Introduction
Lung cancer remains the main cause of cancer-related
deaths worldwide (1). Cases of
non-small cell lung cancer (NSCLC) in young patients (≤50 years
old) represent a small percentage of the total cases, and indeed
this disease typically affects older individuals (>50 years
old), and the incidence rate among elderly patients is increasing
(2). Previous studies have compared
young and aged NSCLC patients, using the range of 40 to 50 years to
define the young group (2–8); however, to date, it is not clear if lung
cancer, particularly adenocarcinoma, in young patients may have
distinct clinicopathological features. In the present paper, liver
kinase B1 (LKB1) and its downstream signalling pathways were
investigated as a therapeutic target in lung adenocarcinoma, a
subtype of NSCLC (9), and compared
between different age groups. LKB1, also known as
serine/threonine kinase 11, is a tumor suppressor gene involved in
cellular responses including growth, polarity and metabolism
(10). LKB1 is a master kinase,
controlling 14 substrates involved in the translation of several
cell growth regulators (11).
LKB1 was been initially identified as the tumor suppressor
responsible for Peutz-Jeghers syndrome, an inherited cancer
predisposition (12). Several
sporadic tumors exhibited LKB1 promoter hypermethylation,
reduced LKB1 expression and somatic LKB1 mutations,
indicating a role of the loss of LKB1 in cancer development and
progression, potentially with additional oncogenic factors
(13). LKB1 may also be
repressed as a result of post-transcriptional regulation by
microRNAs (miRNA/miRs) (10), which
generally serve to repress mRNA translation or promote mRNA
degradation via partial complementary binding to the 3′
untranslated region (3′-UTR) of target mRNAs (14).
The role of LKB1 in NSCLC has previously been
analysed; however, study is made difficult by the fact that the
LKB1 pathway involves multiple substrates that act on metabolism,
apoptosis and the tumor microenvironment. Carretero et al
(15) reported that NSCLC cells with
loss of LKB1 exhibited higher nuclear expression of catenin β-1
(CTNNB1); LKB1, in fact, appears to suppress the Wnt/CTNNB1
pathway, inhibiting the expression of downstream genes, including
cyclin D1 (CCND1) and survivin. Additionally, lysyl
oxidase (LOX) has been reported to be negatively regulated by LKB1
in lung cancer, and yes-associated protein 1 (YAP1) has been
reported to be activated in lung adenocarcinoma as a result of a
lack of LKB1 (16). In the present
study, the mRNA expression of LKB1, CCND1, CTNNB1, LOX, YAP1,
survivin and 15 miRNAs involved in the LKB1 pathway was
investigated using NanoString technology. LKB1 loss has been
reported to be more common within KRAS-mutant lung
adenocarcinomas (17), and therefore,
KRAS mutations were also investigated.
The results presented provide indication that
LKB1 pathway genes, with the involvement of miRNA
regulation, may have a role in lung adenocarcinoma progression,
representing novel potential targets for lung cancer therapy.
Patients and methods
Patients
A total of 88 lung adenocarcinoma patients were
retrospectively selected from patients who were operated between
January 2003 and December 2013 at the Unit of Thoracic Surgery of
the University Hospital of Pisa (Pisa, Italy). Histological
diagnoses were made according to the World Health Organization
classification (9,18,19). Data
on clinicopathological characteristics were collected for all
patients (Table I). The study was
conducted in accordance with the 1964 Helsinki declaration and the
ethical standards of Institutional Research Committee of the
University of Pisa, for the collection of lung cancer samples
following surgery and the related informed consensus for molecular
analysis. Patients ≤50 years old were defined as the younger group
(n=44), and patients >50 years old as the older group
(n=44).
 | Table I.Lung adenocarcinoma patient
characteristics in the two groups. |
Table I.
Lung adenocarcinoma patient
characteristics in the two groups.
| Variable | Young | Old | Total | P-value |
|---|
| Sample size | 44 | 44 | 88 | – |
| Age, years | 46.3±3.9 | 71.5±5.2 | 58.9±13.4 | – |
| Sex | 0.02 |
|
|
|
|
Male | 23 | 33 | 56 |
|
|
Female | 21 | 11 | 32 |
|
| Adenocarcinoma
prevalent pattern |
|
|
| 0.0004 |
|
Lepidic | 13 | 16 | 29 |
|
|
Solid | 9 | 17 | 26 |
|
|
Acinar | 19 | 3 | 22 |
|
|
Papillar | 3 | 8 | 11 |
|
| Tumour grading |
|
|
| 0.07 |
| G1 | 3 | 0 | 3 |
|
| G2 | 30 | 28 | 58 |
|
| G3 | 11 | 16 | 27 |
|
| Stage |
|
|
| 0.79 |
| IA | 8 | 9 | 17 |
|
| IB | 14 | 9 | 23 |
|
|
IIA | 6 | 7 | 13 |
|
|
IIB | 4 | 5 | 9 |
|
|
IIIA | 10 | 13 | 23 |
|
|
IIIB | 1 | 0 | 1 |
|
| IV | 1 | 1 | 2 |
|
| KRAS status |
|
|
| 0.07 |
|
Wild-type | 31 | 23 | 54 |
|
|
Mutant | 13 | 21 | 34 |
|
Target prediction
A total of 15 miRNAs (miRs −93, −96, −34a, −34c,
−214, −33a, −30b, −145, −182, −30c, −183, −29b, −29c, −153 and
−138) were selected based on their involvement in the LKB1 pathway
(20–29). Alignment of miRNAs with target genes
(LKB1, CCND1, CTNNB1, LOX, YAP1 and survivin) was
predicted by using the microRNA target prediction program
(http://www.microrna.org).
DNA and RNA isolation
DNA, RNA and miRNAs were isolated from 5–10 μm
sections of formalin-fixed (buffered formalin, for 24–48 h at room
temperature) and paraffin-embedded (FFPE) resected tissues,
performed immediately following surgery, following manual tumor
macrodissection using a QIAamp DNA Mini kit (Qiagen GmbH, Hilden,
Germany) and a miRNeasy FFPE kit (Qiagen GmbH), respectively,
according to the manufacturer's instructions. The quality and
concentration were assessed using a NanoDrop spectrophotometer
(Thermo Fisher Scientific, Inc., Waltham, MA, USA).
NanoString nCounter® assay,
data normalization and analysis
Expression of the 6 targeted mRNAs and 15 selected
miRNAs was measured using the NanoString nCounter Technology
system, according to the manufacturer's protocol (NanoString
Technologies, Inc., Seattle, WA, USA). The nCounter measures the
total counts of mRNAs/miRNAs through a multiplex hybridization
assay, followed by scanning and digital readout of fluorescent
probes in a high-throughput manner (30). The nCounter custom code set used in
the current study included the 6 targeted genes and 3 housekeeping
genes as references (tubulin β, hypoxanthine
phosphoribosyltransferase and phosphoglycerate kinase 1). Raw
NanoString counts for each gene were subjected to technical and
biological normalization using the positive control probe sets and
three reference genes, respectively. miRNAs were normalized using a
scaling factor based on the 5 miRNAs with the lowest variability
coefficients according to the manufacturer's protocol.
KRAS mutation analysis
Pyrosequencing analysis was performed using the
PyroMark Q96 ID platform (Diatech Pharmacogenetics SRL, Jesi,
Italy) following the manufacturer's instructions in order to
determine KRAS status. Codons 12, 13, 61, 117 and 146 of the KRAS
gene were analysed.
Statistical analysis
The normalized RNA hybridization data, presented as
direct counts of digital reports, were analysed by using nSolver
2.5 analysis software (NanoString Technologies, Inc.). The
χ2 test was applied to analyze lung adenocarcinoma
patient characteristics in the two age groups and to determine the
association between LKB1 and miR-93 expression. Differential
gene expression was determined by applying the non-parametric t
test and analysis of variance. Survival analyses were performed
using the Kaplan-Meier method with the log-rank test and the Cox
proportional hazard model. Statistical analyses were performed
using JMP 10 software (SAS Institute, Inc., Cary, NC, USA), and
two-tailed P<0.05 was considered to indicate statistical
significance.
Results
Comparison of patient characteristics
between the age groups
The current study was conducted in 88 patients with
lung adenocarcinoma (56 males and 32 females). Patients ≤50 years
old were defined as the younger group, and patients >50 years
old as the older group. Among all patients, different histological
subtypes of adenocarcinoma were identified; the most common
histological subtypes were lepidic (29/88, 33.0%), solid (26/88,
29.5%), acinar (22/88, 25.0%), and papillar (11/88, 12.5%). The
median age at diagnosis was 54.5 years old (range, 30–81 years;
mean, 58.9±13.4 years). Regarding grading, 3 tumors (3.4%) were G1,
whereas 58 (65.9%) and 27 (30.7%) were G2 and G3, respectively. The
adenocarcinomas were all invasive, and stages 17 IA, 23 IB, 13 IIA,
9 IIB, 23 IIIA, 1 IIIB, and 2 IV were identified, according to the
World Health Organization classification (9,18,19). The follow-up data, disease-free
interval (DFI) and overall survival (OS) were available for all
patients and were last updated on March 2015. Disease progression
(recurrence/metastasis) was observed in 50 patients (56.8%; data
not shown). Regarding smoking habits, there were 17 non-smokers, 16
former smokers and 23 current smokers; for 33 patients, the smoking
data were not available. Regarding clinicopathological
characteristics, overall gender distribution (P=0.02) as well as
histological subtype distribution (P=0.0004) were significantly
different between the younger and older cases (Table I).
Comparison of survival between the age
groups
The median DFI and OS times of total patients (n=88)
were 21 months (range, 0–148) and 31.5 months (range, 3–148),
respectively.
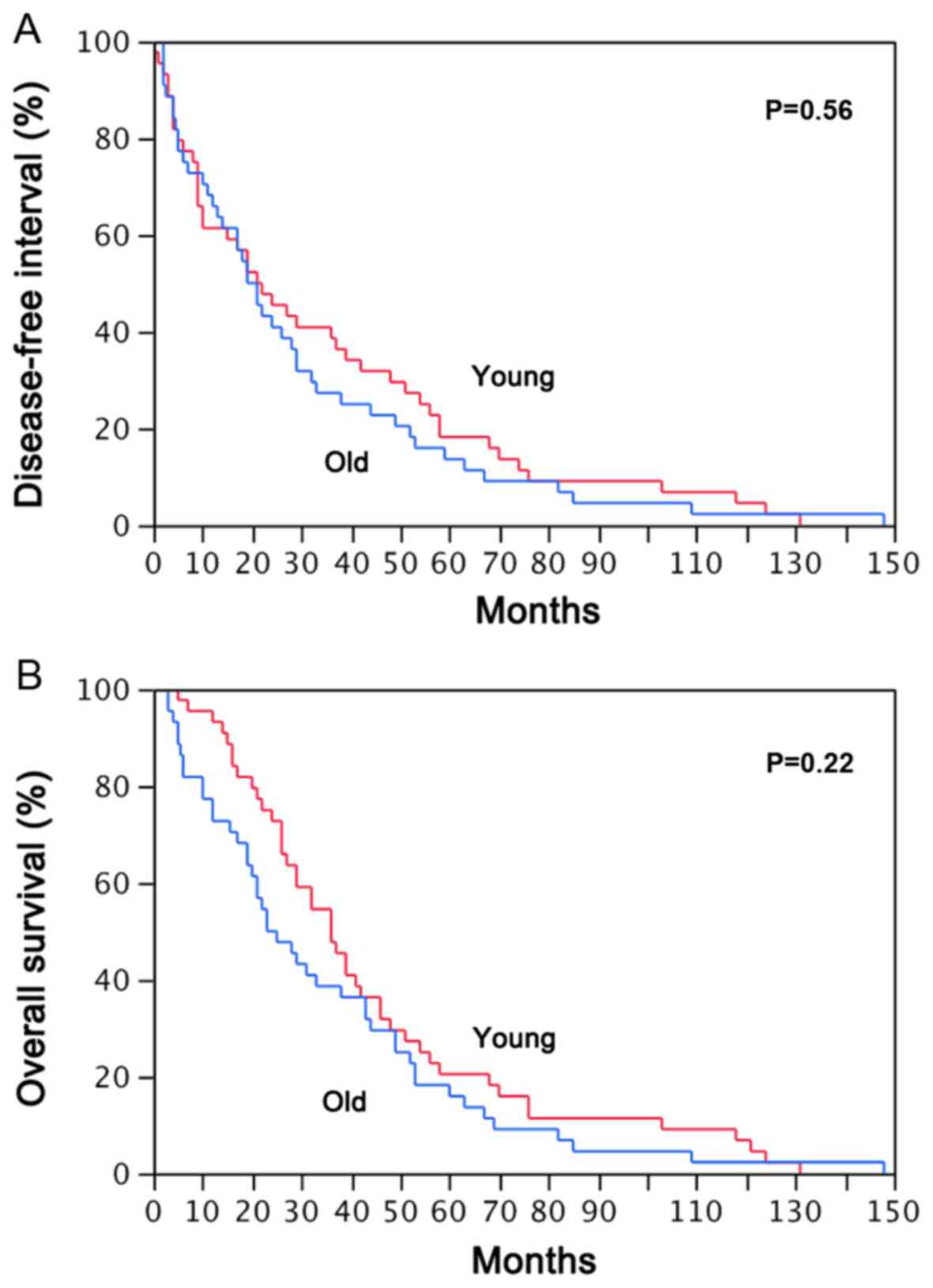
The median DFI times were 22 months in younger
patients and 21 months in older patients; the median OS times were
36 and 23 months in the younger and older groups, respectively.
Survival analysis using the Kaplan-Meier method with DFI and OS as
endpoints did not identify a significant difference between younger
patients and their elderly counterparts (Fig. 1).
LKB1 pathway expression
To investigate the role of the LKB1 pathway in lung
adenocarcinoma, first the levels of LKB1 were screened. As
presented in Table II, there was no
significant association of LKB1 expression with patient age,
prevalent adenocarcinoma pattern or tumor grading; however, the
data indicated a significant association of low LKB1
expression with male gender (P=0.03) and overall clinical stage
(P=0.01) as well as a trend with the solid variant. Next it was
investigated whether the expression of downstream genes and their
regulation was directly affected by LKB1 levels. Low
LKB1 expression was associated with low expression of
CCND1 (P<0.0001), CTNNB1 (P<0.0001) and
YAP1 (P=0.0024; data not shown), suggesting regulation by
LKB1. To test if LOX, one of the other LKB1
network partners, was an important downstream mediator of lung
adenocarcinoma progression, its expression level was also assessed
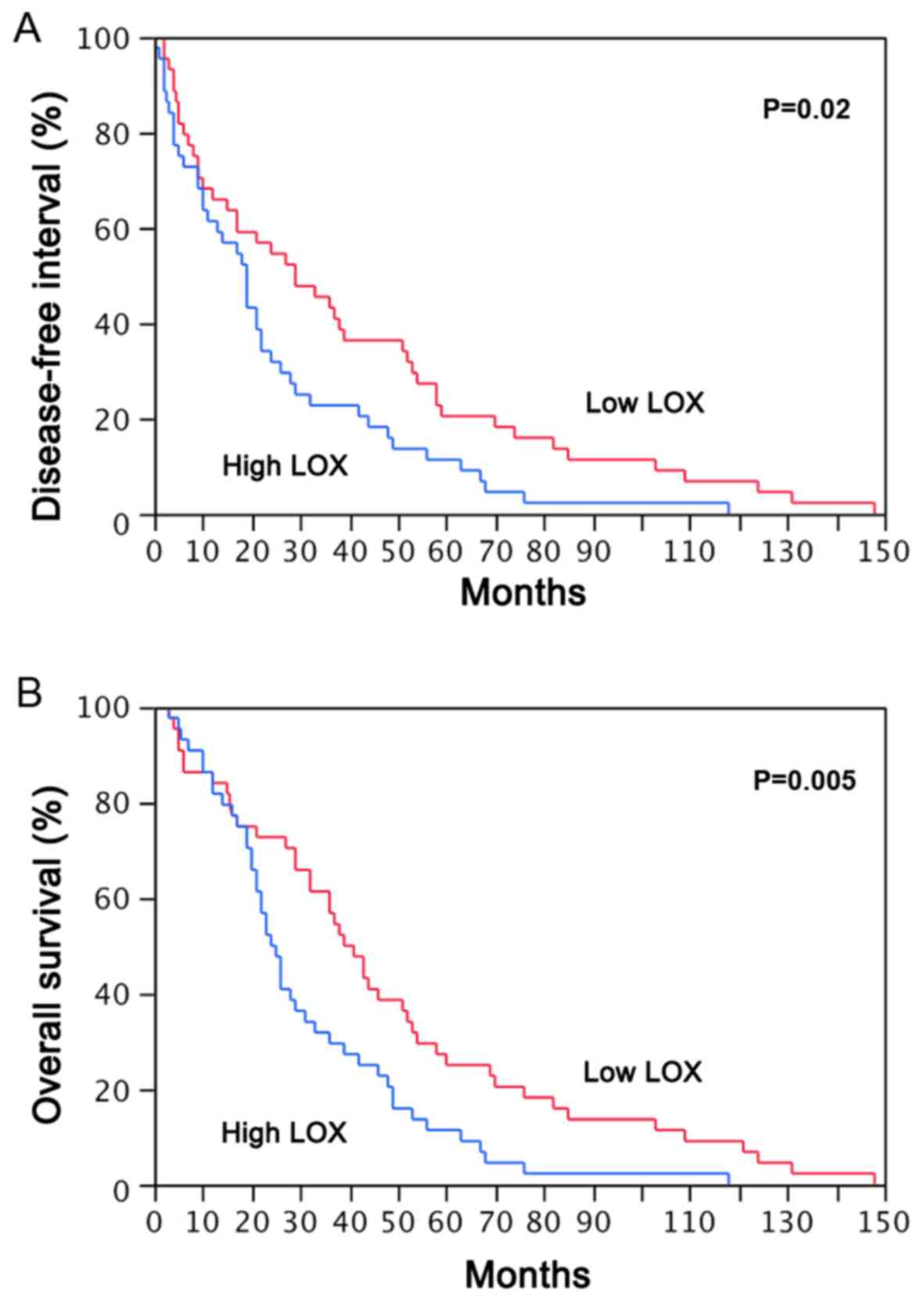
in the present adenocarcinoma series. Notably, LOX levels in
adenocarcinoma patients were significantly associated with
histological subtype (P=0.03), as well as with stage (P<0.0001;
Table III) distribution, indicating
that LOX activation may promote tumor progression. The
samples were divided into high and low LOX expression groups
based on the median LOX fold-change value (median
fold-change, 132; mean, 141.82±88.9). Survival analysis was
performed using the Kaplan-Meier method with the post-operative DFI
and OS times as endpoints in order to evaluate the association
between LOX expression and prognosis in the adenocarcinoma
patients. It was identified that adenocarcinoma cases with high
LOX mRNA expression were associated with significantly
shorter median DFI and OS times compared with the cases with low
LOX expression (P=0.02 and P=0.005, respectively; Fig. 2).
 | Table II.LKB1 expression in the 88 lung
adenocarcinoma patients. |
Table II.
LKB1 expression in the 88 lung
adenocarcinoma patients.
| Variable | LKB1 expression,
mean ± SD | P-value |
|---|
| Age group |
| 0.21 |
| Younger
(≤50 years) | 173.9±13.4 |
|
| Older
(>50 years) | 149.8±13.4 |
|
| Sex |
| 0.03 |
|
Male | 146.9±11.6 |
|
|
Female | 188.0±15.4 |
|
| Prevalent
adenocarcinoma pattern |
| 0.24 |
|
Lepidic | 184.9±16.4 |
|
|
Solid | 137.9±17.3 |
|
|
Acinar | 154.0±18.8 |
|
|
Papillar | 173.1±26.7 |
|
| Tumour grading |
| 0.14 |
| G1 | 244.8±50.9 |
|
| G2 | 166.1±11.5 |
|
| G3 | 143.4±16.9 |
|
| Stage |
| 0.01 |
| I | 192.2±13.5 |
|
| II | 129.8±18.2 |
|
|
III–IV | 142.3±16.7 |
|
 | Table III.LOX expression in the 88 lung
adenocarcinoma patients. |
Table III.
LOX expression in the 88 lung
adenocarcinoma patients.
| Variable | LOX expression,
mean ± SD | P-value |
|---|
| Age group |
| 0.7 |
| Younger
(≤50 years) | 137.6±13.4 |
|
| Older
(>50 years) | 146.1±13.4 |
|
| Sex | 0.05 |
|
|
Male | 156.1±11.6 |
|
|
Female | 116.9±15.4 |
|
| Prevalent
adenocarcinoma pattern |
| 0.03 |
|
Lepidic | 131.0±16.0 |
|
|
Solid | 162.9±17.0 |
|
|
Acinar | 159.6±18.4 |
|
|
Papillar | 85.0±26.1 |
|
| Tumour grading |
| 0.52 |
| G1 | 147.5±51.5 |
|
| G2 | 134.0±11.7 |
|
| G3 | 157.9±17.1 |
|
| Stage |
| <0.0001 |
| I | 102.9±11.9 |
|
| II | 126.4±16.1 |
|
|
III–IV | 214.8±14.8 |
|
Taken together, these data support LKB1
signalling as a key pathway in lung adenocarcinoma, with a
potential relevant role for LOX.
miRNA selection and expression
The microRNA target prediction program (http://www.microrna.org) was used to identify putative
miRNA-mRNA interactions in the LKB1 pathway, and subsequently, the
impact of the 15 selected miRNAs (miRs −93, −96, −34a, −34c, −214,
−33a, −30b, −145, −182, −30c, −183, −29b, −29c, −153 and −138) on
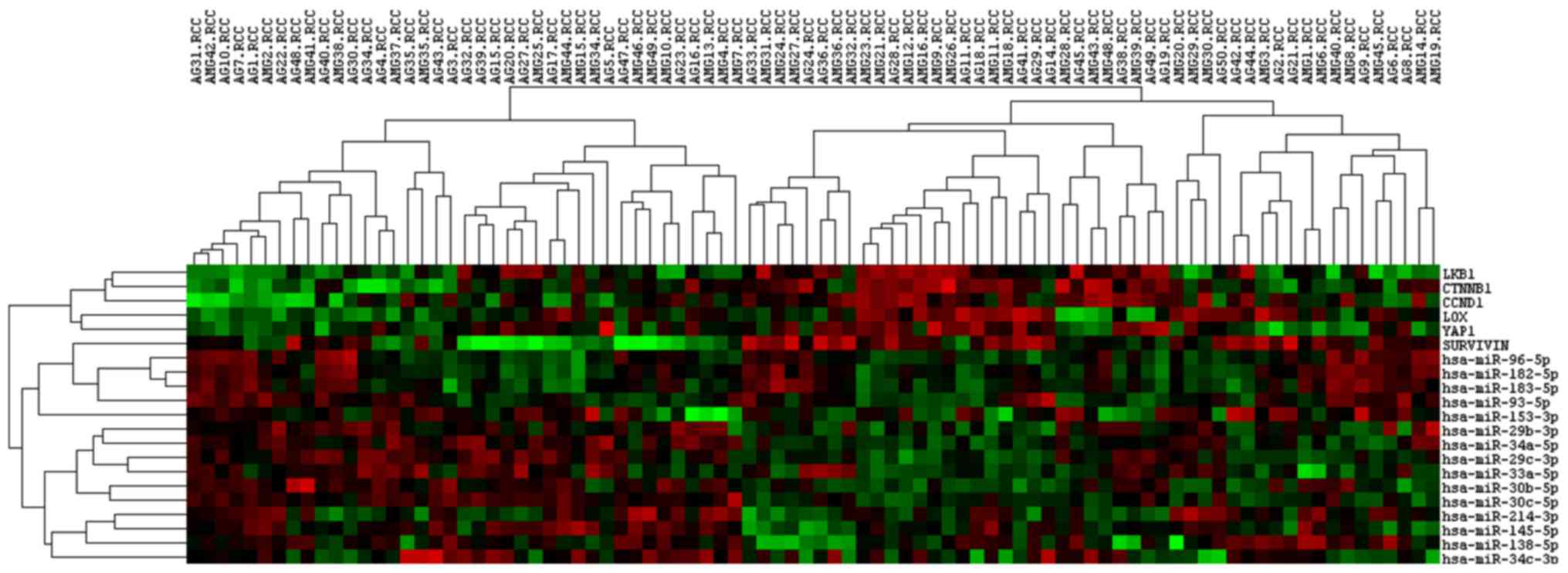
the expression of target genes. Fig.
3 depicts the expression profiles across all the adenocarcinoma
samples in the two age groups. The younger and older patients
shared similar gene expression profiles; any differentially
expressed genes in the LKB1 pathway were associated with
modulated miRNA expression, suggesting that they were the gene
targets of the modulated miRNAs. The seed region of miR-93 was
predicted to bind to one site, position 287, in the human
LKB1 3′-UTR (Fig. 4). The
samples were divided into high and low miR-93 expression groups
based on the median miR-93 fold-change value (median fold-change,
3.933; mean, 4.845.91±3.056); LKB1 expression was reduced in
samples with high miR-93 expression (χ2 test; P=0.0007;
data not shown), indicating that this endogenous miRNA may suppress
LKB1. No statistically significant association was
identified between CCND1 and miR-93 expression, even though
decreased CCND1 levels tended to be observed in cases with
high miR-93 expression (P=0.11) and an alignment at position 1013
of the 3′-UTR was identified (Fig.
4). LOX downregulation was observed in the lung
adenocarcinoma specimens with high miR-30b expression (P=0.04), and
a direct interaction of this miRNA and the 3′-UTR of the LOX
gene at position 594 was identified (Fig.
4). Other potential miRNA binding sites within the LOX
3′-UTR were identified (for miRs −145, −182, −30c, −183, −29b, −29c
and −153); potential miRNA binding sites were also identified in
YAP1 (miR-138) and survivin (miR-214-3p), but neither
of them were indicated to influence the expression levels of their
mRNA targets (data not shown).
KRAS mutation analysis
Pyrosequencing analysis was performed to identify
mutational hot-spots of the KRAS gene. Among the 88 tumor
specimens, 34 tumors (38%) were determined to have point mutations,
13/44 among younger and 21/44 among older patients. However, no
significant difference was identified in KRAS mutation distribution
between the two age groups (P=0.07; Table
I). Concerning the KRAS mutation type, the G12C substitution
was present in 15 samples (7 younger and 8 older patients); the
G12V point mutation in 11 samples (3 younger and 8 older patients);
the G12D mutation in 3 samples (1 younger and 2 older patients);
the G12A mutation in 2 samples from the older cohort; the G12S and
G13D in 1 sample each among younger patients, and Q61L mutations in
only 1 older patient (Table IV).
 | Table IV.KRAS mutation distribution in the two
age groups of lung adenocarcinoma patients. |
Table IV.
KRAS mutation distribution in the two
age groups of lung adenocarcinoma patients.
|
| Cases, n |
|---|
|
|
|
|---|
| KRAS status | Younger (n=44) | Older (n=44) | Total (n=88) |
|---|
| Wild-type | 31 | 23 | 54 |
| Mutated | 13 | 21 | 34 |
|
G12C | 7 | 8 | 15 |
|
G12V | 3 | 8 | 11 |
|
G12D | 1 | 2 | 3 |
|
G12A | 0 | 2 | 2 |
|
G12S | 1 | 0 | 1 |
|
G13D | 1 | 0 | 1 |
|
Q61L | 0 | 1 | 1 |
Discussion
Lung cancer remains the main cause of cancer-related
mortality worldwide, and the age at diagnosis has been decreasing
in recent years (2,4). Younger patients with lung cancer appear
to exhibit distinct clinicopathological features: They are more
commonly non-smokers and female, and present a prevalence for
adenocarcinoma and advanced disease; however, there is controversy
regarding the outcome as it has been reported as improved by
certain studies (4,31) and unaffected by others (31–34). Data
on the management of adenocarcinoma in the elderly are insufficient
(35–39), and thus, whether young lung cancer
patients have specific molecular and pathologic features or
different survival outcomes remain unclear. In the present study,
44 lung adenocarcinoma patients ≤50 years old were selected as the
younger group and 44 cases >50 years as the older group; a
predominance of females was identified in the younger group, and
the acinar pattern was most prevalent, which was in accordance with
previous studies (4,40–42). There
were no significant differences in survival, in terms of DFI and
OS, in young lung adenocarcinoma patients compared with the older
age group. At present, there is no general consensus on the
influence of age on survival, and this issue is open to question.
Any discrepancies between reports may be due to the limited number
of studies and to the specific cut-off age used to separate younger
from older patients; the current study used 50 years old as the
cut-off value, according to several previous reports (2,43,44).
An aim of this retrospective study was also to focus
on the expression pattern of LKB1 and its downstream
signalling pathways, in order to evaluate their associations with
clinicopathological features and prognoses in lung adenocarcinoma,
comparing younger patients with their elderly counterparts.
Although there are currently no drugs in routine
clinical use that specifically target LKB1, there is a growing
number of approaches that may differentially benefit patients with
a dysregulated LKB1 pathway (45–49). A
critical role for LKB1 has been suggested in catenin-beta1
signalling in lung cancer through its modulation of CCND1
and survivin gene expression (50). Additionally, LOX has been
reported to be efficiently suppressed by LKB1, and
YAP1 has been reported to be initially activated by
LKB1 loss in lung ADC.
A mRNA panel was customized of the 6 abovementioned
genes and 15 miRNAs involved in the LKB1 pathway, and expression
profiling was performed with NanoString technology, a recently
developed platform that can make direct multiplexed measurements of
expression through digital readouts of the abundance of mRNA/miRNA
transcripts (51). Several studies
have indicated that the NanoString technique is a reliable and
flexible method for the assessment of gene expression in limited
FFPE tissues, and it has exhibited similar results using
fresh-frozen tissue (52–54). This field appears to be of importance,
since FFPE represents most of the specimens collected in routine
diagnostic pathology, and it will allow for the connection of
clinical follow-up data to large numbers of patients (55); therefore, the NanoString methodology
may be readily adapted clinically as a highly reproducible
alternative to quantitative polymerase chain reaction and
sequencing methods (56).
LKB1 has different molecular targets, and
thus, screening of the LKB1 signalling pathway appears a
necessary step to explain its suppressive function in cancer cell
biology. No differences in LKB1 levels and KRAS
mutation rates were identified between young and older patients,
possibly due to the biological heterogeneity of KRAS-mutant lung
adenocarcinomas, to the relatively small group size, and/or to
other underlying molecular differences such as epidermal growth
factor mutations or anaplastic lymphoma kinase translocations
(18). Notably, low LKB1 expression
was apparent in the solid histological subtype, and high expression
in females and early clinical stage, which suggest an important
role for LKB1 in inhibiting the growth of lung cancer cells,
considering that the solid subtype, male gender and advanced stages
are reportedly survival disadvantages in lung adenocarcinoma
(57). However, it remains unclear
how LKB1 loss contributes to lung carcinogenesis, and the
post-transcriptional regulation of LKB1 may play a central
role. The current results also predicted that miR-93 may be able to
downregulate LKB1 and CCND1, consequently leading to
the loss of LKB1-dependent tumor suppression; such is in
agreement with previous studies reporting that high levels of
miR-93 (58) and low levels of
LKB1 (59) as well as
CCND1 (60) were correlated
with poor survival among lung cancer patients.
The current study also attempted to elucidate the
involvement of LOX. It is reported that the tumor
microenvironment serves a critical role in tumorigenesis (61), and that LOX, due to its
influence on the cellular microenvironment, may be a target for
cancer therapy (62,63). The current data further indicated that
aberrant LOX expression was involved in lung carcinogenesis
and cancer progression, revealing that LOX levels in
adenocarcinoma patients were significantly associated with overall
stage distribution and poor prognosis regardless of age at
diagnosis. Furthermore, the findings indicated that the observed
positive prognostic effect of LOX was associated, at least
in part, to miR-30b regulation, confirming the conclusions by Zhong
et al (64) regarding a
central role of this miRNA in NSCLC suppression. However, the
impact of LOX remains incompletely clear; it is possible
that there exists multiple forms of LOX proteins (65), and it is not known whether the
signalling components downstream of LKB1, including
LOX, may be involved in an LKB1-independent manner;
further studies will be required to reach a conclusive point.
The finding that LKB1 and LOX may be
repressed by specific miRNAs establishes a regulatory link within
the LKB1 tumor suppressor pathway; miRNA-dependent
post-transcriptional regulation of LKB1 may be an
alternative to inactivating LKB1 mutations, which are rare
among sporadic tumors (10).
Additional study on LKB1 pathways and other components of
the LKB1 complex may expand knowledge regarding tumor
metabolism and growth potential in lung adenocarcinoma.
Acknowledgements
Preliminary results of the present study were
presented as a poster at the International Association for the
Study of Lung Cancer 17th World Conference on Lung Cancer, December
4–7, 2012 in Vienna, Austria and published as abstract no.
P1.02-078 in Journal of Thoracic Oncology 12 (Suppl 1): 2017.
Funding
Not applicable.
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LB, MG and GF conceived and designed the
experiments. MG performed the experiments. LB wrote the manuscript.
GF was responsible for lung cancer diagnosis and contributed to
conception and design of the study. FM and ML performed lung
surgery and follow-up and contributed to conception and design of
the study.
Ethics approval and consent to
participate
The current study was conducted in accordance with
the ethical standards of Institutional Research Committee of the
University of Pisa (Pisa, Italy) and with the 1964 Helsinki
declaration; informed consent for the tissue collection and
molecular analysis was collected by the oncologist upon each
patient's first visit from January 2003 to December 2013 to the
University Hospital of Pisa (Pisa, Italy).
Patient consent for publication
Informed consent collected from all patients
permitted the use of their tissues for research purposes.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ramalingam S, Pawlish K, Gadgeel S, Demers
R and Kalemkerian GP: Lung cancer in young patients: Analysis of a
Surveillance, Epidemiology, and End Results database. J Clin Oncol.
16:651–657. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mauri D, Pentheroudakis G, Bafaloukos D,
Pectasides D, Samantas E, Efstathiou E, Kalofonos HP, Syrigos K,
Klouvas G, Papakostas P, et al: Hellenic Coopeprative Oncologic
Group (HeCOG): Non-small cell lung cancer in the young: A
retrospective analysis of diagnosis, management and outcome data.
Anticancer Res. 26(4B): 3175–3181. 2006.PubMed/NCBI
|
|
4
|
Subramanian J, Morgensztern D, Goodgame B,
Baggstrom MQ, Gao F, Piccirillo J and Govindan R: Distinctive
characteristics of non-small cell lung cancer (NSCLC) in the young:
A surveillance, epidemiology, and end results (SEER) analysis. J
Thorac Oncol. 5:23–28. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang J, Chen SF, Zhen Y, Xiang J, Wu C,
Bao P, Luketich J, Hu H, Zhou X, Zhang J, et al: Multicenter
analysis of lung cancer patients younger than 45 years in Shanghai.
Cancer. 116:3656–3662. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Inoue M, Okumura M, Sawabata N, Miyaoka E,
Asamura H, Yoshino I, Tada H, Fujii Y, Nakanishi Y, Eguchi K, et
al: Clinicopathological characteristics and surgical results of
lung cancer patients aged up to 50 years: The Japanese Lung Cancer
Registry Study 2004. Lung Cancer. 83:246–251. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lara MS, Brunson A, Wun T, Tomlinson B, Qi
L, Cress R, Gandara DR and Kelly K: Predictors of survival for
younger patients less than 50 years of age with non-small cell lung
cancer (NSCLC): A California Cancer Registry analysis. Lung Cancer.
85:264–269. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rich AL, Khakwani A, Free CM, Tata LJ,
Stanley RA, Peake MD, Hubbard RB and Baldwin DR: Non-small cell
lung cancer in young adults: Presentation and survival in the
English National Lung Cancer Audit. QJM. 108:891–897. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Travis WD, Brambilla E, Muller-Hemerlink
HK and Harris CC: World Health Organization Classification of
TumoursPathology and Genetics of Tumours of the Lung, Pleura,
Thymus and Heart. IARC press; Lyon: 2004
|
|
10
|
Korsse SE, Peppelenbosch MP and van Veelen
W: Targeting LKB1 signaling in cancer. Biochim Biophys Acta.
1835:194–210. 2013.PubMed/NCBI
|
|
11
|
Beroukhim R, Mermel CH, Porter D, Wei G,
Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J,
Urashima M, et al: The landscape of somatic copy-number alteration
across human cancers. Nature. 463:899–905. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hemminki A, Markie D, Tomlinson I,
Avizienyte E, Roth S, Loukola A, Bignell G, Warren W, Aminoff M,
Höglund P, et al: A serine/threonine kinase gene defective in
Peutz-Jeghers syndrome. Nature. 391:184–187. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Berasain C and Ávila MA: LKB1: Controlling
Quiescence and Genomic Integrity at Home. Trends Endocrinol Metab.
April 5–2018.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fabian MR, Sonenberg N and Filipowicz W:
Regulation of mRNA translation and stability by microRNAs. Annu Rev
Biochem. 79:351–379. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Carretero J, Shimamura T, Rikova K,
Jackson AL, Wilkerson MD, Borgman CL, Buttarazzi MS, Sanofsky BA,
McNamara KL, Brandstetter KA, et al: Integrative genomic and
proteomic analyses identify targets for Lkb1-deficient metastatic
lung tumors. Cancer Cell. 17:547–559. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang W, Gao Y, Li F, Tong X, Ren Y, Han
X, Yao S, Long F, Yang Z, Fan H, et al: YAP promotes malignant
progression of Lkb1-deficient lung adenocarcinoma through
downstream regulation of survivin. Cancer Res. 75:4450–4457. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Calles A, Sholl LM, Rodig SJ, Pelton AK,
Hornick JL, Butaney M, Lydon C, Dahlberg SE, Oxnard GR, Jackman DM,
et al: Immunohistochemical loss of LKB1 is a biomarker for more
aggressive biology in KRAS-mutant lung adenocarcinoma. Clin Cancer
Res. 21:2851–2860. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Travis WD, Brambilla E, Noguchi M,
Nicholson AG, Geisinger KR, Yatabe Y, Beer DG, Powell CA, Riely GJ,
Van Schil PE, et al: International association for the study of
lung cancer/american thoracic society/european respiratory society
international multidisciplinary classification of lung
adenocarcinoma. J Thorac Oncol. 6:244–285. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Travis WD, Brambilla E, Noguchi M,
Nicholson AG, Geisinger K, Yatabe Y, Ishikawa Y, Wistuba I, Flieder
DB, Franklin W, et al: Diagnosis of lung cancer in small biopsies
and cytology: Implications of the 2011 International Association
for the Study of Lung Cancer/American Thoracic Society/European
Respiratory Society classification. Arch Pathol Lab Med.
137:668–684. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang J, Qin L, Han L, Zhao Y, Jing H,
Song W and Shi H: Role of MicroRNA-93 in pathogenesis of left
ventricular remodelling via targeting cyclin-D1. Med Sci Monit.
23:3981–3988. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tang Q, Zou Z, Zou C, Zhang Q, Huang R,
Guan X, Li Q, Han Z, Wang D, Wei H, et al: MicroRNA-93 suppress
colorectal cancer development via Wnt/β-catenin pathway
downregulating. Tumour Biol. 36:1701–1710. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Avtanski DB, Nagalingam A, Bonner MY,
Arbiser JL, Saxena NK and Sharma D: Honokiol activates LKB1-miR-34a
axis and antagonizes the oncogenic actions of leptin in breast
cancer. Oncotarget. 6:29947–29962. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhu XB, Zhang ZC, Han GS, Han JZ and Qiu
DP: Overexpression of miR 214 promotes the progression of human
osteosarcoma by regulating the Wnt/β catenin signaling pathway. Mol
Med Rep. 15:1884–1892. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kang J, Kim W, Lee S, Kwon D, Chun J, Son
B, Kim E, Lee JM, Youn H and Youn B: TFAP2C promotes lung
tumorigenesis and aggressiveness through miR-183- and
miR-33a-mediated cell cycle regulation. Oncogene. 36:1585–1596.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yabushita S, Fukamachi K, Tanaka H, Sumida
K, Deguchi Y, Sukata T, Kawamura S, Uwagawa S, Suzui M and Tsuda H:
Circulating microRNAs in serum of human K-ras oncogene transgenic
rats with pancreatic ductal adenocarcinomas. Pancreas.
41:1013–1018. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang D, Lu G, Shao Y and Xu D: miR-182
promotes prostate cancer progression through activating
Wnt/β-catenin signal pathway. Biomed Pharmacother. 99:334–339.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Beezhold K, Klei LR and Barchowsky A:
Regulation of cyclin D1 by arsenic and microRNA inhibits
adipogenesis. Toxicol Lett. 265:147–155. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wu Z, He B, He J and Mao X: Upregulation
of miR-153 promotes cell proliferation via downregulation of the
PTEN tumor suppressor gene in human prostate cancer. Prostate.
73:596–604. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu X, Lv XB, Wang XP, Sang Y, Xu S, Hu K,
Wu M, Liang Y, Liu P, Tang J, et al: MiR-138 suppressed
nasopharyngeal carcinoma growth and tumorigenesis by targeting the
CCND1 oncogene. Cell Cycle. 11:2495–2506. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Waggott D, Chu K, Yin S, Wouters BG, Liu
FF and Boutros PC: NanoStringNorm: An extensible R package for the
pre-processing of NanoString mRNA and miRNA data. Bioinformatics.
28:1546–1548. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tian DL, Liu HX, Zhang L, Yin HN, Hu YX,
Zhao HR, Chen DY, Han LB, Li Y and Li HW: Surgery for young
patients with lung cancer. Lung Cancer. 42:215–220. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu NS, Spitz MR, Kemp BL, Cooksley C,
Fossella FV, Lee JS, Hong WK and Khuri FR: Adenocarcinoma of the
lung in young patients: The M. D. Anderson experience. Cancer.
88:1837–1841. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kuo CW, Chen YM, Chao JY, Tsai CM and
Perng RP: Non-small cell lung cancer in very young and very old
patients. Chest. 117:354–357. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Skarin AT, Herbst RS, Leong TL, Bailey A
and Sugarbaker D: Lung cancer in patients under age 40. Lung
Cancer. 32:255–264. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Blanco M, García-Fontán E, Rivo JE, Repáaz
JR, Obeso GA and Cañizares MA: Bronchogenic carcinoma in patients
under 50 years old. Clin Transl Oncol. 11:322–325. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Weiss J and Langer C: Treatment of lung
cancer in the elderly patient. Semin Respir Crit Care Med.
34:802–809. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Davidoff AJ, Tang M, Seal B and Edelman
MJ: Chemotherapy and survival benefit in elderly patients with
advanced non-small-cell lung cancer. J Clin Oncol. 28:2191–2197.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Beckett P, Callister M, Tata LJ, Harrison
R, Peake MD, Stanley R, Woolhouse I, Slade M and Hubbard RB:
Clinical management of older people with non-small cell lung cancer
in England. Thorax. 67:836–839. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Meoni G, Cecere FL, Lucherini E and Di
Costanzo F: Medical treatment of advanced non-small cell lung
cancer in elderly patients: A review of the role of chemotherapy
and targeted agents. J Geriatr Oncol. 4:282–290. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ye T, Pan Y, Wang R, Hu H, Zhang Y, Li H,
Wang L, Sun Y and Chen H: Analysis of the molecular and
clinicopathologic features of surgically resected lung
adenocarcinoma in patients under 40 years old. J Thorac Dis.
6:1396–1402. 2014.PubMed/NCBI
|
|
41
|
Kim L, Kim KH, Yoon YH, Ryu JS, Choi SJ,
Park IS, Han JY, Kim JM and Chu YC: Clinicopathologic and molecular
characteristics of lung adenocarcinoma arising in young patients. J
Korean Med Sci. 27:1027–1036. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Pan Y, Zhang Y, Li Y, Hu H, Wang L, Li H,
Wang R, Ye T, Luo X, Zhang Y, et al: ALK, ROS1 and RET fusions in
1139 lung adenocarcinomas: A comprehensive study of common and
fusion pattern-specific clinicopathologic, histologic and cytologic
features. Lung Cancer. 84:121–126. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Radzikowska E, Roszkowski K and Głaz P:
Lung cancer in patients under 50 years old. Lung Cancer.
33:203–211. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Sekine I, Nishiwaki Y, Yokose T, Nagai K,
Suzuki K and Kodama T: Young lung cancer patients in Japan:
Different characteristics between the sexes. Ann Thorac Surg.
67:1451–1455. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Shackelford DB, Abt E, Gerken L, Vasquez
DS, Seki A, Leblanc M, Wei L, Fishbein MC, Czernin J, Mischel PS,
et al: LKB1 inactivation dictates therapeutic response of non-small
cell lung cancer to the metabolism drug phenformin. Cancer Cell.
23:143–158. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Inge LJ, Coon KD, Smith MA and Bremner RM:
Expression of LKB1 tumor suppressor in non-small cell lung cancer
determines sensitivity to 2-deoxyglucose. J Thorac Cardiovasc Surg.
137:580–586. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Carretero J, Medina PP, Blanco R, Smit L,
Tang M, Roncador G, Maestre L, Conde E, Lopez-Rios F, Clevers HC,
et al: Dysfunctional AMPK activity, signalling through mTOR and
survival in response to energetic stress in LKB1-deficient lung
cancer. Oncogene. 26:1616–1625. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Mahoney CL, Choudhury B, Davies H, Edkins
S, Greenman C, Haaften G, Mironenko T, Santarius T, Stevens C,
Stratton MR, et al: LKB1/KRAS mutant lung cancers constitute a
genetic subset of NSCLC with increased sensitivity to MAPK and mTOR
signalling inhibition. Br J Cancer. 100:370–375. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Whang YM, Park SI, Trenary IA, Egnatchik
RA, Fessel JP, Kaufman JM, Carbone DP and Young JD: LKB1 deficiency
enhances sensitivity to energetic stress induced by erlotinib
treatment in non-small-cell lung cancer (NSCLC) cells. Oncogene.
35:856–866. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Jian SF, Hsiao CC, Chen SY, Weng CC, Kuo
TL, Wu DC, Hung WC and Cheng KH: Utilization of liquid
chromatography mass spectrometry analyses to identify LKB1-APC
interaction in modulating Wnt/β-catenin pathway of lung cancer
cells. Mol Cancer Res. 12:622–635. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Geiss GK, Bumgarner RE, Birditt B, Dahl T,
Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, et
al: Direct multiplexed measurement of gene expression with
color-coded probe pairs. Nat Biotechnol. 26:317–325. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Reis PP, Waldron L, Goswami RS, Xu W, Xuan
Y, Perez-Ordonez B, Gullane P, Irish J, Jurisica I and Kamel-Reid
S: mRNA transcript quantification in archival samples using
multiplexed, color-coded probes. BMC Biotechnol. 11:462011.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Mairinger FD, Walter RF, Werner R,
Christoph DC, Ting S, Vollbrecht C, Zarogoulidis K, Huang H, Li Q,
Schmid KW, et al: Activation of angiogenesis differs strongly
between pulmonary carcinoids and neuroendocrine carinomas and is
crucial for carcinoid tumourgenesis. J Cancer. 5:465–471. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Veldman-Jones MH, Brant R, Rooney C, Geh
C, Emery H, Harbron CG, Wappett M, Sharpe A, Dymond M, Barrett JC,
et al: Evaluating robustness and sensitivity of the NanoString
Technologies nCounter platform to enable multiplexed gene
expression analysis of clinical samples. Cancer Res. 75:2587–2593.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Walter RF, Mairinger FD, Wohlschlaeger J,
Worm K, Ting S, Vollbrecht C, Schmid KW and Hager T: FFPE tissue as
a feasible source for gene expression analysis - a comparison of
three reference genes and one tumor marker. Pathol Res Pract.
209:784–789. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Chen L, Engel BE, Welsh EA, Yoder SJ,
Brantley SG, Chen DT, Beg AA, Cao C, Kaye FJ, Haura EB, et al: A
Sensitive NanoString-Based Assay to Score STK11 (LKB1) Pathway
Disruption in Lung Adenocarcinoma. J Thorac Oncol. 11:838–849.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ujiie H, Kadota K, Chaft JE, Buitrago D,
Sima CS, Lee MC, Huang J, Travis WD, Rizk NP, Rudin CM, et al:
Solid Predominant Histologic Subtype in Resected Stage I Lung
Adenocarcinoma Is an Independent Predictor of Early, Extrathoracic,
Multisite Recurrence and of Poor Postrecurrence Survival. J Clin
Oncol. 33:2877–2884. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Du L, Zhao Z, Ma X, Hsiao TH, Chen Y,
Young E, Suraokar M, Wistuba I, Minna JD and Pertsemlidis A:
miR-93-directed downregulation of DAB2 defines a novel oncogenic
pathway in lung cancer. Oncogene. 33:4307–4315. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Xiao J, Zou Y, Chen X, Gao Y, Xie M, Lu X,
Li W, He B, He S, You S, et al: The Prognostic Value of Decreased
LKB1 in Solid Tumors: A Meta-Analysis. PLoS One. 11:e01526742016.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Xu P, Zhao M, Liu Z, Liu Y, Chen Y, Luo R
and Fang W: Elevated nuclear CCND1 expression confers an
unfavorable prognosis for early stage lung adenocarcinoma patients.
Int J Clin Exp Pathol. 8:15887–15894. 2015.PubMed/NCBI
|
|
61
|
Deep G and Agarwal R: Targeting tumor
microenvironment with silibinin: Promise and potential for a
translational cancer chemopreventive strategy. Curr Cancer Drug
Targets. 13:486–499. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Gao Y, Xiao Q, Ma H, Li L, Liu J, Feng Y,
Fang Z, Wu J, Han X, Zhang J, et al: LKB1 inhibits lung cancer
progression through lysyl oxidase and extracellular matrix
remodeling. Proc Natl Acad Sci USA. 107:18892–18897. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Hou X, Du H, Quan X, Shi L, Zhang Q, Wu Y,
Liu Y, Xiao J, Li Y, Lu L, et al: Silibinin Inhibits NSCLC
Metastasis by targeting the EGFR/LOX pathway. Front Pharmacol.
9:212018. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zhong K, Chen K, Han L and Li B:
MicroRNA-30b/c inhibits non-small cell lung cancer cell
proliferation by targeting Rab18. BMC Cancer. 14:7032014.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
de la Cueva A, Emmerling M, Lim SL, Yang
S, Trackman PC, Sonenshein GE and Kirsch KH: A polymorphism in the
lysyl oxidase propeptide domain accelerates carcinogen-induced
cancer. Carcinogenesis. March 22–2018.(Epub ahead of print).
View Article : Google Scholar : PubMed/NCBI
|