Introduction
Acute myeloid leukemia (AML) is a hematopoietic stem
cell disorder characterized by genetic alterations in precursor
cells within the bone marrow (BM), leading to the proliferation of
neoplastic clonal myeloid stem cells (1). AML presents a considerable global
health burden, with ~500 new cases in adults annually in Taiwan
alone (2). As the most prevalent
form of acute leukemia in adults, AML accounts for the highest
proportion of leukemia-related deaths (3). The dysregulation of transcription
factors and signaling pathways in hematopoiesis contributes to the
development of malignant phenotypes such as AML and myelodysplastic
syndromes (MDS), ultimately leading to the formation of leukemic
stem cells (4).
Prognostic stratification in AML is predominantly
based on the World Health Organization (WHO) classification, which
takes into account cytogenetic and molecular genetic aberrations
(5-7).
These genetic alterations can serve as potent prognostic markers
for AML (8). Additionally,
epigenetic alterations have gained significance as contributors to
AML pathogenesis (9).
Consequently, AML is classified into favorable, intermediate and
adverse risk groups based on newly recognized cytogenetic and
molecular subsets, each displaying distinct responses to standard
therapies (4). However, the
translation of these novel biomarkers as tools for effective
treatment decisions remains limited in clinical practice. Thus, the
identification of molecular markers for risk-adapted therapies
represents a critical goal in improving the clinical outcomes of
AML (10).
Among the epigenetic mechanisms, genomic imprinting
emerges as an interesting regulatory process that restricts gene
expression to one of the two parental chromosomes. While the
majority of the genes in the genome are expressed equally
regardless of parental origin, a small subset of genes displays
genomic imprinting (11). As of
December 2018, ~165 imprinted genes in humans and 197 imprinted
genes in mice have been reported (12). Although the exact mechanisms of
imprinting remain elusive, the vital roles of imprinted genes in
regulating cell proliferation and fetal growth are widely
recognized (12,13). Consequently, dysregulated
expression of imprinted genes has been implicated in tumorigenesis,
exemplified by the upregulation of H19 imprinted maternally
expressed transcript (H19) (10,14),
insulin like growth factor 2 (IGF2) (14), maternally expressed 3 (MEG3)
(15,16) and Zinc finger protein 215
(ZNF215) (14) genes in
AML.
Significant progress has been achieved in the
genomic profiling of AML, enabling the development of targeted
therapies based on genomic characteristics. Nonetheless, the
dysregulation of imprinted genes in AML remains relatively
unexplored. A previous study conducted by the authors demonstrated
that altered expression of imprinted genes and loss of ZNF215
imprinting can function as prognostic indicators for poor five-year
survival in patients with cytogenetically abnormal (CA)-AML
(14). Building upon these
findings, the present study investigated whether the expression of
imprinted genes is similarly altered in patients with
cytogenetically normal (CN)-AML. The primary aim of the authors was
to establish a possible correlation between this disease subtype
and loss of imprinting.
Materials and methods
Patients and samples
Patients diagnosed with CN-AML were recruited from
the Division of Hematology-Oncology, Department of Internal
Medicine, Kaohsiung Medical University Hospital
(Kaohsiung, Taiwan) during the period spanning from
August 1999 to October 2012. At the hospital, the standard protocol
for diagnosis of AML included cytogenetic examination and
mutational analysis. Immunophenotyping was conducted for the
diagnosis of lymphocytic leukemia. The present study received
ethical approval from the Institutional Review Board (IRB) of the
Kaohsiung Medical University Hospital Ethical Committee (approval
no. KMU-IRB-20130129; Kaohsiung, Taiwan). Written informed consent
was provided by all participants.
BM samples were collected from 64 patients with
CN-AML, while peripheral blood (PB) samples were obtained from 85
healthy adult volunteers. Within 1 h of collection, red blood cell
(RBC) lysis buffer (MilliporeSigma) was added to the samples to
deplete RBCs from both BM and PB samples. The isolated total
leukocytes were then preserved in TRIzol reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) and stored in a -80˚C deep freezer
until RNA extraction.
Cytogenetic analysis
Cytogenetic analysis was conducted on BM aspirate
samples during the initial diagnosis using conventional G-banding
cytogenetic techniques. Short-term unstimulated cell cultures were
established for one to three days in RPMI-1640 medium supplemented
with 20% fetal bovine serum (both from Invitrogen; Thermo Fisher
Scientific, Inc.). A total of 20 metaphase cells with G-banded
patterns were karyotyped and described according to the ISCN 2013
guidelines (17,18).
Selection of imprinted genes
Candidate imprinted genes were identified based on
the Catalogue of Imprinted Genes- University of Otago from
2001(19), followed by a thorough
evaluation using the Geneimprint Imprinted gene database
(https://www.geneimprint.com/site/genes-by-species).
Genes listed as either ‘predicted’ or ‘not imprinted’ were excluded
from the study. Subsequently, a total of 16 human imprinted genes
were selected, previously reported to be involved in various
malignancies (14,20). Next, for primer design purposes,
the sequences of these genes were acquired from the EnsEMBL human
archive (Ensembl 54 NCBI 36; https://may2009.archive.ensembl.org/Homo_sapiens/Info/Index).
Expression analysis of imprinted
genes
The expression analysis of imprinted genes adhered
to the protocol is described in previous studies (14,20).
Briefly, total RNA was isolated using TRIzol reagent (Invitrogen,
Thermo Scientific, Inc.), followed by cDNA synthesis using the High
Capacity cDNA Reverse Transcription Kit (Applied Biosystems; Thermo
Scientific, Inc.) according to the manufacturer's protocols. Gene
expression analysis was executed using SYBR® Green.
Sequences of the forward and reverse primers of the 16 imprinted
genes analyzed are listed in Table
I. The primer sequences of reference gene ACTB (β-actin)
were as follows: forward, 5'-GGCCAACCGCGAGAAGAT-3' and reverse,
5'-CGTCACCGGAGTCCATCAC-3'. In each reaction, 50 ng of cDNA, along
with 200 nM of each primer, and 5 µl of 2x Power SYBR®
Green PCR Master Mix (Applied Biosystems; Thermo Scientific, Inc.)
were combined to a final volume of 10 µl. The PCR test was
performed using a 7500 Fast Real-Time System (Applied Biosystems;
Thermo Scientific, Inc.), employing the following thermal cycling
parameters: An initial denaturation step at 95˚C for 10 min,
followed by 40 cycles of PCR reaction at 95˚C for 20 sec and 60˚C
for 60 sec. The expression of the ACTB gene in the same
sample was used as an endogenous control. The ΔCq value for each
imprinted gene was calculated using the equation ΔCq=(Cq of
imprinted gene-Cq of ACTB gene) (21). Higher-ΔCq values indicate a higher
expression level of the imprinted gene, while lower-ΔCq values
indicate a lower expression level.
 | Table IList of imprinted genes and
oligonucleotide primers for real-time quantitative polymerase chain
reaction analysis. |
Table I
List of imprinted genes and
oligonucleotide primers for real-time quantitative polymerase chain
reaction analysis.
| Gene | Location | Expressed
allele | GenBank Accession
No. | Amplicon Size
(bp) | Sequence
(5→3') | Primer
location |
|---|
| C15orf2 | 15q11-q13 | Unknown | AB527113.1 | 61 | F: GTG ACA GCA TTG
CCT CAG C | 3062-3080 |
| | | | | | R: GGT CTC CTA TCT
GCC TGT GC | 3103-3122 |
| COPG2 | 7q32 | Paternal | NM_012133.4 | 67 | F: TTC CAG ATG AGG
ATG GGT ATG | 2303-2323 |
| | AS | | | | R: TGG TCA GAC ACA
GTC ACT TCG | 2349-2369 |
| CPA4 | 7q32 | Maternal | NM_016352.3 | 85 | F: GTC GGG CAC TGA
GTA CCA A- | 1082-1100 |
| | | | | | R: GTT GTC ATA CGC
CCA GTC G | 1148-1166 |
| GABRB3 | 15q11.2-q12 | Paternal | NM 000814.5 | 72 | F: GGG TGT CCT TCT
GGA TCA ATT A | 899-920 |
| | AS | | | | R: GTT GTC AGC ACA
GTT GTG ATC C | 950-970 |
| H19 | 11p15.5 | Maternal | NR_002196.1 | 84 | F: TTA CTT CCT CCA
CGG AGT CG | 3050-3069 |
| | AS | | | | R: GCT GGG TAG CAC
CAT TTC TT | 4570-4589 |
| IGF2 | 11p15.5 | Paternal | NM_000612.4 | 101 | F: ACA CCC TCC AGT
TCG TCT GT | 868-887 |
| | AS | | | | R: GAA ACA GCA CTC
CTC AAC GA | 949-968 |
| INPP5F | 10q26.11 | Paternal | NM_014937.3 | 87 | F: TTC TTG ATA TGA
AGT GGT GTT GG | 1514-1536 |
| | | | | | R: GGC AGT CCA TAC
AAT TAA CAC G | 1579-1600 |
| L3MBTL | 20q13.12 | Paternal | NM_015478.6 | 72 | F: AGC GCA GGG AAT
ACC AGA G | 564-582 |
| | | | | | R: TTC CTT CTTCTT
GCT TCT CCA | 615-635 |
|
PEG3-AS1 | 19q13.4 | Paternal | NR_023847.2 | 76 | F: GGG TCA AGT CCT
AGG TGA AGG | 4802-4822 |
| | AS | | | | R: CGC CAG ACA CCA
GAA TAC C | 4747-4765 |
| PPP1R9A | 7q21.3 | Maternal | NM_001166160.1 | 76 | F: GCC CAA AAC ATC
ACT GGA G | 4141-4159 |
| | | | | | R: GGG ATG CTG TCA
TTC CAA G | |
| PRIM2 | 6p12-p11.1 | Biallelic | NM_000947.2 | 68 | F: GCC TGC TGT GCA
GTC TGA T | 798-816 |
| | AS | | | | R: GGC CAG TGT AGG
AAT GAC TGA | 845-865 |
| RTL1 | 14q32.31 | Paternal | NM_001134888.2 | 60 | F: CGC AGA GAA TTC
CAC GAG TT | 687-706 |
| | | | | | R: TCT TGG GTA GCT
CTG TAA GGT CA | 724-746 |
| SLC22A3 | 6q26-q27 | Maternal | NM_021977.3 | 71 | F: CCA CCA TCG TCA
GCG AGT | 460-477 |
| | | | | | R: CAG GAT GGC TTG
GGT GAG | 513-530 |
| SNURF | 15q12 | Paternal | NM_022804.2 | 103 | F: CTC ACT GAG CAA
CCA AGA GTG T | 219-240 |
| | | | | | R: AGC TAA GAA TGC
CTG CCT CA | 302-321 |
| TCEB3C | 18q21.1 | Maternal | NM_145653.3 | 82 | F: GGC CAA GAC GCC
TTA TGA T | 1601-1619 |
| | AS | | | | R: TGG CTC CAT CTC
TCC ATT TC | 1663-1682 |
| ZNF215 | 11p15.4 | Maternal | NM_013250.2 | 75 | F:TGT CCA AGA CAG
CGA TTC C- | 220-238 |
| | | | | | R: TGC AAG TAG CTT
AAG TGG CAA A | 273-294 |
Statistical analysis
Statistical analyses were performed using SPSS 22.0
software (IBM Corp.) and GraphPad Prism 7.04 (Dotmatics). Results
were presented as the mean ± SEM (standard error of mean).
Variations in the expression of each imprinted gene between healthy
subjects and CN-AML patients were assessed using the unpaired
Student's t-test or Mann-Whitney test, depending on data
distribution. Pearson correlation analysis was used to evaluate the
correlation between two variables. To ascertain whether the
expression of a specific imprinted gene could function as a
predictor of survival in CN-AML, multivariate analysis using the
Cox proportional hazards regression model was employed. The
predictive capacity of the gene for the survival of the patients
was evaluated by constructing a receiver operating characteristic
(ROC) curve, with the calculation of the area under the curve
(AUC). For comparison of patient survival across groups,
Kaplan-Meier survival analysis and log-rank test were performed.
Statistical analyses for expression analysis of imprinted genes
were based on the -ΔCq values, with P<0.05 signifying
statistical significance in all assessments.
Results
Patient characteristics
The present study included a total of 64 patients
with CN-AML, consisting of 38 males and 26 females. Additionally,
85 healthy subjects, consisting of 47 males and 38 females, were
enrolled in the present study. The age of the participants ranged
from 23 to 57 years, with a median age of 36.3 years. Detailed
information on the clinical characteristics of the patients, such
as sex, age, laboratory data, and French-American-British (FAB)
subtypes are presented in Table
II. The diagnosis and subtyping of AML were determined based on
the classification systems of FAB and WHO (15). To examine the karyotypes of
patients at the time of initial diagnosis, conventional G-banding
cytogenetic analysis was performed on BM aspirate samples.
 | Table IICharacteristics of patients newly
diagnosed with cytogenetically normal-acute myeloid leukemia
(n=64). |
Table II
Characteristics of patients newly
diagnosed with cytogenetically normal-acute myeloid leukemia
(n=64).
| | Mutational
analysis |
|---|
| Characteristic | Values | ACEBP
(%) | DNMT3A
(%) | FLT3-ITD
(%) | FLT3-TKD
(%) | IDH1
(%) | IDH2
(%) | MLL (%) | NPM1
(%) |
|---|
| Sex, n (%) | | | | | | | | | |
|
Male | 38 (59.4%) | | | | | | | | |
|
Female | 26 (40.6%) | | | | | | | | |
| Age, year, median
(range) | 53.5 (22-86) | | | | | | | | |
| Laboratory data,
median (range) | | | | | | | | | |
|
WBCs, per
µl | 39.37
(0.10-328.30) | | | | | | | | |
|
Hemoglobin,
g/dl | 8.45
(4.10-15.60) | | | | | | | | |
|
Platelets,
x1,000/µl | 43.00
(8.00-369.00) | | | | | | | | |
|
Blasts in PB
(%) | 66.25
(2.00-97.50) | | | | | | | | |
|
Blasts in BM
(%) | 74.90
(3.40-96.60) | | | | | | | | |
|
LDH,
U/l | 444 (52-4,023) | | | | | | | | |
| FAB subtype, n | | | | | | | | | |
|
M0 | 2 | 2(100)a | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1(50) | 0 (0) | 0 (0) |
|
M1 | 20 | 9(45) | 0 (0) | 5(25) | 1(5) | 0 (0) | 1(5) | 1(5) | 9(45) |
|
M2 | 23 | 8(35) | 3(13) | 8(35) | 1(4) | 1(4) | 4(17) | 4(17) | 7(30) |
|
M3 | 1 | 0 (0%) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
|
M4 | 16 | 3(19) | 5(31) | 6(38) | 1(6) | 1(6) | 1(6) | 1(6) | 10(63) |
|
M5 | 2 | 1(50) | 2(100) | 0 (0) | 1(50) | 1(50) | 0 (0) | 0 (0) | 1(50) |
|
M6 | 0 | | | | | | | | |
|
M7 | 0 | | | | | | | | |
Expression of imprinted genes in
patients with CN-AML
In a previous study conducted by the authors,
altered expressions of imprinted genes in patients with CA-AML were
observed (14). In the present
study, the objective was to determine whether these genes were also
altered in patients with CN-AML. For analysis, the same panel of 16
human imprinted genes was selected, namely C15orf2,
COPG2, CPA4, GABRB3, H19, IGF2,
INPP5F, L3MBTL, PEG3-AS, PPP1R9A,
PRIM2, RTL1, SLC22A3, SNURF,
TCEB3C and ZNF215. To initially screen the expression
of these 16 genes, a cohort of 25 patients with AML, were randomly
chosen regardless of their karyotypes, alongside 19 healthy
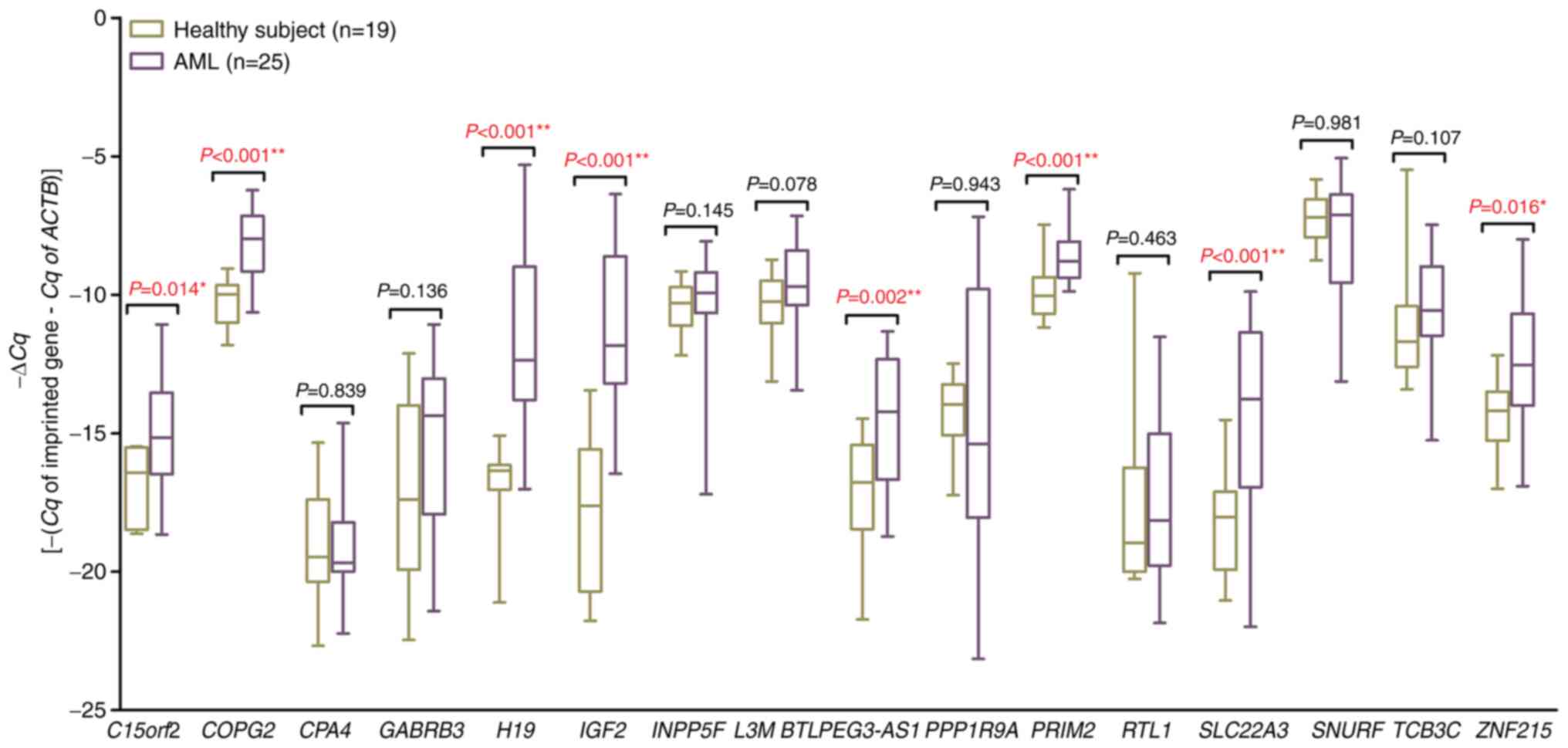
subjects. The findings revealed that 8 of the genes exhibited
upregulation in patients compared with the healthy subjects
(Fig. 1). After the initial
screening of 25 patients regardless of their karyotypes, the
results were analyzed by segregating the cohort into CN-AML and
CA-AML groups. The results from the broader 25-patient group
aligned consistently with those exclusively from CN-AML patients.
Therefore, the apprehension about discarding genes uniquely
regulated in patients with CN-AML rather than patients with CA-AML
during the initial screening was effectively addressed.
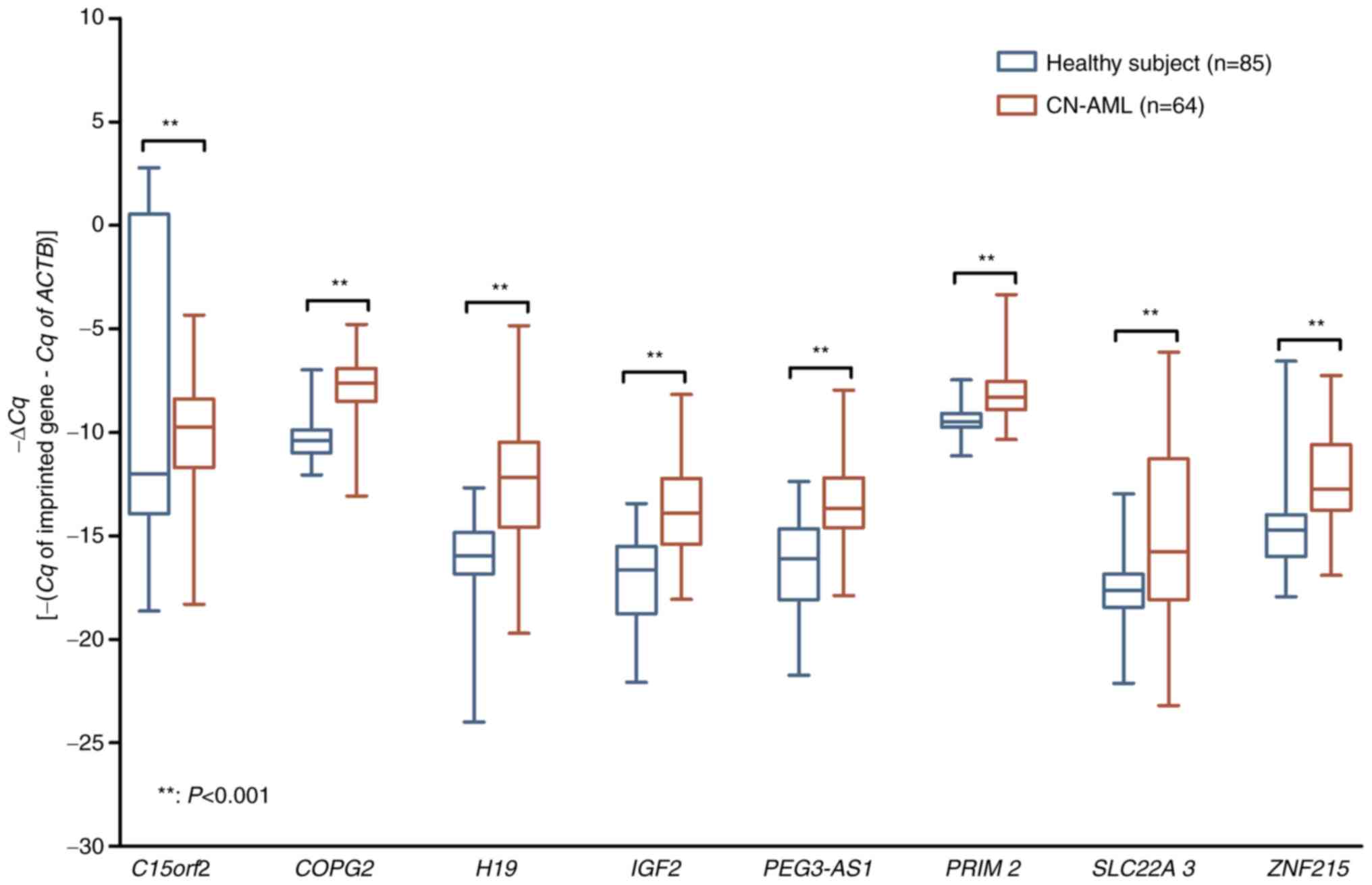
Subsequently, for further validation, the focus shifted to a larger
group comprising 64 patients with CN-AML and 85 healthy subjects.
From the 8 genes previously identified (excluding C15orf2),
the remaining 7 imprinted genes (COPG2, H19,
IGF2, PEG3-AS1, PRIM2, SLC22A3 and
ZNF215) that were upregulated in patients with CA-AML were
similarly observed to be significantly upregulated in patients with
CN-AML (P<0.001) when compared with healthy subjects (Fig. 2).
Correlation between expression of
imprinted genes and clinical parameters of patients
To determine a possible association, Pearson
correlation analysis was performed on the altered expression levels
of seven imprinted genes in patients with CN-AML. The results,
presented in Table III, revealed
several significant correlations. Specifically, the expression
levels of COPG2, PEG3AS, PRIM2 and
ZNF215 were positively correlated with the percentage of
blasts in PB (P<0.05). Finally, the expression level of
H19 was positively correlated with the level of lactic
dehydrogenase (P=0.040) in patients with CN-AML.
 | Table IIICorrelation between expression of the
7 imprinted genes and clinical parameters in patients with
cytogenetically normal-acute myeloid leukemia. |
Table III
Correlation between expression of the
7 imprinted genes and clinical parameters in patients with
cytogenetically normal-acute myeloid leukemia.
| Gene | Blasts in BM
(%) | Blasts in PB
(%) | WBC (/µl) | Hb (g/dl) | Platelets
(x1,000/µl) | LDH (U/l) |
|---|
| COPG2 | 0.173 (0.187) | 0.344 (0.011a) | 0.198 (0.130) | -0.026 (0.845) | 0.061 (0.642) | -0.098 (0.475) |
| H19 | 0.238 (0.067) | 0.141 (0.310) | 0.184 (0.160) | -0.235 (0.071) | 0.059 (0.655) | 0.277
(0.040a) |
| IGF2 | -0.146 (0.267) | -0.112 (0.418) | -0.113 (0.390) | 0.129 (0.326) | 0.159 (0.224) | 0.050 (0.715) |
| PEG3-AS | -0.077 (0.567) | 0.337
(0.017a) | 0.067 (0.625) | 0.077 (0.574) | 0.277
(0.039a) | 0.043 (0.759) |
| PRIM2 | -0.072 (0.597) | 0.525
(<0.001b) | 0.038 (0.781) | 0.175 (0.196) | 0.358
(0.007b) | -0.038 (0.788) |
| SLC22A3 | 0.081 (0.541) | 0.187 (0.181) | 0.107 (0.419) | -0.054 (0.687) | 0.165 (0.211) | -0.009 (0.949) |
| ZNF215 | -0.031 (0.820) | 0.281
(0.048a) | -0.017 (0.900) | 0.277
(0.039a) | 0.311
(0.020a) | 0.064 (0.646) |
Correlation, regression and survival
analysis of expression of imprinted genes in patients with
CN-AML
The correlation between the expression levels of the
seven altered imprinted genes and the survival rate of patients
with CN-AML was additionally explored. Pearson correlation analysis
was conducted, revealing that among these seven genes, solely the
expression level of H19 exhibited a negative correlation
with the survival rate of the patients (P=0.018) (Table IV). Furthermore, the binary
logistic analysis indicated that H19 could function as a
predictive factor for the survival of patients with CN-AML
[β=142.90, 95% Confidence interval (CI): 30.23-255.28,
P=0.014].
 | Table IVCorrelation between expression of the
7 imprinted genes and survival days in patients with
cytogenetically normal-acute myeloid leukemia. |
Table IV
Correlation between expression of the
7 imprinted genes and survival days in patients with
cytogenetically normal-acute myeloid leukemia.
| Imprinted gene | Survival days |
|---|
| COPG2 | -0.013 (0.920) |
| H19 | -0.295
(0.018a) |
| IGF2 | 0.070 (0.581) |
| PEG-3AS | -0.171 (0.192) |
| PRIM2 | -0.062 (0.637) |
| SLC22A3 | -0.074 (0.566) |
| ZNF215 | -0.126 (0.338) |
To assess the impact of H19 on survival, a
Cox proportional hazard model was utilized. The hazard ratio of
H19 for two-year survival was computed as 0.852 (95.0% CI:
0.774-0.938, P=0.001), and for five-year survival, it stood at
0.882 (95.0% CI: 0.808-0.963, P=0.005).
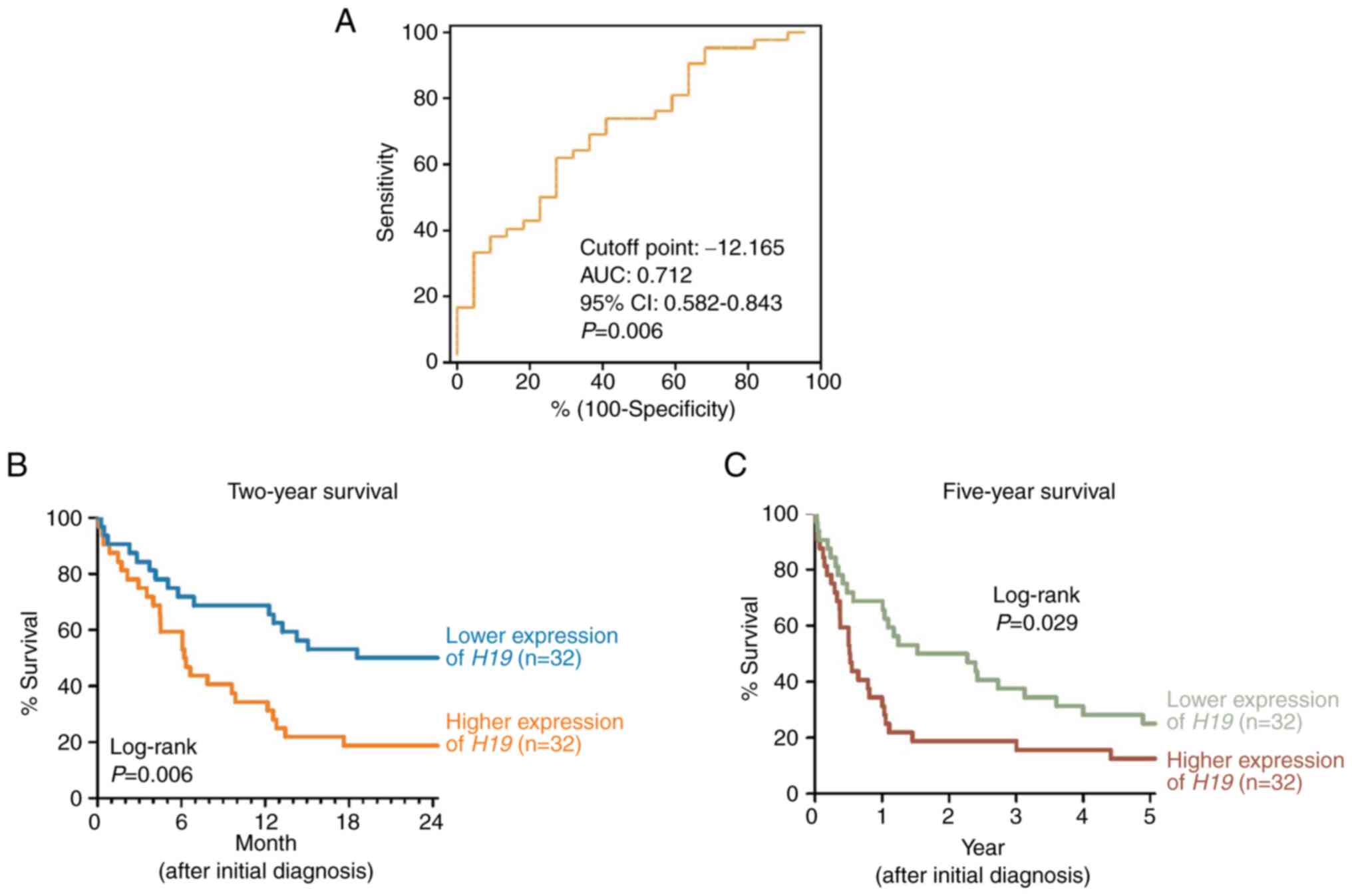
Additionally, ROC analysis was performed to evaluate
the area under the ROC curve (AUC) for two-year survival (AUC:
0.712, 95% CI: 0.582-0.843, P=0.006) (Fig. 3A). Using the cut-off point of
H19 expression (-12.165), Kaplan-Meier survival analysis
revealed that patients with lower H19 expression had a
higher two-year survival rate (log-rank P=0.006; Fig. 3B) and improved five-year survival
rate (log-rank P=0.029; Fig. 3C)
compared with those with higher H19 expression.
Discussion
CN-AML constitutes a significant proportion (40-50%)
of newly diagnosed AML cases and the identification of somatically
acquired genetic alterations in both CA-AML and CN-AML groups is
crucial for effective risk stratification. Despite presenting with
a normal karyotype, patients with CN-AML exhibit various mutations,
rendering them a heterogeneous group characterized by distinctive
clinical outcomes (22). In the
present study, by investigating the expression of imprinted genes,
the authors aimed to identify possible prognostic biomarkers that
could enhance treatment decisions for CN-AML.
Genomic imprinting plays a vital role in regulating
growth and development, with its disruption frequently observed in
the early stages of tumorigenesis (23,24).
Consistent with the previous findings in CA-AML (14), significantly higher expression was
observed in 7 out of the 16 imprinted genes analyzed in CN-AML.
These results underscore the significance of imprinting and its
dysregulation in AML, manifesting in cases with both normal and
abnormal karyotypes.
The novel finding of the present study is the
significant correlation between the H19 expression and the
survival of patients with CN-AML. H19, the first reported
human imprinted gene (25), has
been demonstrated to promote leukemogenesis across diverse types of
leukemia. In AML, increased H19 expression has been
associated with poor prognosis (10,14,25,26)
by inhibiting apoptosis through the targeting of miR-29a-3p
(26) and miR-19a/b (27). Furthermore, loss of imprinting in
the IGF2-H19 cluster has also been observed in a high
percentage of patients with MDS and AML (28). Elevated H19 expression has
been documented in acute lymphoblastic leukemia (29,30)
and chronic myeloid leukemia (31,32),
correlating with adverse outcomes in both cases. The oncogenic
properties of H19, including inhibition of apoptosis,
promotion of tumor cell proliferation, invasion, metastasis and
chemoresistance, have been demonstrated across multiple solid
tumors (33). Downstream targets
of H19 in cancer include Akt2, β-catenin,
FOXM1, RUNX1, STAT3 and miRNAs. Additionally,
H19 can be transmitted to cancer cells via exosomes to
promote tumor progression (34).
Thus, H19 emerges as a potential biomarker and therapeutic
target in both leukemia and solid tumors.
In addition to H19 and IGF2, the loss
of imprinting of COPG2, PEG3-AS, PRIM2,
SLC22A3 and ZNF215 genes in patients with CN-AML was
observed. These genes have been associated with various
malignancies and disorders, as discussed in the previous study
conducted by the authors (14).
The present study detected correlations between the expression of
COPG2, H19, PEG3-AS, PRIM2,
ZNF215 and clinical parameters in patients with CN-AML.
However, only H19 expression was negatively correlated with
patient survival. These results differ from the observations made
in patients with CA-AML, further supporting the critical role of
the loss of imprinting in the leukemogenesis of AML. The findings
of the present study also indicated that specific imprinted genes
are influenced under CA or CN conditions, leading to different
clinical outcomes.
Over the past decade, numerous gene mutations in AML
have been identified through genomics technologies (for example,
RUNX1, IDH1, IDH2, DNMT3A, WT1,
TET2, MLL, ASXL1, CBL, NRAS,
KRAS and TP53 genes) (5,35).
Patients with CN-AML, being the largest subgroup of AML, can be
further classified into molecular subgroups based on these
mutations. Targeted therapies are being developed to address these
genetic and epigenetic changes, including demethylating agents and
tyrosine kinase inhibitors (36).
Although mutations in several genes (ACEBP, DNMT3A,
FLT3-ITD, FLT3-TKD, IDH1, IDH2,
MLL and NPM1) in the patients of the present study
with CN-AML were analyzed, the presence or absence of these
mutations did not significantly contribute to the prediction of
patient survival (data not shown). Therefore, despite the inclusion
of H19 as a prognostic marker, the application of novel
biomarkers for treatment decisions remains limited. Future goals
include improving the specificity of diagnostic classification to
facilitate more efficacious targeted therapies and the development
of personalized treatment strategies.
Similar to the previous study of the authors on
CA-AML, the present study on CN-AML included certain limitations.
Firstly, the samples obtained from healthy subjects were PB rather
than BM. Secondly, the patient subgroups were relatively modest in
size, posing constraints on more rigorous correlation analysis.
Lastly, an inadequacy of post-treatment cases hindered the
validation of the relationship between imprinted gene expression
and disease status. Additionally, although novel and clinically
relevant findings were observed, the functions of the altered
imprinted genes in CN-AML were not examined. Studying H19
comes with its share of limitations and challenges. While it is
established that H19 plays crucial roles in both normal
development and disease, its functional complexity adds a layer of
difficulty to comprehending its mechanisms. Moreover, the functions
of H19 may vary among different cell types and tissues,
highlighting the need to consider cell type specificity.
Consequently, the effects observed in a specific context may not
universally apply to all cell types or biological circumstances.
H19 has been implicated in cancers. However, disease
progression can involve numerous factors. Isolating the specific
contribution of H19 among these factors can be intricate.
Furthermore, it shows promise as a potential biomarker or
therapeutic target in various diseases. Translating these findings
into clinical applications can be a complex process involving
validation, safety assessments and regulatory approvals.
Despite the extensive identification of cytogenetic
and molecular biomarkers for AML, their application for diagnosis,
prediction and treatment decisions remains limited. A recent study
designed diagnostic models for diverse cancer types using
measurements of elevated expression of imprinted genes
(GNAS, GRB10 and SNRPN genes) (37). In combination with other molecular
biomarkers such as gene mutations and expression of histone
modifying genes, the integration of a panel of biomarkers could
significantly enhance prediction sensitivity and specificity. In
pursuit of this goal, forthcoming research should focus on
elucidating the functions of altered imprinted genes in CA-AML and
CN-AML, establishing histone modifications and exploring their
clinical applications.
In conclusion, the present study revealed
upregulated expression of 7 out of the 16 examined imprinted genes
in BM samples from patients with CN-AML, indicating the involvement
of loss of imprinting in the leukemogenesis of CN-AML. Notably, a
significant negative correlation was observed between the
H19 expression and the survival rate of CN-AML patients,
highlighting its potential as a prognostic predictor for two- and
five-year survival. The upregulated H19 expression observed
in CN-AML implies its potential involvement in modulating cell
proliferation, apoptosis and differentiation. Additionally, its
interactions with other genes and regulatory factors may contribute
to the intricate molecular pathways underlying CN-AML development
and progression. By aligning the findings of the present study in
CN-AML with the previous study on CA-AML, a substantiated
connection between imprinted genes and AML has been established.
Furthermore, the results suggest that expression of different
imprinted genes are influenced by distinct cytogenetic conditions,
leading to divergent clinical outcomes. To gain a deeper
understanding of the roles these imprinted genes play in the
leukemogenesis of both CA-AML and CN-AML, further functional
studies are needed.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Chang Gung
Memorial Hospital (grant nos. CMRPG6J0251, CMRPD8E0171, CMRPD8J0011
and CMRPG6M0361). Further funding was provided by the National
Science and Technology Council of Taiwan (grant nos. NSTC
103-2314-B-037-066-MY2 and NSTC 104-2314-B-037-035).
Availability of data and materials
The datasets used and analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MYY conceptualized and supervised the study,
acquired funding, wrote the original draft, wrote, reviewed and
edited the manuscript. CMH conceptualized the study, acquired
funding, wrote, reviewed and edited the manuscript. PML conducted
investigation and developed methodology. CHY and MLH conducted
investigation and project administration. IYC curated and validated
data, performed formal analysis, and conducted software analysis.
SFL conceptualized the study, acquired funding, wrote, reviewed and
edited the manuscript. MYY and SFL confirm the authenticity of all
the raw data. All authors read and approved the final version of
the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Review Board of the Kaohsiung Medical University Hospital Ethical
Committee (approval no. KMU-IRB-20130129; Kaohsiung, Taiwan).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Pelcovits A and Niroula R: Acute myeloid
leukemia: A review. R I Med J (2013). 103:38–40. 2020.PubMed/NCBI
|
|
2
|
Department of Health Promotion
Administration, Ministry of Health and Welfare. Cancer Registry
Annual Report, Taiwan, 2016.
|
|
3
|
Wu SJ, Chiang CJ, Lin CT, Tien HF and Lai
MS: A nationwide population-based cross-sectional comparison of
hematological malignancies incidences between Taiwan and the United
States of America. Ann Hematol. 95:165–167. 2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Doulatov S, Notta F, Laurenti E and Dick
JE: Hematopoiesis: A human perspective. Cell Stem Cell. 10:120–136.
2012.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Marcucci G, Haferlach T and Döhner H:
Molecular genetics of adult acute myeloid leukemia: Prognostic and
therapeutic implications. J Clin Oncol. 29:475–486. 2011.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Čolović N, Denčić-Fekete M, Peruničić M
and Jurišić V: Clinical characteristics and treatment outcome of
hypocellular acute myeloid leukemia based on WHO classification.
Indian J Hematol Blood Transfus. 36:59–63. 2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Jurisić V, Pavlović S, Colović N,
Djordjevic V, Bunjevacki V, Janković G and Colović M: Single
institute study of FLT3 mutation in acute myeloid leukemia with
near tetraploidy in Serbia. J Genet. 88:149–152. 2009.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Rowe JM: The increasing genomic complexity
of acute myeloid leukemia. Best Pract Res Clin Haematol.
27:209–213. 2014.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Li S, Mason CE and Melnick A: Genetic and
epigenetic heterogeneity in acute myeloid leukemia. Curr Opin Genet
Dev. 36:100–106. 2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Zhang TJ, Zhou JD, Zhang W, Lin J, Ma JC,
Wen XM, Yuan Q, Li XX, Xu ZJ and Qian J: H19 overexpression
promotes leukemogenesis and predicts unfavorable prognosis in acute
myeloid leukemia. Clin Epigenetics. 10(47)2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Wrzeska M and Rejduch B: Genomic
imprinting in mammals. J Appl Genet. 45:427–433. 2004.PubMed/NCBI
|
|
12
|
Tucci V, Isles AR, Kelsey G and
Ferguson-Smith AC: Erice Imprinting Group. Genomic imprinting and
physiological processes in mammals. Cell. 176:952–965.
2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Ferguson-Smith AC and Bourc'his D: The
discovery and importance of genomic imprinting. Elife.
7(e42368)2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Yang MY, Lin PM, Yang CH, Hu ML, Chen IY,
Lin SF and Hsu CM: Loss of ZNF215 imprinting is associated with
poor five-year survival in patients with cytogenetically
abnormal-acute myeloid leukemia. Blood Cells Mol Dis.
90(102577)2021.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Sellers ZP, Bolkun L, Kloczko J,
Wojtaszewska ML, Lewandowski K, Moniuszko M, Ratajczak MZ and
Schneider G: Increased methylation upstream of the MEG3 promotor is
observed in acute myeloid leukemia patients with better overall
survival. Clin Epigenetics. 11(50)2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Yu Y, Kou D, Liu B, Huang Y, Li S, Qi Y,
Guo Y, Huang T, Qi X and Jia L: LncRNA MEG3 contributes to drug
resistance in acute myeloid leukemia by positively regulating ALG9
through sponging miR-155. Int J Lab Hematol. 42:464–472.
2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Shaffer LG, McGowan-Jordan J and Schmid M:
ISCN 2016: An International System for Human Cytogenetic
Nomenclature. Basel, Switzerland, Karger, 2013.
|
|
18
|
Campbell LJ and White JS: Cytogenetic
analysis in acute myeloid leukaemia. Methods Mol Biol. 730:63–77.
2011.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Morison IM, Paton CJ and Cleverley SD: The
imprinted gene and parent-of-origin effect database. Nucleic Acids
Res. 29:275–276. 2001.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Hsu CM, Lin PM, Lin HC, Lai CC, Yang CH,
Lin SF and Yang MY: Altered expression of imprinted genes in
squamous cell carcinoma of the head and neck. Anticancer Res.
36:2251–2258. 2016.PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Walker A and Marcucci G: Molecular
prognostic factors in cytogenetically normal acute myeloid
leukemia. Expert Rev Hematol. 5:547–558. 2012.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Sun H, Pan Y, He B, Deng Q, Li R, Xu Y,
Chen J, Gao T, Ying H, Wang F, et al: Gene therapy for human
colorectal cancer cell lines with recombinant adenovirus 5 based on
loss of the insulin-like growth factor 2 imprinting. Int J Oncol.
46:1759–1767. 2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Pan Y, He B, Li T, Zhu C, Zhang L, Wang B,
Xu Y, Qu L, Hoffman AR, Wang S and Hu J: Targeted tumor gene
therapy based on loss of IGF2 imprinting. Cancer Biol Ther.
10:290–298. 2010.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zhang Y and Tycko B: Monoallelic
expression of the human H19 gene. Nat Genet. 1:40–44.
1992.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zhao TT and Liu X: LncRNA-H19 inhibits
apoptosis of acute myeloid leukemia cells via targeting miR-29a-3p.
Eur Rev Med Pharmacol Sci. 23(3 Suppl):224–231. 2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zhao TF, Jia HZ, Zhang ZZ, Zhao XS, Zou
YF, Zhang W, Wan J and Chen XF: LncRNA H19 regulates ID2 expression
through competitive binding to hsa-miR-19a/b in acute myelocytic
leukemia. Mol Med Rep. 16:3687–3693. 2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Hofmann WK, Takeuchi S, Frantzen MA,
Hoelzer D and Koeffler HP: Loss of genomic imprinting of
insulin-like growth factor 2 is strongly associated with cellular
proliferation in normal hematopoietic cells. Exp Hematol.
30:318–323. 2002.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Mofidi M, Rahgozar S and Pouyanrad S:
Increased level of long non coding RNA H19 is correlated with the
downregulation of miR-326 and BCL-2 genes in pediatric acute
lymphoblastic leukemia, a possible hallmark for leukemogenesis. Mol
Biol Rep. 48:1531–1538. 2021.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Asadi M, Gholampour MA, Kompani F and
Alizadeh S: Expression of long non-coding RNA H19 in acute
lymphoblastic leukemia. Cell J. 25:1–10. 2023.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Zhou JD, Lin J, Zhang TJ, Ma JC, Li XX,
Wen XM, Guo H, Xu ZJ, Deng ZQ, Zhang W and Qian J:
Hypomethylation-mediated H19 overexpression increases the risk of
disease evolution through the association with BCR-ABL transcript
in chronic myeloid leukemia. J Cell Physiol. 233:2444–2450.
2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Yang J, Yin Z, Li Y, Liu Y, Huang G, Gu C
and Fei J: The identification of long non-coding RNA H19 target and
its function in chronic myeloid leukemia. Mol Ther Nucleic Acids.
19:1368–1378. 2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Liu FT, Pan H, Xia GF, Qiu C and Zhu ZM:
Prognostic and clinicopathological significance of long noncoding
RNA H19 overexpression in human solid tumors: Evidence from a
meta-analysis. Oncotarget. 7:83177–83186. 2016.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Hashemi M, Moosavi MS, Abed HM, Dehghani
M, Aalipour M, Heydari EA, Behroozaghdam M, Entezari M,
Salimimoghadam S, Gunduz ES, et al: Long non-coding RNA (lncRNA)
H19 in human cancer: From proliferation and metastasis to therapy.
Pharmacol Res. 184(106418)2022.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Colovic M, Jurisic V, Pavlovic S, Terzic T
and Colovic N: FLT3/D835 mutation and inversion of chromosome 16 in
leukemic transformation of myelofibrosis. Eur J Intern Med.
17:434–435. 2006.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Döhner H, Estey EH, Amadori S, Appelbaum
FR, Büchner T, Burnett AK, Dombret H, Fenaux P, Grimwade D, Larson
RA, et al: Diagnosis and management of acute myeloid leukemia in
adults: Recommendations from an international expert panel, on
behalf of the European LeukemiaNet. Blood. 115:453–474.
2010.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Shen R, Cheng T, Xu C, Yung RC, Bao J, Li
X, Yu H, Lu S, Xu H, Wu H, et al: Novel visualized quantitative
epigenetic imprinted gene biomarkers diagnose the malignancy of ten
cancer types. Clin Epigenetics. 12(71)2020.PubMed/NCBI View Article : Google Scholar
|