Introduction
Ovarian cancer accounts for the highest fatality
among gynecological malignancies, with an increasing number of
patients worldwide (1). Ninety
percent of ovarian cancers are epithelial cell types, encompassing
various histologic types with diverse molecular alterations,
clinical behaviors, and therapeutic outcomes. The remaining 10%
comprises non-epithelial ovarian cancers, such as exceedingly rare
tumors, primarily germ cell tumors, sex cord stromal tumors, and
small cell carcinomas (2). Ovarian
cancer accounts for 2.5% of all malignancies in women but 5% of all
cancer deaths because four out of five patients are diagnosed at an
advanced stage (3). Patients with
advanced-stage ovarian cancer have achieved the best outcomes via
complete resection of the diseased tissues and combination
chemotherapy (4). DNA damage
repair (DDR) defects are prevalent in various cancer types, and
these alterations can be strategically utilized for therapeutic
purposes. Epithelial ovarian cancer (EOC) stands out as one of the
tumor types with the highest percentage of hereditary cases.
Mutations occurring within DNA repair pathways elevate the risk of
developing resistance to chemotherapy. Considering the substantial
occurrence of homologous recombination deficiency in ovarian clear
cell carcinoma, it becomes susceptible to PARP inhibitor therapy.
Notably, the U.S. Food and Drug Administration (FDA) and/or the
European Medicines Agency (EMA) have approved olaparib, rucaparib,
and niraparib, among the PARP inhibitors, for use in EOC across
different treatment contexts (5).
However, the 5-year survival rate of advanced
ovarian cancer (stages III and IV) was only approximately 20% and
was considered to have the poorest prognosis among female genital
malignancies (6).
There are two recognized types of EOCs. Type I EOC
is believed to be relatively slow-growing and genetically stable,
often originating from identifiable precursor lesions like
endometriosis or borderline tumors with low malignant potential. In
contrast, type II EOC are proposed to be biologically aggressive
tumors from their outset, with a tendency for metastasis even from
small primary lesions (7).
Histopathologically, ovarian cancer comprises five major subgroups:
clear cell, endometrioid, mucinous, high-grade serous, and
low-grade serous. High-grade serous is the predominant subtype of
EOC, comprising approximately 75% of all cases; it follows the type
II pathway of development and is characterized by the presence of
p53 and BRCA mutations (7). Among
them, clear cell carcinoma is common in Japanese patients, but the
etiology is still unclear. This group of ovarian cancer is more
resistant to the standard platinum and paclitaxel chemotherapy than
the other advanced serous ones (8). Hence, the prognosis for advanced
ovarian clear cell cancer is poor compared to that of early-stage
ovarian cancer (6). This
necessitates the early diagnosis of ovarian clear cell carcinoma.
To date, there has been no specific test for the diagnosis of
early-stage ovarian cancer. The patients suspected of ovarian
cancer are conventionally tested using transvaginal sonography and
tumor markers, such as CA125. CA125 assessment is the standard
method for diagnosis, following response to treatment, for
predicting the prognosis of ovarian cancer like clear cell
carcinoma (9). The level of this
marker does not increase at an early stage and is not increased in
ovarian clear cell carcinoma, as reported previously (10). To date, no studies have screened
any candidates specifically for clear cell carcinoma. This economic
aspect is also evident in the research conducted on cost-effective
approaches for the early detection and prevention of ovarian cancer
in the past decade. Clearly, the cost of treatment per patient with
ovarian cancer remains the highest among all cancer types. For
instance, the average initial cost in the first year can reach
approximately USD 80,000, with the final year cost potentially
escalating to USD 100,000(11).
Extracellular RNA, including serum microRNAs
(miRNAs), has received much attention recently. miRNAs comprise
small non-coding RNAs of 20-25 nucleotides that regulate gene
expression in cells by suppressing the translation of the target
gene or by degrading the target mRNA (12). The miRNAs secreted from cells are
stably present in body fluids in extracellular vesicles containing
exosomes or bind to the proteins or lipids (13) playing an important role in
cell-cell communication (14).
Many studies have reported serum miRNAs as promising biomarkers for
various diseases because they reflect physiological and
pathological states (15-17).
There are some ovarian clear cell carcinoma-specific
miRNAs. Recently, Yokoi et al reported some miRNAs to be
specific to ovarian cancer (18).
On the other hand, histopathological examination revealed
endometriosis prevalent in middle-aged women to be associated with
the risk of ovarian cancer. Similarly, ovarian endometrioma is
associated with the risk of endometriosis-associated ovarian
cancer, especially clear cell carcinoma (19,20).
Clinically, it is difficult to distinguish between
ovarian endometriosis and clear cell carcinoma because of the
evident similarities on ultrasound and increasing CA125 levels in
both endometrioma and clear cell carcinoma. In this report, we
independently investigated and explored the specific miRNAs in
clear cell carcinoma compared to ovarian endometrioma and healthy
patients.
Materials and methods
Study design
The present study was approved by the internal
review boards of Tokyo Medical University (Tokyo, Japan; approval
no. 3769). Written informed consent was obtained from all the
patients before the collection of specimens, according to the
Declaration of Helsinki. The patient backgrounds were obtained
through interviews. The blood samples were collected before
operations, chemotherapy, and radiation therapy. The ovarian clear
cell carcinoma and endometriosis were diagnosed based on the
histological examinations.
A total of 64 patients participated in the research
conducted at the Tokyo Medical University Hospital from February
2010 to January 2019 and at the Jikei University School of Medicine
between August 2008 and November 2011. Twenty-nine patients had
ovarian clear cell carcinoma, 17 had endometriosis, and 18 were
healthy. The patients were diagnosed with ovarian clear cell
carcinoma, 6 with stage Ia, 5 with stage Ic1, 5 with stage IC2, 2
with stage IC3, 1 with stage IIa, 2 with stage IIb, 1 with stage
IIIa1, 4 with stage IIIb, 2 with stage IIIc, and 1 with stage
IVb.
Serum preparation and total RNA
extraction
The blood samples were collected from patients with
ovarian clear cell carcinoma and endometriosis, as well as from the
healthy controls. We measured the CA125 levels in patients with
endometriosis and ovarian cancer before surgery. The blood serum
was separated by centrifugation at 1,800 rpm for 10 min and stored
at -80˚C. The total RNA was extracted from the serum using the
miRNeasy Serum/Plasma Advanced Kit (Qiagen, Hilden, Germany)
according to the manufacturer's protocol.
Search for candidate miRNAs with
TaqMan Array Human microRNA Cards
We used TaqMan™ Array Human MicroRNA A+B
Cards Set v3.0 (Thermo Fisher Scientific, Inc.) to search for
candidate miRNAs in 20 samples (16 samples of ovarian clear cell
carcinoma and four healthy control samples). Using a volcano plot,
we identified target miRNAs with significantly different expression
levels in control and ovarian clear cell carcinoma patients. Then,
using an amplification plot, we narrowed down the miRNAs that were
amplified in almost all targets. Finally, we determined the target
miRNAs based on relative gene expression.
miRNA expression analysis by
quantitative polymerase chain reaction (qPCR) and receiver
operating characteristic curves
Four miRNAs (miR-146a-5p, miR-191-5p, miR-484, and
miR-574-3p) were analyzed by the TaqMan miRNA expression analysis
(Thermo Fisher Scientific, Inc.) and reverse transcription-qPCR
(RT-qPCR). The expression analyses were performed using the TaqMan
Advanced miRNA assays (Thermo Fisher Scientific, Inc.) for human
miR-146-5p (478399_mir), miR-191-5p (477952_mir), miR-484
(478308_mir), miR-574-3p (478163_mir), and miR-16 (477860_mir) as
an endogenous control (21). The
cDNA was synthesized using the TaqMan Advanced miRNA cDNA Synthesis
Kit (Thermo Fisher Scientific, Inc.).
qPCR was performed with RT primers using the
Universal Master Mix and specific miRNAs using the Applied
Biosystems StepOnePlus™ real-time PCR system (Thermo
Fisher Scientific, Inc.). The sequence detection was performed
according to the manufacturer's protocol.
The reaction mixtures were incubated at 95˚C for 2
min, followed by 40 cycles at 95˚C for 15 sec and 60˚C for 1 min.
The miRNA expression levels in the participants with ovarian clear
cell carcinoma and endometriosis compared to healthy controls were
calculated using the comparative 2-ΔΔCq method (22). Receiver operating characteristic
(ROC) curves were generated using the miR-146a miR-191 expression
profile. The graphical plots of the true and false positive rates
are shown. The area under the ROC curve represents the
identification accuracy.
Statistical analysis
The statistical analyses of the causal association
between the clinical background, the expression level of the
miRNAs, and the ROC curve analysis were performed using SPSS-27
software. The statistical significance was determined by the
Kruskal-Wallis test (between healthy controls, endometriosis, and
ovarian clear cell carcinoma) followed by Dunn's post hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
MiRNA 146a-5p and miRNA 191-5p
analyzed using MiRTarBase
Subsequent to the identification of differentially
expressed miRNAs, the predicted target genes for these altered
miRNAs were subjected to experimental validation using the
miRNA-target interaction database MiRTarBase (http://mirtarbase.cuhk.edu.cn/php/index.php) (23).
Results
Characteristics of the
participants
Of the 64 participants, 18 were healthy (control),
17 had endometriosis, and 29 had ovarian cancer. The median age of
healthy patients was 47.5 years (range 31-82 years), median age for
patients with endometriosis was 35 years (range 22-56 years), and
median age for patients with ovarian clear cell carcinoma was 53
years (range 31-81 years). Table I
shows the clinical characteristics and the values of CA125 in
patients with ovarian clear cell carcinoma (One patient did not
check CA125 before the operation).
 | Table ICharacteristics of patients with
ovarian clear cell carcinoma. |
Table I
Characteristics of patients with
ovarian clear cell carcinoma.
| Characteristics | Ovarian clear cell
carcinoma (n=29) |
|---|
| Age, years | |
|
Median | 53 |
|
Range | 31-81 |
| Clinical stage | |
|
IA | 6 |
|
IC1 | 5 |
|
IC2 | 5 |
|
IC3 | 2 |
|
IIA | 1 |
|
IIB | 2 |
|
IIIA1 | 1 |
|
IIIB | 4 |
|
IIIC | 2 |
|
IVB | 1 |
| Serum CA125
antigen, ng/ml | |
|
Median | 407 |
|
Range | 13-5,877 |
Table II shows the
clinical characteristics and the value of CA125 in patients with
endometriosis. CA125 varied differently in each endometriosis and
ovarian clear cell carcinoma patient.
 | Table IICharacteristics of the patients with
ovarian endometriosis. |
Table II
Characteristics of the patients with
ovarian endometriosis.
|
Characteristics | Endometriosis
(n=17) |
|---|
| Age, years | |
|
Median | 35 |
|
Range | 22-56 |
| BMI,
kg/m2 | |
|
Median | 21.6 |
|
Range | 17.7-34.7 |
| Tumor size, cm | |
|
Median | 62 |
|
Range | 30-150 |
| Serum CA125
antigen, ng/ml | |
|
Median | 55.1 |
|
Range | 10.7-555.6 |
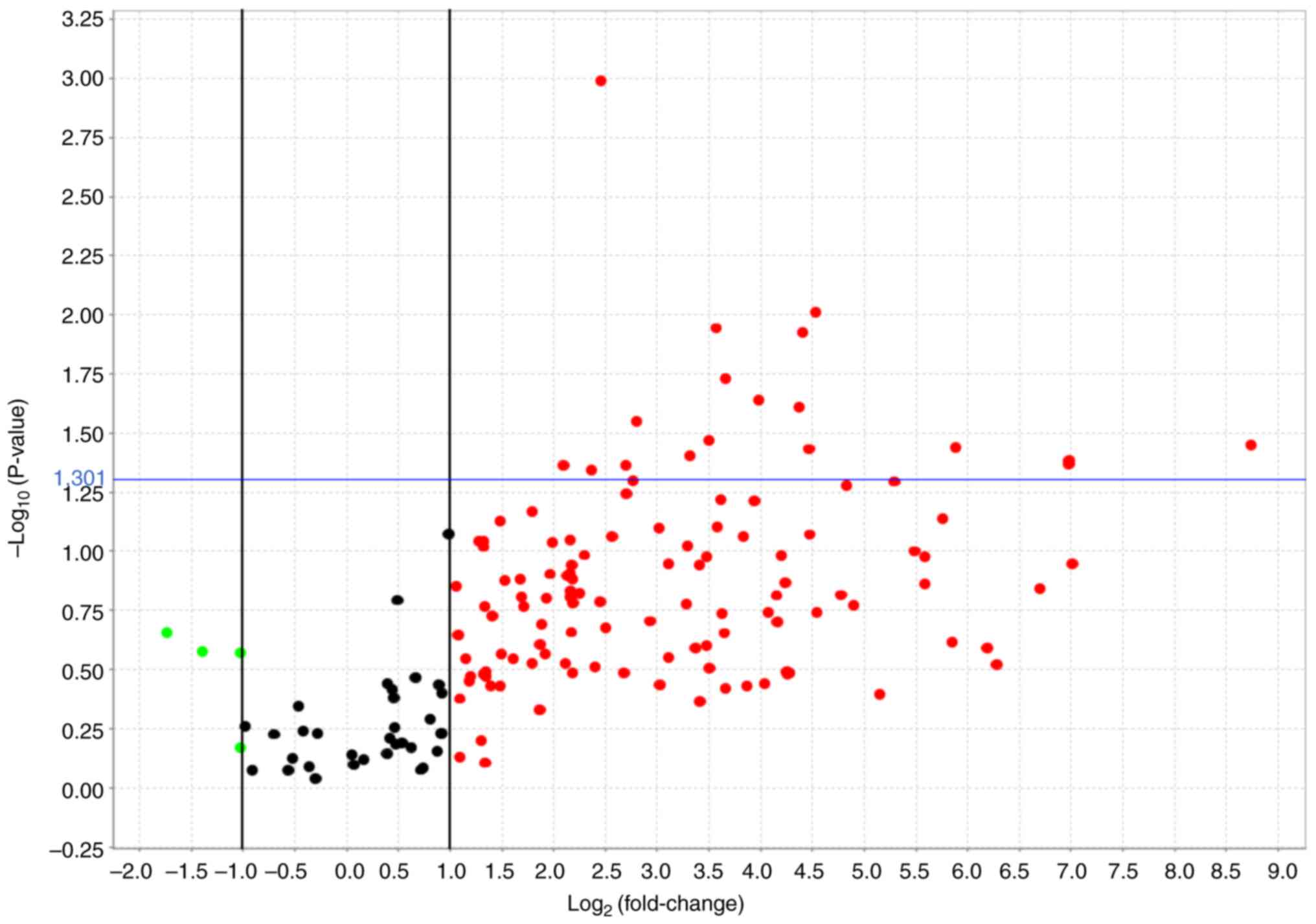
Identifying the candidate miRNAs
Based on the volcano plot, 18 miRNAs were identified
(Fig. 1). In the amplification
plot, 7 miRNAs (mir-146a-5p, mir-191-5p, mir-223-3p, mir-24-3p,
mir-320a-3p, mir-484, 574-3p) were confirmed as amplified. The
results of gene expression analysis showed that hsa-miR-191-5p and
hsa-miR-574-3p were more than 100-fold differentially expressed in
patients with carcinoma compared to the controls. Differential
expression was also observed for hsa-miR-146a-5p and hsa-miR-24-3p
(Fig. 2). Among the 16 samples of
ovarian clear cell carcinoma, 12 samples with similar miRNA
amplification were again analyzed using a volcano plot, and four
miRNAs (mir-146a-5p, mir-191-5p, mir-484, 574-3p) were listed.
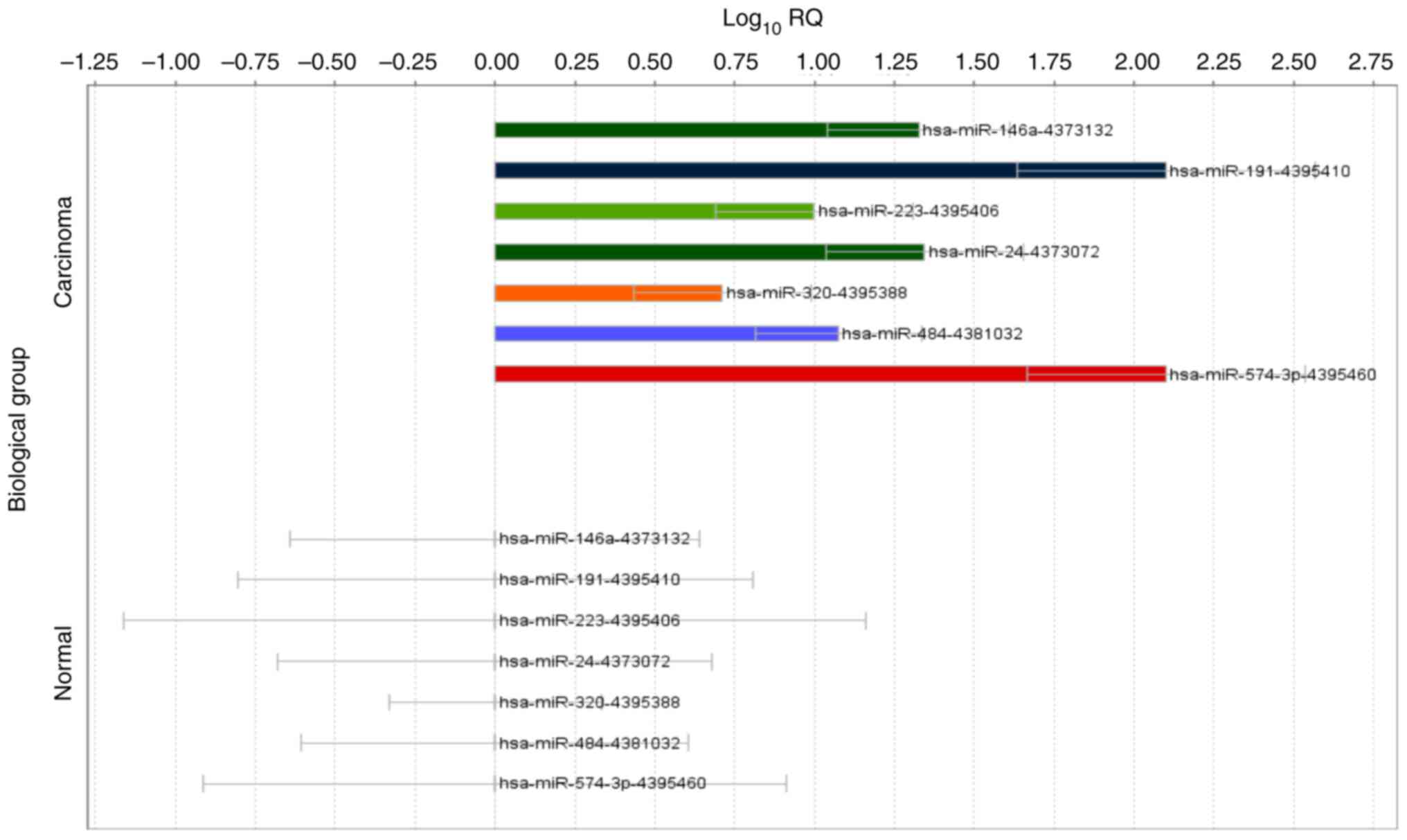
miRNA expression status in ovarian
clear cell carcinoma
The expression of miR-484 and miR-574-3p were not
different among the three groups. However, the miR-146a-5p and
miR-191-5p expression levels were significantly increased in the
serum samples from the participants with ovarian clear cell
carcinoma compared to the healthy controls but not in the
participants with endometriosis (P<0.05).
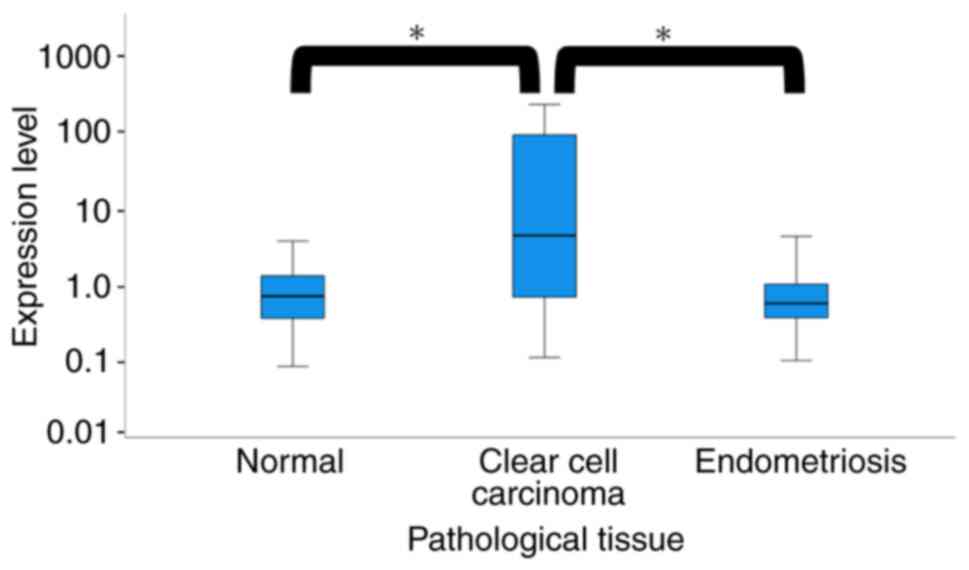
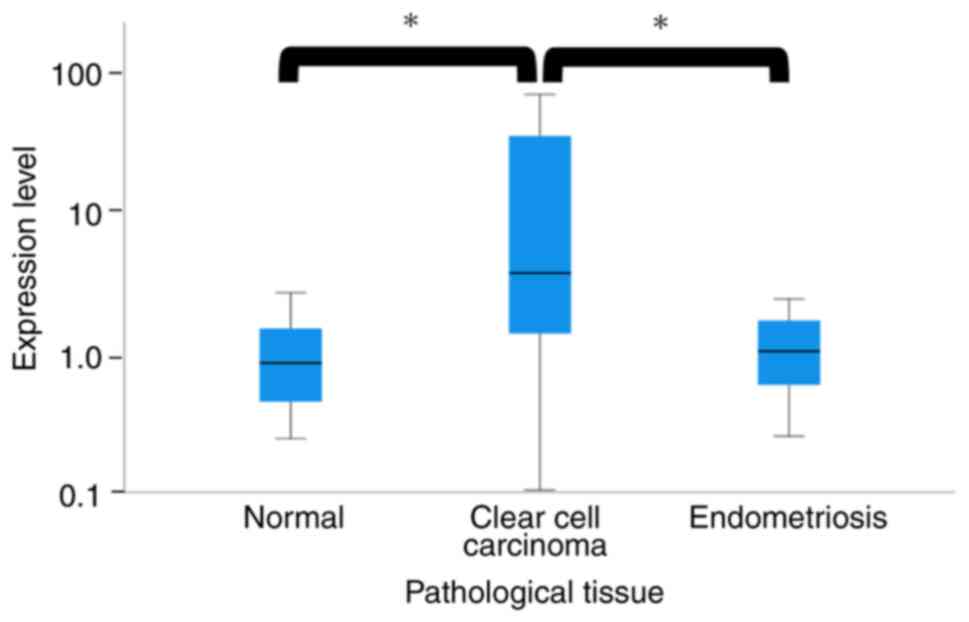
The median serum miR-146a-5p expression level was
0.72 in the healthy patients, 0.57 in the patients with
endometriosis, and 4.42 in patients with ovarian clear cell
carcinoma, respectively (P<0.01, Fig. 3; Kruskal-Wallis test).
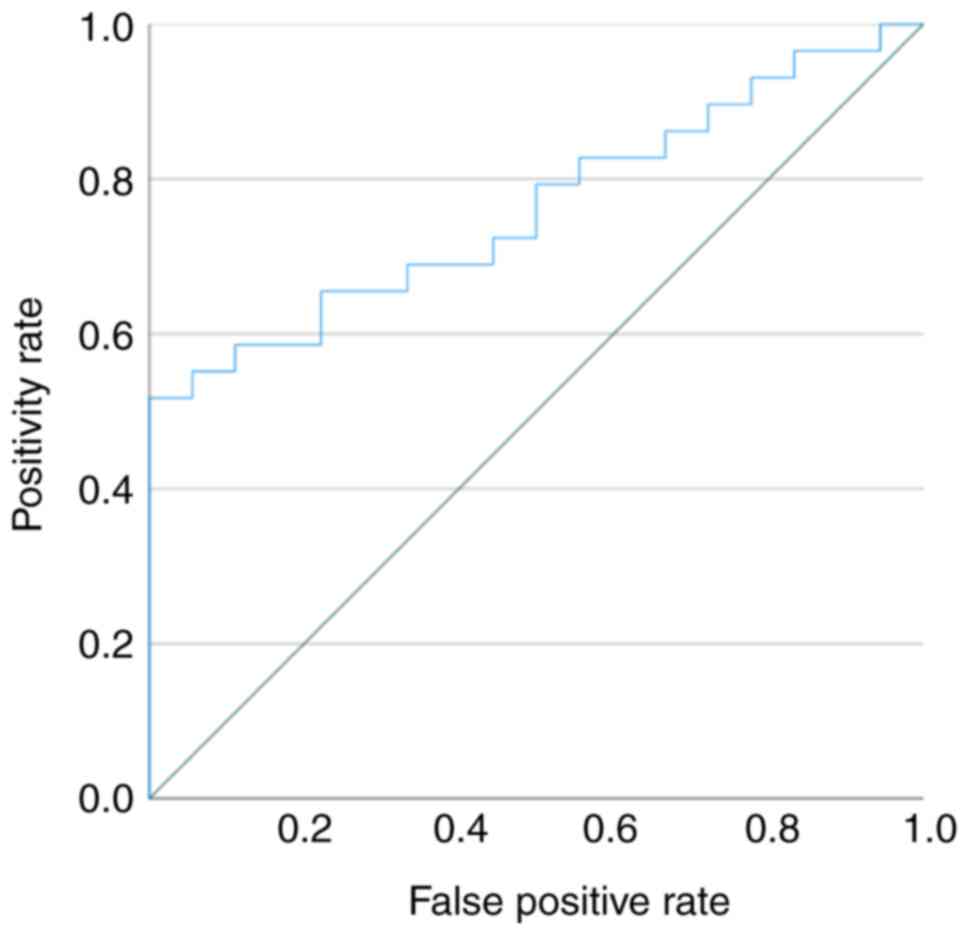
The ROC curve showed that the miR-146a-5p serum
levels may differentiate patients with ovarian clear cell carcinoma
from the healthy controls, and the ROC curve area was 0.762 (95%
confidence interval: 0.629-0.896; Fig.
4).
When the cut-off value was 0.652 (relative
expression value), miR-146a-5p was 79.3% sensitive to ovarian clear
cell carcinoma and 50.0% specific compared to the healthy controls.
In contrast, the median serum miR-191-5p expression level was 0.833
in the healthy participants, 1.00 in the patients with
endometriosis, and 3.58 in patients with ovarian clear cell
carcinoma (P<0.01, Fig. 5;
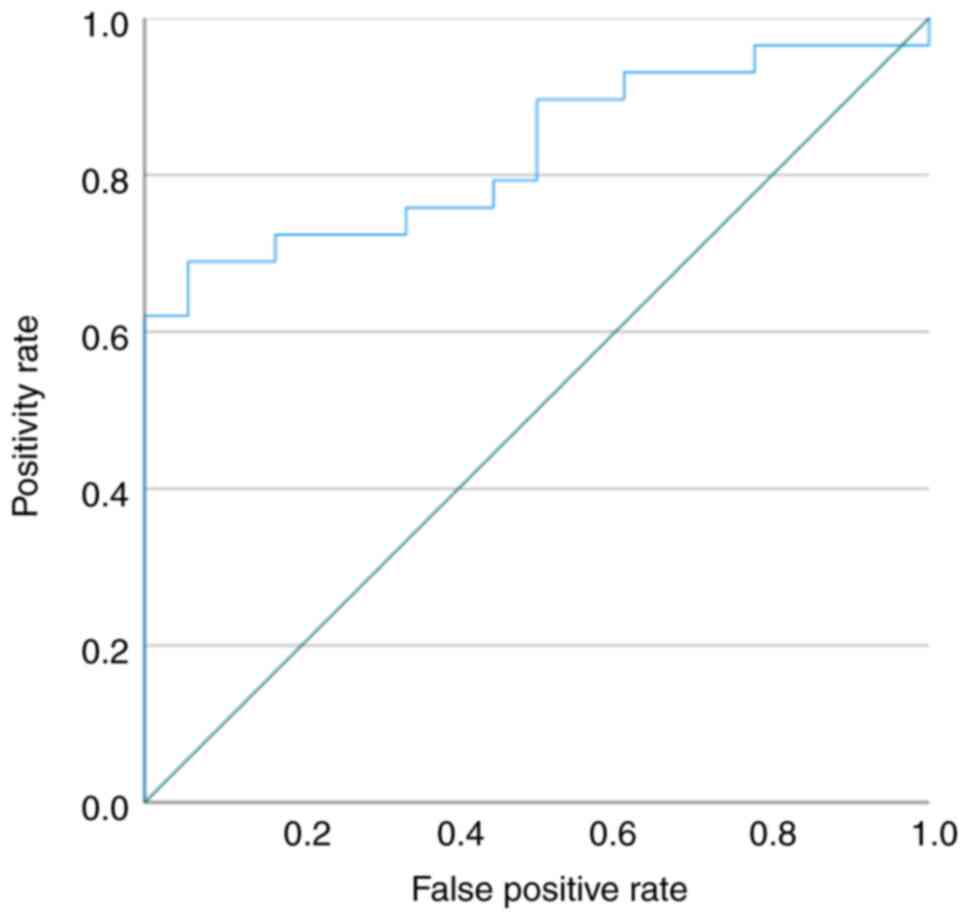
Kruskal-Wallis test). The ROC curve showed that the miR-191-5p
serum levels may differentiate patients with ovarian clear cell
carcinoma from healthy controls, and the ROC curve area was 0.830
(95% confidence interval: 0.714-0.945) (Fig. 6). When the cut-off value was 0.723
(relative expression value), the miR-191-5p was 89.7% sensitive to
ovarian clear cell carcinoma and 50.0% specific compared to the
healthy controls. Compared to the cancer stage, no difference was
observed in the expression of miR-146a-5p and miR-191-5p.
The MiRTarBase was used to identify the predicted
target genes of miR-146a-5p and miR-191-5p to determine their
biological significance. More than 50 target genes were extracted
by the MiRTarBase, and the target genes that showed strong evidence
are summarized in Fig. S1. The
CCND2 and NOTCH2 genes were the candidate targets of
miR-146a-5p and miR-191-5p.
Discussion
The early detection of cancer may contribute to
improved patient survival rates. BRCA1/2 germline mutations
represent the most potent identified genetic risk factors for EOC
and are detected in 6-15% of women diagnosed with EOC. Determining
the BRCA1/2 status can aid in providing patients with counseling
regarding their anticipated survival outcomes. It is noteworthy
that BRCA1/2 carriers with EOC tend to exhibit more favorable
responses to platinum-based chemotherapies compared to non-carriers
(24).
Biomarkers that can be used to detect cancer at an
early stage are important for the diagnosis and prognosis of
cancer. Targeted proteomics serves as a crucial technique for
validating and confirming discovered biomarkers. It works in
conjunction with untargeted proteomics to complete the biomarker
discovery and validation cycle. Additionally, peptidomics, a newly
established subdivision of proteomics, can provide insights into
novel biomarkers. Peptidomics focuses on studying peptides to
determine their specific forms, and like proteomics, it aids in
identifying new peptides present in tissues. Lastly, exosomes play
a vital role in intercellular communication and have emerged as
promising diagnostic and prognostic biomarkers for ovarian clear
cell carcinoma. They have the potential to transport certain
tumor-associated proteins (25).
This study aimed to investigate the novel miRNAs in
ovarian clear cell carcinoma. As previously reported, CA125 levels
were not distinguishable between endometriosis and clear cell
carcinoma (26). The expression
levels of miR-146a-5p and miR-191-5p were significantly elevated in
patients with ovarian clear cell carcinoma in the three groups. In
the ROC analysis, miR-146-5p and miR-191-5p revealed around 0.8
sensitivity for ovarian clear cell carcinoma. This indicated that
miR-146-5p and miR-191-5p were useful for exclusion diagnosis.
Using bioinformatics analysis, MiRTarBase showed
that the CCND2 and NOTCH2 genes were the candidate
targets of miR-146a-5p and miR-191-5p (Fig. S1). CCND2 belongs to the cyclin
family, which functions in cell cycle progression (27). CCND2 forms a complex with the
cyclin-dependent kinase CDK4 or CDK6 and functions as the
regulatory subunit of the complex, whose activity is required for
the cell cycle G1/S transition (28). As CCND2 shortens the G1 phase and
participates in cell progression, the CCND2 gene is suspected to be
involved in cancer cell growth (29).
Several studies have demonstrated that CCND2 is
associated with tumorigenesis (30). Chang et al (31) revealed that CCND2 is involved in
stimulating the proliferation, cell cycle progression, migration,
and invasion of ovarian cancer cells. NOTCH2 promotes cell
proliferation and epithelial-mesenchymal transition in the EOC cell
lines (32). MiR-146a-5p and
MiR-191-5p may be upregulated in patients with ovarian cancer to
inhibit the function of NOTCH2 and prevent the progression of
ovarian cancer.
Our results showed that CCND2 and NOTCH2 are the
candidates for both miRNAs. Thus, it was hypothesized that in
patients with ovarian clear cell carcinoma, miR-146a-5p and
miR-191-5p were upregulated to inhibit the function of CCND2 and
NOTCH2. A recent report has revealed that ovarian clear cell
carcinoma exhibits a unique genetic profile characterized by a
lower p53 mutation rate (25%) and a lower BRCA1/2 mutation rate
(6.3%) compared to high-grade serous ovarian cancer. However, it
demonstrates higher mutation rates in genes such as ARID1A,
PIK3CA, and PTEN. This highlights the genetic
differences between ovarian clear cell carcinoma and high-grade
serous ovarian cancer (33). A
major limitation of this study was the small sample size.
Our results showed that miR-146a-5p and miR-191-5p
may be useful as early and non-invasive diagnostic tools in the
search for ovarian clear cell cancer. These miRNAs can also
distinguish between ovarian clear cell carcinoma and ovarian
endometrioma.
Supplementary Material
Target genes of mir-146a and mir-191
MiRTarBase showed the candidate targets of miR-146a-5p and
miR-191-5p.
Acknowledgements
Not applicable.
Funding
Funding: This study was funded by JSPS KAKENHI (grant no.
15K10733).
Availability of data and materials
The microarray datasets generated and/or analyzed
during the current study are available in the Gene Expression
Omnibus repository, (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE239685).
All other data generated or analyzed during this study are included
in this published article.
Authors' contributions
ST, AO and HN confirm the authenticity of all the
raw data. ST, JK, HN, TU, SH, MK, OA and TM performed data
analysis. ST, JK, HN, AO and SH explained the present study to
patients and obtained informed consent. ST performed the
experiments. ST and JK wrote the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The patient data were used according to the ethical
principles of The Declaration of Helsinki. The study protocol was
approved by the internal review boards of Tokyo Medical University
(Tokyo, Japan; (approval no. 3769), and all patients provided
written informed consent before participation.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Webb PM and Jordan SJ: Epidemiology of
epithelial ovarian cancer. Best Pract Res Clin Obstet Gynaecol.
41:3–14. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Cheung A, Shah S, Parker J, Soor P, Limbu
A, Sheriff M and Boussios S: Non-epithelial ovarian cancers: How
much do we really know? Int J Environ Res Public Health.
19(1106)2022.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Howlader N, Noone AM, Krapcho M, Miller D,
Brest A, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR (eds), et
al: SEER cancer statistics review, 1975-2017, National Cancer
Institute. Bethesda, MD, 2020. https://seer.cancer.gov/csr/1975_2017.
|
|
4
|
Landrum LM, Java J, Mathews CA, Lanneau GS
Jr, Copeland LJ, Armstrong DK and Walker JL: Prognostic factors for
stage III epithelial ovarian cancer treated with intraperitoneal
chemotherapy: A gynecologic oncology group study. Gynecol Oncol.
130:12–18. 2013.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Boussios S, Rassy E, Moschetta M, Ghose A,
Adeleke S, Sanchez E, Sheriff M, Chargari C and Pavlidis N: BRCA
mutations in ovarian and prostate cancer: Bench to bedside. Cancers
(Basel). 14(3888)2022.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Trimble EL, Christan MC and Korsay C:
Surgical debulking plus paclitaxel-based adjuvant chemotherapy
superior to previous ovarian cancer therapies. Oncology.
13(1068)1999.
|
|
7
|
Pavlidis N, Rassy E, Vermorken JB, Assi T,
Kattan J, Boussios S and Smith-Gagen J: The outcome of patients
with serous papillary peritoneal cancer, fallopian tube cancer, and
epithelial ovarian cancer by treatment eras: 27 Years data from the
SEER registry. Cancer Epidemiol. 75(102045)2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
del Carmen MG, Birrer M and Schorge JO:
Clear cell carcinoma of the ovary: A review of the literature.
Gynecol Oncol. 126:481–490. 2012.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Duffy MJ, Bonfrer JM, Kulpa J, Rustin GJ,
Soletormos G, Torre GC, Tuxen MK and Zwirner M: CA125 in ovarian
cancer: European group on tumor markers guidelines for clinical
use. Int J Gynecol Cancer. 15:679–691. 2005.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Tian C, Markman M, Zaino R, Ozols RF,
McGuire WP, Muggia FM, Rose PG, Spriggs D and Armstrong DK: CA-125
change after chemotherapy in prediction of treatment outcome among
advanced mucinous and clear cell epithelial ovarian cancers: A
gynecologic oncology group study. Cancer. 115:1395–1403.
2009.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Ghose A, Bolina A, Mahajan I, Raza SA,
Clarke M, Pal A, Sanchez E, Rallis KS and Boussios S: Hereditary
ovarian cancer: Towards a cost-effective prevention strategy. Int J
Environ Res Public Health. 19(12057)2022.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Kim VN, Han J and Siomi MC: Biogenesis of
small RNAs in animals. Nat Rev Mol Cell Biol. 10:126–139.
2009.PubMed/NCBI View
Article : Google Scholar
|
|
13
|
Kosaka N, Yoshioka Y, Fujita Y and Ochiya
T: Versatile roles of extracellular vesicles in cancer. J Clin
Invest. 126:1163–1172. 2016.PubMed/NCBI View
Article : Google Scholar
|
|
14
|
Kim VN: MicroRNA biogenesis: Coordinated
cropping and dicing. Nat Rev Mol Cell Biol. 6:376–385.
2005.PubMed/NCBI View
Article : Google Scholar
|
|
15
|
Pritchard CC, Cheng HH and Tewari M:
MicroRNA profiling: approaches and considerations. Nat Rev Genet.
13:358–369. 2012.PubMed/NCBI View
Article : Google Scholar
|
|
16
|
Cortez MA, Bueso-Ramos C, Ferdin J,
Lopez-Berestein G, Sood AK and Calin GA: MicroRNAs in body
fluids-the mix of hormones and biomarkers. Nat Rev Clin Oncol.
8:467–477. 2011.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Nagamitsu Y, Nishi H, Sasaki T, Takaesu Y,
Terauchi F and Isaka K: Profiling analysis of circulating microRNA
expression in cervical cancer. Mol Clin Oncol. 5:189–194.
2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Yokoi A, Matsuzaki J, Yamamoto Y, Yoneoka
Y, Takahashi K, Shimizu H, Uehara T, Ishikawa M, Ikeda SI, Sonoda
T, et al: Integrated extracellular microRNA profiling for ovarian
cancer screening. Nat Commun. 9(4319)2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Gurung A, Hung T, Morin J and Gilks CB:
Molecular abnormalities in ovarian carcinoma: Clinical,
morphological and therapeutic correlates. Histopathology. 62:59–70.
2013.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Grandi G, Toss A, Cortesi L, Botticelli L,
Volpe A and Cagnacci A: The association between endometriomas and
ovarian cancer: Preventive effect of inhibiting ovulation and
menstruation during reproductive life. Biomed Res Int.
2015(751571)2015.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Schrauder MG, Strick R, Schulz-Wendtland
R, Strissel PL, Kahmann L, Loehberg CR, Lux MP, Jud SM, Hartmann A,
Hein A, et al: Circulating micro-RNAs as potential blood-based
markers for early stage breast cancer detection. PLoS One.
7(e29770)2012.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Ohyashiki K, Umezu T, Yoshizawa SI, Ito Y,
Ohyashiki M, Kawashima H, Tanaka M, Kuroda M and Ohyashiki JH:
Clinical impact of down-regulated plasma miR-92a levels in
non-Hodgkin's lymphoma. PLoS One. 6(e16408)2011.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Huang HY, Lin YC, Li J, Huang KY, Shrestha
S, Hong HC, Tang Y, Chen YG, Jin CN, Yu Y, et al: miRTarBase 2020:
Updates to the experimentally validated microRNA-target interaction
database. Nucleic Acids Res. 48 (D1):D148–D154. 2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Shah S, Cheung A, Kutka M, Sheriff M and
Boussios S: Epithelial ovarian cancer: Providing evidence of
predisposition genes. Int J Environ Res Public Health.
19(8113)2022.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Ghose A, Gullapalli SVN, Chohan N, Bolina
A, Moschetta M, Rassy E and Boussios S: Applications of proteomics
in ovarian cancer: Dawn of a new era. Proteomes.
10(16)2022.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Taniguchi F: New knowledge and insights
about the malignant transformation of endometriosis. J Obstet
Gynaecol Res. 43:1093–1100. 2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Hua M, Qin Y, Sheng M, Cui X, Chen W,
Zhong J, Yan J and Chen Y: miR-145 suppresses ovarian cancer
progression via modulation of cell growth and invasion by targeting
CCND2 and E2F3. Mol Med Rep. 19:3575–3583. 2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Kato JY and Sherr CJ: Inhibition of
granulocyte differentiation by G1 cyclins D2 and D3 but not D1.
Proc Natl Acad Sci USA. 90:11513–11517. 1993.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Song H, Hogdall E, Ramus SJ, Dicioccio RA,
Hogdall C, Quaye L, McGuire V, Whittemore AS, Shah M, Greenberg D,
et al: Effects of common germ-line genetic variation in cell cycle
genes on ovarian cancer survival. Clin Cancer Res. 14:1090–1095.
2008.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Zhu H, Dougherty U, Robinson V, Mustafi R,
Pekow J, Kupfer S, Li YC, Hart J, Goss K, Fichera A, et al: EGFR
signals downregulate tumor suppressors miR-143 and miR-145 in
Western diet-promoted murine colon cancer: Role of G1 regulators.
Mol Cancer Res. 9:960–975. 2011.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Chang L, Guo R, Yuan Z, Shi H and Zhang D:
LncRNA HOTAIR regulates CCND1 and CCND2 expression by sponging
miR-206 in ovarian cancer. Cell Physiol Biochem. 49:1289–1303.
2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Lu S, Liu W, Shi H and Zhou H: Exosomal
miR-34b inhibits proliferation and the epithelial-mesenchymal
transition by targeting Notch2 in ovarian cancer. Oncol Lett.
20:2721–2728. 2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Revythis A, Limbu A, Mikropoulos C, Ghose
A, Sanchez E, Sheriff M and Boussios S: Recent insights into PARP
and immuno-checkpoint inhibitors in epithelial ovarian cancer. Int
J Environ Res Public Health. 19(8577)2022.PubMed/NCBI View Article : Google Scholar
|