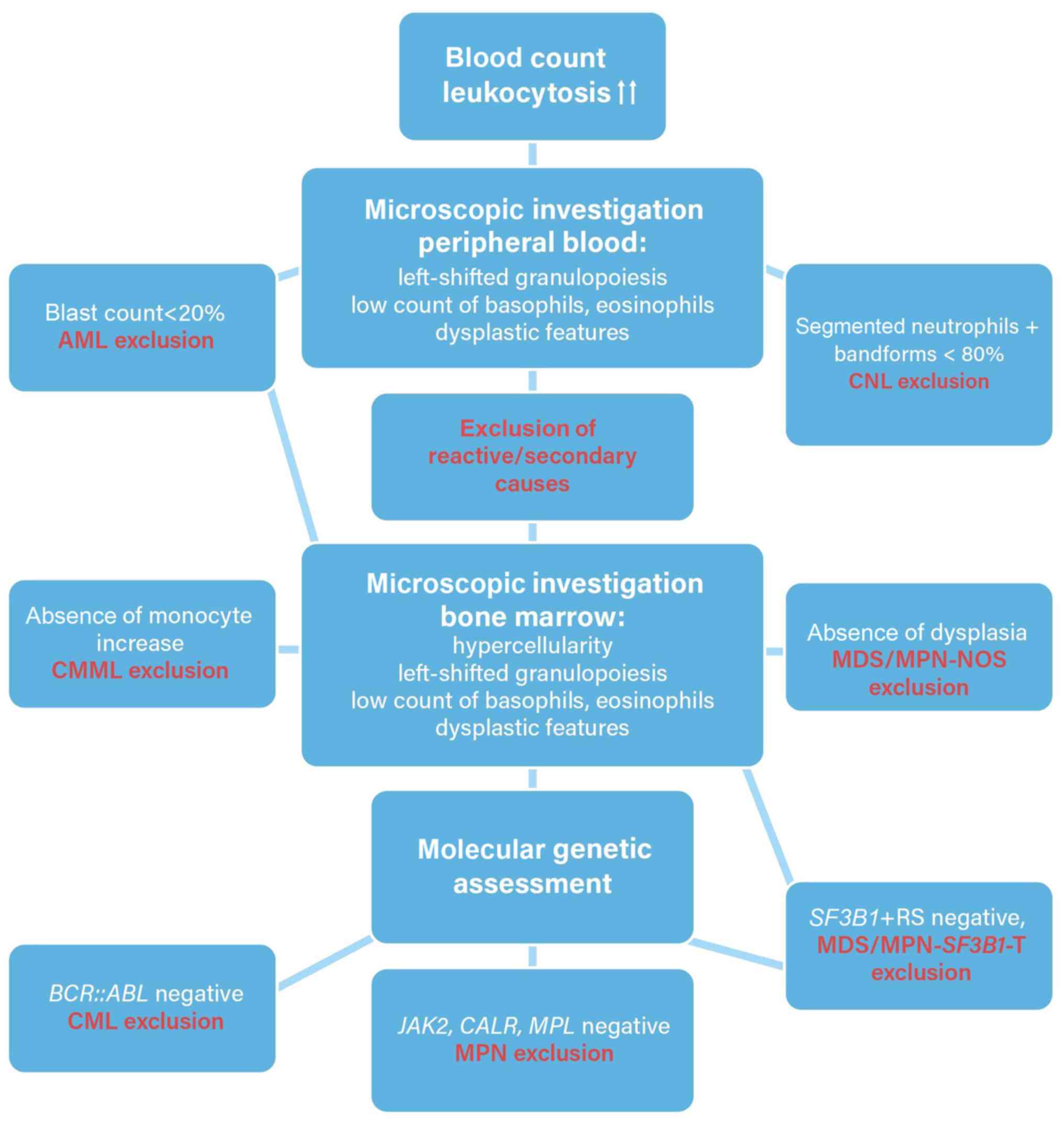
Microscopic differential blood examination showed a
picture of proliferatively dominating myelopoiesis that was
pathologically left shifted with a myelocyte peak. A bone marrow
puncture was performed, the results of which corresponded with that
of the peripheral blood smear. The bone marrow was hypercellular,
with a dominating granulopoiesis that was left shifted with a
distribution in favor of myelocytes and a blast count of 15%.
Basophilic granulocytes and eosinophilic granulocytes were
massively underrepresented in the hematopoiesis (<2%). When
considering classical chronic myeloid leukemia (CML), this
microscopic appearance with low counts of basophilic granulocytes
and eosinophilic granulocytes is very atypical (7,8).
Second, the erythropoietic precursor cells accounted for between
27-31% of the hematopoiesis, as assessed by three different
investigators. Moreover, such a high percentage of erythropoiesis
in an untreated patient with de novo CML in the chronic
phase implied an atypical disease course (9). Ultimately, the hematopoiesis showed
trilinear dysplasia. Secondary causes of hematopoietic dysplasia
were evaluated and excluded (10);
the patient had no history of alcohol abuse, his
carbohydrate-deficient transferrin value was not increased, and
vitamin B12, folic acid, and iron levels were within the reference
ranges, excluding deficiencies. The patient's medical records did
not contain any prescribed cytotoxic medications or prior radiation
therapy, and a congenital disorder was implausible because the male
patient was 69 years old. In addition, the patient's clinical
status and subsequent laboratory assessment did not indicate an
infectious disease.
Molecular biological assessment
Conventional cytogenetic assessment was performed,
and karyotype analyses on unstimulated and stimulated (24 and 48 h)
cultures showed no aberrations (karyotype: 46, XY). PCR analysis of
BCR::ABL1 major and minor breakpoints produced negative
results, as did fluorescence in situ hybridization.
In summary, the hypercellular bone marrow in
combination with significant dysplasia of hematopoiesis and
BCR::ABL1 negativity led to the diagnosis of MDS/MPN-N
(1). Finally, next-generation
sequencing (NGS) was performed with a myeloid solution panel,
including 30 gene sections of ABL1, ASXL1,
BRAF, CALR, CBL, CEBPA, CSF3R,
DNMT3A, ETV6, EZH2, FLT3, HRAS,
IDH1, IDH2, JAK2, KIT, KRAS,
MPL, NPM1, NRAS, PTPN11, RUNX1,
SETBP1, SF3B1, SRSF2, TET2,
TP53, U2AF1, WT1, and ZRSR2. The
detected mutations that matched with the final diagnosis of
MDS/MPN-N (11,12) are presented in Table I.
 | Table IMutations in our patient detected via
next-generation sequencing. |
Table I
Mutations in our patient detected via
next-generation sequencing.
| Gene | DNA sequence
change | Amino acid
change | Exon | Type of
mutation | VAF (%) |
|---|
| ASXL1 | c.1892_1938del | p.
(His631Profs*11) | 13 | Frameshift | 43.90 |
| RUNX1 | c.1256_1262dup | p.
(Glu422Glyfs*180) | 9 | Frameshift | 48.60 |
| SRSF2 | c.284C>T | p. (Pro95Leu) | 1 | Missense | 50.40 |
| TET2 | c.5618T>C | p.
(IIe1873Thr) | 11 | Missense | 49.50 |
| TET2 | c.3782G>A | p.
(Arg1261His) | 6 | Missense | 49.10 |
TET2, SRSF2 and ASXL1 mutations are
the most frequent reported mutations in MDS/MPN-N. Thus, the
mutational profile of our patient confirmed the diagnosis of
MDS/MPN-N (13-21).
In our patient the dysplastic features, in particular the
granulopoiesis accounted for MDS/MPN-N, albeit the most specific
mutations of MDS/MPN-N in ETNK1 and SETBP1 genes were
not analyzed or detected (13,15).
After one year of follow up, a progression of MDS/MPN-N (e.g.
transformation to acute leukemia) was not observed in our patient,
even though ASXL1 mutations and ≥3 mutations are associated
with an adverse clinical outcome (20,21).
Haemato-oncological differential
diagnoses
As MDS/MPN-N was diagnosed, it was differentiated
from other MPN/MDS neoplasms in adults (1,2).
CMML. This possibility was excluded due to
the absence of monocytosis in peripheral blood: A persistent
absolute (≥0.5x109/ l) and relative (≥10% of white blood
cells) monocytosis is required according to the 5th Edition of the
WHO classification of hematolymphoid tumors (1).
MDS/MPN neoplasm with SF3B1 mutation and
thrombocytosis. This possibility was excluded as SF3B1
mutations were not detected by NGS analysis. In addition,
persistent thrombocytosis with a platelet count
≥450x109/l was not detected (1).
MDS/MPN neoplasm not otherwise specified
(NOS). This entity remains a diagnosis of exclusion in MPN/MDS
overlap syndromes. In the present case report, the drastic
myelodysplasia determined MDS/MPN-N (1).
In addition to MDS/MPN overlap syndromes, a CML as a
relevant differential diagnosis was considered; however, as the
molecular-biological analyses yielded negative BCR::ABL1
results, a classical CML could not be confirmed (22). One of the most important
differential diagnoses is chronic neutrophilic leukemia (CNL).
However, diagnosing CNL requires >80% banded and segmented
neutrophils (1,23). Further differential diagnoses
included acute myeloid leukemia (AML) as the blast count was 15% in
the bone marrow. In the 5th Edition of the WHO classification of
hematolymphoid tumors, the 20% blast requirement for most AML types
with recurrent genetic abnormalities was eliminated. However, in
the present case report, molecular-biological assessment did not
yield those specific rearrangements, nor the corresponding
translocations (1,24). Primary myelofibrosis was also
considered, as leukocytosis is a possible presentation of
myelofibrotic disease, and the patient exhibited
leucoerythroblastosis on the peripheral blood smear. Primary
myelofibrosis (or even pre-fibrotic or post-essential
thrombocythemia or post-polycythemia vera myelofibrosis) was
excluded, as the patient's bone marrow did not exhibit substantial
fibrosis, which is essential for the diagnosis of myelofibrotic
diseases (1,25). A flowchart for diagnosing MDS/MPN-N
is shown in Fig. 1.
Discussion
A PubMed/Medline search was performed by three
different investigators with the MeSH terms ‘atypical CML’ AND
‘BCR/ABL negative CML’ AND ‘MDS/MPN-neoplasm with neutrophilia’.
More than 400 articles were found; however, all reviews and case
reports, clinical letters with <10 reported patients with
MDS/MPN-N, and clinical trials that lacked reproducible molecular
genetic data of the mutations were all excluded. The literature
search on articles published was limited to January 2013 and
December 2022 to gain more detailed information of the molecular
genetic profile of the disease after the advent of NGS, with the
first exome sequencing trial of MDS/MPN-N by Piazza et al
(16) in 2013. An overview of the
identified studies and mutation prevalence is presented in Table II.
 | Table IIMutation frequencies of
MDS/MPN-N. |
Table II
Mutation frequencies of
MDS/MPN-N.
| First name,
year | Mutations |
MDS/MPN-Na | (Refs.) |
|---|
| Maxson et
al, 2013 | 42.1% CSF3R | 19 | (17) |
| Montalban-Bravo
et al, 2021 | 86% ASXL; 63%
SRSF2; 56% SETBP1; 34% TET2; 20-30% GATA, NRAS and CBL; 10-20%
RUNX1, NF1 and JAK2; <10% Miscellaneous | 65 | (20) |
| Piazza et
al, 2013 | 25% TET2; 23%
ASXL1; 23-24% SETBP1; 15% EZH2; 10% N/KRAS |
18/61/70b | (16) |
| Wang et al,
2014 | 35% N/KRAS; 7.3%
JAK2 | 65 | (18) |
| Meggendorfer et
al, 2013 | 31.7% SETBP1 | 60 | (13) |
| Patnaik et
al, 2017 | 28% ASXL1; 16% TET2
and NRAS; 12% SETBP1 and RUNX1 | 25 | (21) |
| Meggendorfer et
al, 2014 | 66% ASXL1; 41%
TET2; 40% SRSF2; 33% SETBP1; 10% CBL; 3% CSF3R; ≤3% JAK2, CALR and
MPL | 58 | (14) |
|
Gambacorti-Passerini et al,
2015 | 27% NRAS and
SETBP1; 20% EZH and ASXL1; 13 and 9% ETNK1; 13% U2AF1 | 15/68c | (15) |
| Zhang et al,
2019 | 81% ASXL1; 37%
SRSF2 and TET2; 20-30% EZH2, CSF3R and NRAS | 27 | (19) |
In 2013, Piazza et al (16) performed the first whole-genome
sequencing of 8 MDS/MPN-N cases and identified SETBP1 as a
common mutation. Accordingly, targeted sequencing of SETBP1 in 70
MDS/MPN-N samples was performed. During those analyses,
SETBP1 mutations were detected in 17 of 70 patients, which
resulted in a frequency of 24%. Most of the SETBP1 mutations
occurred between codons 858 and 871 and were reported as being
similar to mutations in Schinzel-Giedion syndrome. SETBP1
was assumed to be a mutation that was predominately enriched in
MDS/MPN-N and related disorders as the researchers were unable to
detect this mutation in 458 individuals with other hematological
neoplasms, nor in 344 cell lines representative of lymphomas and
the most common solid tumors. Distinctively, patients with
SETBP1 mutations showed higher leukocyte counts at diagnosis
compared with patients with wildtype SETBP1 status and
MDS/MPN-N (median of 81.0 vs. 38.5x109 cells/l,
P=0.008). In addition, SETBP1 mutations were associated with
an adverse clinical course in MDS/MPN-N patients, as the overall
survival was significantly worse compared with patients who lacked
the mutation (median survival=22 vs. 77 months, P=0.01, hazard
ratio=2.27). This study also provided NGS data covering 15 gene
sections of 61 MDS/MPN-N individuals. The most frequent mutation
was in the TET2 gene at 25%, followed by SETBP1 and
ASXL1, both at 23%. This study decisively discovered
SETBP1 mutations as a possible recurrent mutation in
MDS/MPN-N that also played a causative role in the entity's
pathophysiology and provided a comprehensive overview of the
mutational profile of MDS/MPN-N patients (16).
A clinical trial in 2013 with the goal of additional
molecular genetic characterization of MDS/MPN-N was conducted by
Meggendorfer et al (13),
as the previous discovery of SETBP1 as a novel molecular
marker for MDS/MPN-N increased interest in this scientific field.
This study group analyzed the SETBP1 mutational status of
1,130 patients with MPN and MPN/MDS overlap neoplasms. Meggendorfer
et al (13) demonstrated a
dominance in the MDS/MPN cohort (9.4% vs. 3.8% in MPN), with the
highest frequencies in MDS/MPN-N (31.7%; 19/60) and MDS/MPN-NOS
(9.3%; 20/240). Furthermore, SETBP1 mutations were
associated with significantly higher leukocyte counts, lower
thrombocyte counts, and hemoglobin levels, and a more dysplastic
phenotype (dysplasia of granulopoiesis and megakaryopoiesis) on
cytomorphological assessment. The effect of SETBP1 mutations
in leukemogenesis of different diseases has been well described in
prior publications; overexpression of SETBP1 leads to the
protection of the molecule SET from proteolytic cleavage and, in
terms of quantitative increase of the SET protein, a complex is
formed comprising of SETBP1, SET, and protein phosphatase 2,
which is responsible for the proliferation of leukemic cells.
Unexpectedly, in that context, it must be mentioned that mutation
of SETBP1 did not significantly alter the overall survival.
However, the authors addressed a relevant limitation of the study;
the relatively short median follow-up time of 17.1 months in
MDS/MPN-N and 12 months in CMML patients. Moreover, a pattern of
concomitant occurrence of SETBP1 and ASXL1 mutation
was described. Of note, this study was the first to discover
ASXL1 mutations in MDS/MPN-N, which has been reported in
recent publications as one of the most frequent molecular
abnormalities in MDS/MPN-N (13).
In the same year, Maxson et al (17) simultaneously conducted a trial to
discover more regarding the clonal nature of MDS/MPN-N. They
investigated MDS/MPN-N and CNL as, at the time of the study, little
was known regarding the mutations in those diseases and both
entities lacked knowledge of specific cytogenetic aberrations. They
identified CSF3R as a potential driver mutation of those
diseases as CSF3R is a receptor of colony-stimulating factor
3, which is hypothesized to play a pivotal role in the growth and
differentiation of granulocytes. Previous reports have described
CSF3R mutations, amongst others, as a contributor to severe
congenital neutropenia, which frequently evolves into AML. While
the study population was small, with 9 CNL patients and 18 patients
with MDS/MPN-N, this trial revealed an association between
CSF3R mutations and those entities. The high frequency of
CSF3R mutations in leukemia with neutrophilic expansion was
consistent with its function as a receptor that promotes
neutrophilic differentiation and proliferation. However, it must be
noted that CSF3R mutations occurred in ~89% of CNL cases and
only 44% of MDS/MPN-N cases. Therefore, this study identified
CSF3R mutations in MDS/MPN-N and CNL; however, this mutation
was determined to be more specific to CNL than MDS/MPN-N. This was
an important finding, as the discrimination of these entities had
previously relied more or less on hematological parameters, such as
leukocyte counts (>25x109/l for CNL and
>13x109/l for MDS/MPN-N), the percentage of immature
precursor leukocytes in the total white cell population (<10%
for CNL and >10% for MDS/MPN-N), and the presence of
dysgranulopoiesis in MDS/MPN-N. Consequently, consecutive studies
could not confirm the distinctive frequency of CSF3R
mutations in MDS/MPN-N but promoted it as a typical marker of CNL.
This study group also investigated different types of CSF3R
mutations with different susceptibilities to tyrosine kinase
inhibitors. CSF3R truncation mutations are preferentially
activators of SRC family-TNK2 kinase signaling, with a sensitivity
to dasatinib, whereas CSF3R membrane proximal mutations
resulted in activation of the JAK signaling pathway and should be
treated with JAK1/2 inhibitors (such as ruxolitinib).
Notably, the T618I variant was the most detected commonly
CSF3R mutation of the proximal membrane mutations (17).
In the following year, Meggendorfer et al
(14) performed a trial including
14 patients with CNL, 68 with MDS/MPN-N, and 146 with CMML to
enable improved differentiation within this group of common MDS/MPN
overlapping malignancies based on molecular genetic markers.
Importantly, this was the first trial to describe ASXL1
mutations as the most frequent mutation in an MDS/MPN-N cohort.
Nevertheless, its value as a differentiation marker of other
MPN/MDS neoplasms could not be determined, as ASXL1
mutations were also detected with comparable prevalence in CNL
(57%) and CMML (66%). A novel observation was made, as they
discovered SRSF2 mutations with 40% prevalence in MDS/MPN-N.
Interestingly, MDS/MPN-N patients with SETBP1 mutations
presented with higher hemoglobin levels than wild-type patients.
Importantly, CSF3R was often mutated in CNL (43%), but
rarely in MDS/MPN-N or CMML (1-3%) which supported previous data
suggesting that CSF3R was a molecular genetic marker of CNL
(14).
Wang et al (18) compared the clinical outcomes of
MDS/MPN-N patients with patients diagnosed with MDS/MPN-NOS. In
addition, they provided data on the detected mutations, thereby
contributing to an improved understanding of the molecular nature
of those diseases. This previous study clearly highlighted adverse
features, inferior overall survival, and inferior AML-free survival
of patients with MDS/MPN-N compared with MDS/MPN-NOS. There was
controversy surrounding studies on CSF3R, as certain
publications reported a strong association with MDS/MPN-N while
others reported no association. In 27 patients with MDS/MPN-N,
CSF3R mutations were not detected. Therefore, they proposed
that an initial diagnosis of MDS/MPN-N should be reconsidered when
CSF3R analysis was positive; instead, a diagnosis of CNL
should be considered (18).
In 2015, Gambacorti-Passerini et al (15) performed whole-exome sequencing on
15 MDS/MPN-N cases. They detected a groundbreaking somatic
ETNK1 mutation for the first time in cancer in two patients.
ETNK1 encodes an ethanolamine kinase that catalyzes the
biosynthesis of phosphatidylethanolamine, a molecule that is
involved in the regulation of the transmembrane domains of membrane
proteins, the progression of cytokinesis during cell division, and
the activation of the respiratory complex in mitochondria. The
discovery of ETNK1 mutations in MDS/MPN-N prompted the study
group to sequence 515 cases of several hematologic diseases.
ETNK1 mutations were detected exclusively in MDS/MPN-N (9%,
6/68) and CMML (2.6%, 2/77) (15).
In 2017, Patnaik et al (21) performed an MDS/MPN-N trial with an
extended panel of 29 genes and analyzed bone marrow specimens.
Based on prior publications, they were also interested in clinical
outcomes. The most mutated gene was ASXL1. Notably,
ASXL1 mutations did not adversely impact overall survival in
contrast to NRAS (P=0.04), TET2 (P=0.03), PTPN11
(P=0.02), and ≥3 myeloid mutations. However, in two patients,
leukemic transformation was documented. One of these patients
harbored an ASXL1 separate from JAK2, and the second
was positive for TET2 and PTPN11 mutations (21).
Zhang et al (19) analyzed specimens from 158 patients
with MDS/MPN neoplasms (27 MDS/MPN-N) and CNL by whole exome and
RNA sequencing. In these rare leukemic diseases, an increased
variant allele frequency of mutations in signal-transduction genes
was observed; this may indicate a preferential pharmaceutical
target. In >50% of the patients with either MDS/MPN neoplasms or
CNL, ≥3 or more co-occurring pathway mutations involving genes of
chromatin modification, epigenetic regulator genes, signaling
pathway genes, or genes of the splicing complex were observed. In
contrast, in MPN, only mutations of signal-transduction genes were
predominant, whereas in MDS, mutations of the splicing complex
typically predominated. In conclusion, this trial contributed to an
improved understanding of the differentiation of MDS/MPN neoplasms,
including MDS/MPN-N, from other myeloid malignancies; the authors
also stated that malignancies classified as MDS/MPN neoplasms more
often represent a group of related diseases than discrete
diagnostic entities (19).
In 2021, the most recent clinical trial
investigating the mutational architecture of MDS/MPN-N was
conducted. The study included 68 MDS/MPN-N patients from 2005-2020,
and NGS data were available for 35 patients. One major strength of
this study was the long follow-up time (median 35.6 months).
Transformation to AML was observed in 28% of patients. The genes
that contributed to AML transformation were ASXL1,
PTPN11, N/KRAS, NF1, CEBPA,
ETV6, and FLT3-ITD. The median leukemia-free survival
was 19.8 months, and the median post-transformation survival was
8.9 months. One of the most important key messages of this study
was that MDS/MPN-N is a disease prone to transformation to AML
(20).
In the present case report, the mutational profile
with ASXL1, 2x TET2, SRSF2, and RUNX1
mutations in combination with the significant dysgranulopoiesis
accounted for MDS/MPN-N.
MDS/MPN-N is a hematological neoplasm with a
relatively low incidence. However, a systematic review of studies
showed that in 251 patients, a comprehensive molecular genetic
analysis by whole-genome sequencing or targeted sequencing,
including a broad spectrum of myeloid genes was performed. Our
analysis identified ASXL1, TET2, and SRSF2
mutations as the most frequent molecular genetic alterations in
MDS/MPN-N. Mutations in transcriptional and epigenetic regulator
genes and genes encoding the spliceosome are typical in MDS/MPN
overlapping neoplasms and are responsible for the phenotype of
these entities including cell proliferation and myelodysplasia.
Therefore, cytomorphological investigation remains a pivotal
diagnostic procedure in the differentiation of MDS/MPN-neoplasms.
In addition, the analysis revealed a heterogeneous picture of
several myeloid gene mutations with lower prevalence in MDS/MPN-N.
However, the sample size of all included studies was low and the
reproducibility was limited due to the different molecular genetic
approaches (such as different gene panels) applied in the reviewed
trials. Thus, the evidence of low frequent mutations in MDS/MPN-N
remains insufficient, and isolated quantitative analysis of those
infrequent mutations is indistinct except for ETNK1 and
SETBP1 mutations. In those genes, the mutational mechanism
of leukemogenesis was assessed. The analyzed data indicated that
ETNK1 and SETBP1 mutations were highly specific for
MDS/MPN neoplasms, particularly for MDS/MPN-N. In the absence of
dysgranulopoiesis ETNK1 and SETBP1 mutations were
associated with MDS/MPN-NOS. Moreover, MDS/MPN-N remains a
diagnosis of exclusion to a certain degree: BCR::ABL1
fusions are the indispensable diagnostic hallmark of CML,
CSF3R mutations are known driver mutations of CNL, and
SF3B1 mutations are highly indicative of MDS or
MDS/MPN-SF3B1-T.
In conclusion, MDS/MPN-N (formerly known as atypical
CML) is a rare disease accounting for 5% of all CML cases (26). The information surrounding
distinctive mutations in MDS/MPN-N was scarce 10-15 years ago and
was predominantly based on data from small case-controlled studies.
However, in the last decade, major improvements in understanding
the nature of the disease and the underlying mutations have been
achieved. Initial deep-sequencing trials identified CSF3R as
one of the most common recurrent mutations of these diseases;
however, following additional study of MDS/MPN-N, it was determined
that the initial CSF3R prevalence was overestimated. To
comply with several recommendations, the prevalence of CSF3R
should justify the revision of MDS/MPN-N diagnosis, instead of
considering a diagnosis of CNL. As NGS-based techniques have
improved and their applications have expanded with larger gene
panels, the discovery of more mutations in more genes has
accelerated. Although the epidemiology of MDS/MPN-N allows for
relatively small study cohorts, the tendency for recurrent
mutations (such as ASXL1, SETBP, TET2,
SRSF2, and ETNK1) has been shown. The latest data
support the notion that MDS/MPN-N is an entity with more adverse
clinical outcomes than other MDS/MPN neoplasms. In addition, high
AML-transformation susceptibility has been observed. Completing
whole-genome sequencing studies of MDS/MPN-N will be beneficial,
allowing further steps to be made in this field of research.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The molecular genetic data of NGS analysis that were
generated during the current study are available in the ClinVar
repository https://www.ncbi.nlm.nih.gov/clinvar under accession
numbers SCV003929446-SCV003929450. The non-NGS data are available
from the corresponding author upon reasonable request.
Authors' contributions
All authors contributed to the conception and design
of the study. BS wrote the manuscript. BS, MG and AH carried out
microscopic investigations. BS, RS and SH carried out
molecular-genetic assessment. BS, RS and MG participated in the
literature analysis for the review section. SH, JT and AH
contributed to revising the manuscript. BS and AH confirm the
authenticity of all the raw data. All authors agree to be
accountable for all aspects of the work. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Written informed consent was obtained from the
patient for the publication of anonymized data.
Competing interests
The authors declare that they have no competing
interest.
References
|
1
|
Khoury JD, Solary E, Abla O, Akkari Y,
Alaggio R, Apperley JF, Bejar R, Berti E, Busque L, Chan JKC, et
al: The 5th edition of the World Health Organization Classification
of Haematolymphoid Tumours: Myeloid and Histiocytic/Dendritic
Neoplasms. Leukemia. 36:1703–1719. 2022.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Thota S and Gerds AT: Myelodysplastic and
myeloproliferative neoplasms: Updates on the overlap syndromes.
Leuk Lymphoma. 59:803–812. 2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Urrechaga E, Boveda O and Aguirre U:
Improvement in detecting sepsis using leukocyte cell population
data (CPD). Clin Chem Lab Med. 57:918–926. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Vanderlinde RE: Measurement of total
lactate dehydrogenase activity. Ann Clin Lab Sci. 15:13–31.
1985.PubMed/NCBI
|
|
5
|
Chabot-Richards DS and George TI:
Leukocytosis. Int J Lab Hematol. 36:279–288. 2014.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Riley LK and Rupert J: Evaluation of
patients with leukocytosis. Am Fam Physician. 92:1004–1011.
2015.PubMed/NCBI
|
|
7
|
Langabeer SE, Bhreathnach U, Cahill MR,
Elhassadi E, Ní Loingsigh S, Conneally E and Enright H: Can
absolute basophilia distinguish e1a2 BCR-ABL1 chronic myeloid
leukemia from chronic myelomonocytic leukemia? Blood Cells Mol Dis.
87(102521)2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Dhakal P, Gundabolu K, Amador C, Rayamajhi
S and Bhatt VR: Atypical chronic myeloid leukemia: A rare entity
with management challenges. Future Oncol. 14:177–185.
2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Thiele J, Kvasnicka HM, Schmitt-Graeff A,
Zirbes TK, Birnbaum F, Kressmann C, Melguizo-Grahmann M,
Frackenpohl H, Sprungmann C, Leder LD, et al: Bone marrow features
and clinical findings in chronic myeloid leukemia-a comparative,
multicenter, immunohistological and morphometric study on 614
patients. Leuk Lymphoma. 36:295–308. 2000.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Shekhar R, Srinivasan VK and Pai S: How I
investigate dysgranulopoiesis. Int J Lab Hematol. 43:538–546.
2021.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Dao KH and Tyner JW: What's different
about atypical CML and chronic neutrophilic leukemia? Hematology Am
Soc Hematol Educ Program. 2015:264–271. 2015.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Crisa E, Nicolosi M, Ferri V, Favini C,
Gaidano G and Patriarca A: Atypical chronic myeloid leukemia: Where
are we now? Int J Mol Sci. 21(6862)2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Meggendorfer M, Bacher U, Alpermann T,
Haferlach C, Kern W, Gambacorti-Passerini C, Haferlach T and
Schnittger S: SETBP1 mutations occur in 9% of MDS/MPN and in 4% of
MPN cases and are strongly associated with atypical CML, monosomy
7, isochromosome i(17)(q10), ASXL1 and CBL mutations. Leukemia.
27:1852–1860. 2013.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Meggendorfer M, Haferlach T, Alpermann T,
Jeromin S, Haferlach C, Kern W and Schnittger S: Specific molecular
mutation patterns delineate chronic neutrophilic leukemia, atypical
chronic myeloid leukemia, and chronic myelomonocytic leukemia.
Haematologica. 99:e244–e246. 2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Gambacorti-Passerini CB, Donadoni C,
Parmiani A, Pirola A, Redaelli S, Signore G, Piazza V, Malcovati L,
Fontana D, Spinelli R, et al: Recurrent ETNK1 mutations in atypical
chronic myeloid leukemia. Blood. 125:499–503. 2015.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Piazza R, Valletta S, Winkelmann N,
Redaelli S, Spinelli R, Pirola A, Antolini L, Mologni L, Donadoni
C, Papaemmanuil E, et al: Recurrent SETBP1 mutations in atypical
chronic myeloid leukemia. Nat Genet. 45:18–24. 2013.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Maxson JE, Gotlib J, Pollyea DA,
Fleischman AG, Agarwal A, Eide CA, Bottomly D, Wilmot B, McWeeney
SK, Tognon CE, et al: Oncogenic CSF3R mutations in chronic
neutrophilic leukemia and atypical CML. N Engl J Med.
368:1781–1790. 2013.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wang SA, Hasserjian RP, Fox PS, Rogers HJ,
Geyer JT, Chabot-Richards D, Weinzierl E, Hatem J, Jaso J,
Kanagal-Shamanna R, et al: Atypical chronic myeloid leukemia is
clinically distinct from unclassifiable
myelodysplastic/myeloproliferative neoplasms. Blood. 123:2645–2651.
2014.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Zhang H, Wilmot B, Bottomly D, Dao KT,
Stevens E, Eide CA, Khanna V, Rofelty A, Savage S, Reister Schultz
A, et al: Genomic landscape of neutrophilic leukemias of ambiguous
diagnosis. Blood. 134:867–879. 2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Montalban-Bravo G, Kanagal-Shamanna R,
Sasaki K, Masarova L, Naqvi K, Jabbour E, DiNardo CD, Takahashi K,
Konopleva M, Pemmaraju N, et al: Clinicopathologic correlates and
natural history of atypical chronic myeloid leukemia. Cancer.
127:3113–3124. 2021.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Patnaik MM, Barraco D, Lasho TL, Finke CM,
Reichard K, Hoversten KP, Ketterling RP, Gangat N and Tefferi A:
Targeted next generation sequencing and identification of risk
factors in World Health Organization defined atypical chronic
myeloid leukemia. Am J Hematol. 92:542–548. 2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Soverini S, Abruzzese E, Bocchia M,
Bonifacio M, Galimberti S, Gozzini A, Iurlo A, Luciano L, Pregno P,
Rosti G, et al: Next-generation sequencing for BCR-ABL1 kinase
domain mutation testing in patients with chronic myeloid leukemia:
A position paper. J Hematol Oncol. 12(131)2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Gotlib J, Maxson JE, George TI and Tyner
JW: The new genetics of chronic neutrophilic leukemia and atypical
CML: Implications for diagnosis and treatment. Blood.
122:1707–1711. 2013.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Arber DA, Orazi A, Hasserjian RP, Borowitz
MJ, Calvo KR, Kvasnicka HM, Wang SA, Bagg A, Barbui T, Branford S,
et al: International consensus classification of myeloid neoplasms
and acute leukemias: Integrating morphologic, clinical, and genomic
data. Blood. 140:1200–1228. 2022.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Monte-Mor Bda C, Ayres-Silva Jde P,
Correia WD, Coelho AC, Solza C, Daumas AH, Bonamino MH, Santos FP,
Datoguia TS, Pereira Wde O, et al: Clinical features of JAK2V617F-
or CALR-mutated essential thrombocythemia and primary
myelofibrosis. Blood Cells Mol Dis. 60:74–77. 2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Cross NCP: Update on CML-Like Disorders.
Clin Lymphoma Myeloma Leuk. 20 (Suppl 1):S101–S102. 2020.PubMed/NCBI View Article : Google Scholar
|