Introduction
Hepatic fibrosis (HF) is an important health
condition linked to chronic liver injury and HF is a disease
associated with high morbidity (1). HF is a pathological process
involving structural or functional abnormalities of the liver
mainly attributed to the excessive accumulation of extracellular
matrix components in hepatic tissues, which can lead to liver
cirrhosis and liver cancer (2).
The morbidity and mortality of HF-induced liver cirrhosis and liver
cancer rank sixth globally and are increasing year-on-year in China
(3). Previous studies reported
that drugs for liver fibrosis treatment mainly inhibit hepatocyte
apoptosis (emricasan) (4) and
hepatic stellate cell (HSC) activation (PRI-724) (5), reduce fibrotic scar evolution and
contraction (simtuzumab) (6), and
regulate the immune response (cenicriviroc) (7). However, there is currently a lack of
effective strategies and drugs to directly treat this disease
(8,9). Numerous studies have demonstrated
that the initiation and development of HF are primarily elicited by
factors including viral infections, alcohol toxins and metabolites
(Such as cholesterol). Furthermore, the activation and
proliferation of HSCs also serve as key drivers of HF (10,11). HSC activation is a complicated
process controlled by multiple molecules and signaling pathways
(12), yet its underlying
mechanisms remain largely undetermined. Exploring the molecular
mechanisms related to the function of HSCs in order to identify new
diagnostic and therapeutic molecular targets could be instrumental
in developing new possibilities for the treatment of HF.
Long non-coding (lnc)RNAs are a set of
non-protein-coding transcripts with a length of over 200 nt
(13). A better understanding of
HF could support the use of novel concepts and avenues in the
targeted therapy of HF (14). A
recent study by Zhang et al (15) reported that lnc-Lfar1 was closely
associated with HF and modulated the macrophage activation evoked
via lipopolysaccharide and IFN-γ to influence the occurrence and
development of fibrosis via the NF-κB signaling pathway.
Furthermore, Liao et al (16) recently reported that the lncRNA
Gpr137b-ps could bind to and impair microRNA (miRNA/miR)-200a-3p
causing inhibition of chemokine (C-X-C motif) ligand 14, which may
promote the pathological process of HF via alteration of the
activation and proliferation of HSCs. A whole-transcriptome
sequencing analysis by Gong et al (17) revealed that lncRNA LOC102553417
(Genbank ID: XR_595047) was expressed at a high level in a rat
carbon tetrachloride-induced HF model. LOC102553417 is a lncRNA of
rats, which can be queried through the UCSC database
(genome-asia.ucsc.edu). However, the role of LOC102553417 in the
occurrence and development of HF remains undetermined. In the
present study, LOC102553417 silencing was used to investigate its
role in HSC activation via the miR-30e/metadherin (MTDH) axis. An
in-depth understanding of the functions and mechanisms of
LOC102553417 in HF could provide a theoretical basis for the
clinical use of LOC102553417 as a diagnostic biomarker and
treatment for HF.
Materials and methods
Culture of rat HSC-T6 cells
The rat HSC-T6 cell line was purchased from The Cell
Bank of Type Culture Collection of The Chinese Academy of Sciences.
Cells were cultured at 37°C with 5% CO2 in DMEM (cat.
no. PM150210; Procell Life Science & Technology Co., Ltd.),
which was supplemented with 10% fetal bovine serum (cat. no.
164210-500; Procell Life Science & Technology Co., Ltd.) and 1%
penicillin-streptomycin (cat. no. PB180120; Procell Life Science
& Technology Co., Ltd.). The cells were passaged at a 1:3 ratio
and the medium was renewed three times per week. For TGF-β1
induction, HSC-T6 cells were stimulated with 10 ng/ml TGF-β1 (cat.
no. HY-P7118; MedChemExpress) for 24 h at 37°C or treated with an
equal amount of double-distilled H2O as the control.
Cell transduction and
transfection
The LOC102553417 silencing lentiviral vector (vector
no. GV492) was constructed by Guangzhou Anernor Biotechnology Co.,
Ltd. Briefly, GV492 plasmid vector (100 nM) and packaging plasmid
(psPaX2; third generation lentiviral packaging system (vector:
packaging vector: envelope ratio, 10:3:1; Promega Corporation) were
mixed with lipo3000 (L3000-015; Invitrogen; Thermo Fisher
Scientific, Inc.) were co-transfected into 1×106 293T
cells (Cell Bank of the Chinese Academy of Sciences). When 293T
cells were cultured for 4 days, the cell supernatant was collected,
and then the collected cell supernatant was centrifuged at 251.55 ×
g for 5 min at 4°C, and the supernatant was filtered with a 0.22 µm
membrane. The targeting sequences used for the knockdowns (KDs) and
negative control (NC, non-targeting.) were as follows: KD1,
5′-GACCCTGCATCAGAGTCTTCCAGA-3′; KD2,
5′-GGACTCTTCCTGGACTACCATAAAT-3′; KD3,
5′-CATACATGCTAGGGCAGCCATTGCA-3′; and NC, 5′-TTCTCCGAACGTGTCACGT-3′.
HSC-T6 cells were transduced at 37°C (multiplicity of infection,
30) for 96 h and green fluorescence was observed via fluorescence
microscopy to demonstrate transduction (data not shown).
LOC102553417 expression was determined via reverse
transcription-quantitative (RT-q)PCR. The vector with the highest
silencing efficiency was used for subsequent experimentation at 24
h post-transduction.
The miR-30e mimic (sense,
5′-UGUAAACAUCCUUGACUGGAAG-3′, double chain; antisense
5′-CUUCCAGUCAAGGAUGUUUACA-3′), miR-30e inhibitor
(5′-CUUCCAGUCAAGGAUGUUUACA-3′), mimic NC
(5′-UUUGUACUACACAAAAGUACUG-3′) and inhibitor NC
(5′-CAGUACUUUUGUGUAGUACAAA-3′) vectors were synthesized by Huzhou
Hippo Biotechnology Co., Ltd. Lipofectamine® 3000 kit
(cat. no. L3000015; Invitrogen; Thermo Fisher Scientific, Inc.) was
employed for transfection(final concentration of nucleic acid:
20nM, cell density, 50%, 37°C) for 48 h, subsequent experiments
were performed 48 h after transfection.
RT-qPCR
TRIzol® reagent (cat. no. 15596-026;
Invitrogen; Thermo Fisher Scientific, Inc.) was used to extract
total cellular RNA. RT-PCR was performed using an RT-PCR kit (cat.
no. RT-02011; Chengdu Foregene Biotechnology Co., Ltd.), which
consisted of 10 µl SYBR-Green Mix, 2 µl forward (F) primer, 2 µl
reverse (R) primer, 1 µl cDNA and 5 µl RNase-free deionized water.
After the purity and concentration tests, RNA was reverse
transcribed into cDNA (70°C, 5 min; 42°C, 60 min; 70°C, 10 min).
Primer Premier 5.0 software (Premier, Inc.) was employed for primer
design; each primer was composed of 20–22 bases and the product
size was 70–200 bp. The reaction conditions were set as follows:
Pre-denaturation for 15 sec at 95°C; 40 cycles of denaturation for
5 sec at 95°C, annealing and extension at 60°C for 30 sec; and for
the melting curve, amplification at 55–95°C for 10 sec (81 cycles
in total). Three parallel wells were used for each group with a
blank control and a NC group at the same time. After determining
the cycle threshold, the relative expression of the target gene
normalized to the internal control(GAPDH was used for lncRNA and
mRNA, U6 was used for miRNA) was calculated using 2−ΔΔCq
analysis (18). The primer
sequences were synthesized by Sangon Biotech Co., Ltd. as follows:
LOC102553417 (F), AGTCCTGCCCACACTGCTTTT; LOC102553417 (R),
AACAGAGGCCTGAAATAGAC; MTDH (F), AGCGGGAGGAGGTGACCCCGCC; MTDH (R),
ATTTGGTTTGGGCTTTTCA; miR-30e-RT, GTCGTATCCAGTGCAGGGTCCG
AGGTATTCGCACTGGATACGACCTTCCAGT; miR-30e (F), GCCGAGTGTAAACATCCT;
miR-30e (R), GTCGTATCCAGTGCGAATAC; GAPDH (F), TGTGAACGGATTTGGCCGTA;
GAPDH (R), GATGGTGATGGGTTTCCCGT; U6 (F), CTCGCTTCGGCAGCACA; and U6
(R), AACGCTTCACGAATTTGCGT.
Western blotting
Cells were lysed using RIPA lysis buffer (cat. no.
P0013B; Beyotime Institute of Biotechnology). Following
homogenization, lysis and centrifugation at 200 × g for 5 min at
4°C, total protein was extracted and the concentration was assessed
using the BCA method. Following separation via SDS-PAGE (5%
stacking gel, 12% running gel, 20 µg total protein per lane).
Electrophoresed proteins were transferred to a PVDF membrane
(constant current of 200 mA for 1 h and a constant current of 250
mA for 2 h). The membrane was then blocked with TBS-Tween (Tween
0.1% in TBS) solution containing 5% skimmed milk powder for 1 h and
was incubated with primary antibodies against the following
proteins: Akt (1:500; cat. no. ab18785; Abcam), phosphorylated
(p)-Akt (1:500; cat. no. ab8933; Abcam), p53 (1:333; cat. no. ab26;
Abcam), MTDH (1:10,000; cat. no. ab124789; Abcam), β-actin
(1:20,000; cat. no. 66009-1-Ig; ProteinTech Group, Inc.), Cleaved
Caspase-3 (1:500; cat. no. ab32042; Abcam) and caspase-3 (1:1,000;
cat. no. 19677-1-AP; ProteinTech Group, Inc.). After 1 h of
agitation at room temperature, overnight incubation was performed
with the primary antibodies at 4°C. After washing, the membrane was
incubated with a secondary antibody (HRP; anti-rabbit 1:5000, cat.
no. 074-1506; anti-mouse 1:5000, cat. no. 074-1807; KPL, Inc.) for
1 h at 37°C. The bands were visualized using ECL kit (P0018S;
Beyotime Institute of Biotechnology), followed by pressing,
exposure and fixation. Grayscale analysis of the images was
performed using ImageJ software 1.8.0 (National Institutes of
Health).
Cell Counting Kit-8 (CCK-8)
HSC-T6 cell suspension was prepared and seeded onto
a 96-well plate (100 µl/well) at 1×103 cells/well with
three parallel wells for each group. On each of 5 consecutive days,
10 µl CCK-8 reagent (cat. no. FC101-03; TransGen Biotech Co., Ltd.)
was added to each well. Following incubation in 37°C and 5%
CO2 incubator for 2 h, absorbance was quantified at 450
nm using an ELx800 Absorbance Microplate Reader (BioTek Instruments
Inc.).
Flow cytometry
HSC-T6 cells were transferred directly into a 10-ml
centrifuge tube at 106/ml cells per sample, total 1ml.
Following centrifugation at 200 × g for 5 min, the supernatant was
discarded. Subsequently, cells (100 µl) were suspended in a 5 ml
flow tube and incubated with 5 µl Annexin V-PE and 5 µl PI (cat.
no. 640914; Biolegend, Inc.) at ambient temperature for 15 min. The
samples were then loaded onto a CytoFLEX Flow Cytometer (Beckman
Coulter, Inc.) for apoptosis analysis with FlowJo Software V10
(FlowJo, LLC).
Bioinformatics analysis of binding
sites
It has been reported that miR-30e is a miRNA
associated with HF using the RegRNA2 database
(regrna2.mbc.nctu.edu.tw/) (19),
predicted that wild-type (WT) LOC102553417 contained a binding site
with miR-30e. The binding site was mutated into the original
complementary sequence and designed as LOC102553417 mutant (MUT).
The TargetScan database, version 8.0 (http://www.targetscan.org/vert_80/) was used to
identify potential target genes selecting the total context++ score
<-0.8, aggregate PCT ≥90 and selecting for genes that
have been reported to be related to liver disease. miR-30e was
predicted to have a binding site with MTDH and based on the binding
site, the binding site was mutated into the original complementary
sequence and designed as MTDH MUT.
Dual-luciferase reporter assay
Log-phase HSC-T6 cells were seeded into a 24-well
plate and cultured for 24 h. Cell transfection by lipo3000
(L3000-015; Invitrogen; Thermo Fisher Scientific, Inc.) was
performed using MTDH 3′UTR or LOC102553417 (wild or mutant type)
psicheck2 luciferase reporter gene plasmid (Shanghai Genechem
Technology Co., Ltd), Renilla control plasmid,
aforementioned miR-30e mimic or mimic-NC. Three parallel wells were
used in each group. After culture at 37°C and 5% CO2 for
24 h, the medium was discarded and the residual medium was removed
via two washes in ice-cold PBS. Immediately following transduction,
activity measurement was performed. According to the manufacturers'
protocol, 100 µl 1X Passive Lysis Buffer from the Dual-Luciferase
Reporter Assay System kit (cat. no. E1910; Promega Corporation) was
added to each well. Following gentle shaking, the cells were
maintained at ambient temperature for 15 min. The lysate (20 µl)
was placed into a 96-well plate and the luminescence was quantified
using a Lux-T020 Ultra-sensitive Tube Luminometer (Guangzhou
Biolight Biotechnology Co., Ltd.) for statistical analysis of
relative luciferase activity normalized to Renilla
luciferase activity.
RNA-binding protein
immunoprecipitation (RIP)
According to the manufacturer's instructions of RIP
kit (cat. no. Bes5101; Guangzhou Boxin Biotechnology Co., Ltd.),
RIP buffer and Argonaute RNA-induced silencing complex catalytic
component 2 (Ago2) antibodies 3ug (cat. no. ab233727; Abcam) were
equilibrated at ambient temperature for 10 min. The harvested
HSC-T6 cells were lysed with RIP lysis buffer at 4°C for 10 min,
washed with 2 ml PBS and centrifuged at 200 × g for 5 min at room
temperature to collect cell lysate. The cell lysate (100 µl) was
incubated with RIP buffer (900 µl) containing antibody-labeled A +
G magnetic beads (20 µl) at 4°C for 16 h (overnight). IgG (cat. no.
ab172730; Abcam) served as a NC and the supernatant was harvested
as ‘Input’ as a control. The immunoprecipitated complex was
isolated and detached with 150 µl proteinase K buffer to isolate
the RNA at 55°C for 1 h. RT-qPCR was employed to analyze the
relative expression of miR-30e and LOC102553417 as
aforementioned.
Statistical analysis
All experimental data were analyzed using Prism 8.0
software (GraphPad Software Inc.). Statistics was obtained from
three repeat experiments. Data are presented as the mean ± standard
deviation. Comparisons between two groups were performed using
unpaired Student's t-test, whereas one-way ANOVA followed by
Tukey's post-hoc test was used for comparisons among three or more
groups. P<0.05 was considered to indicate a statistically
significant difference.
Results
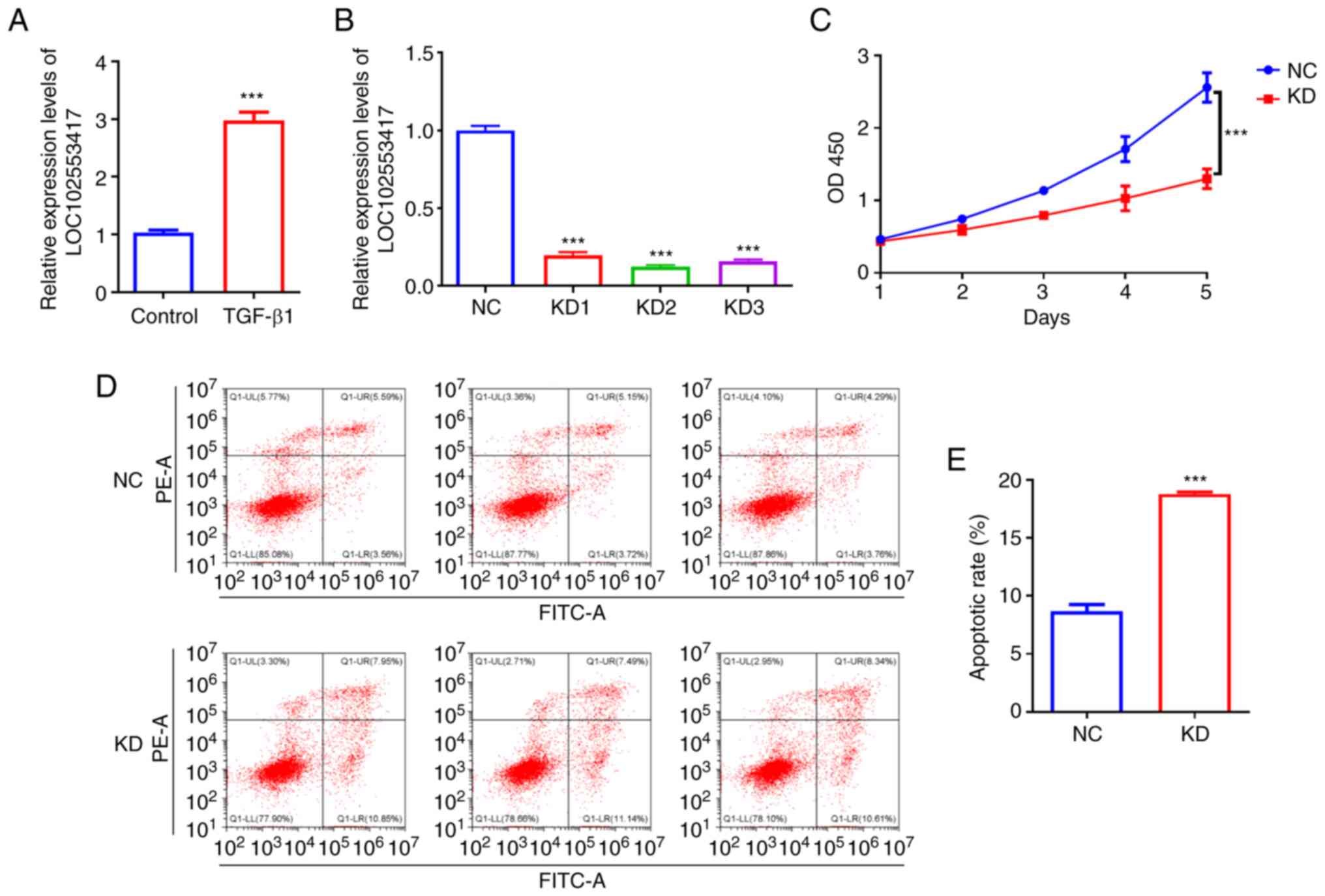
LOC102553417 silencing decreases
HSC-T6 cell proliferation and increases their apoptosis
To ascertain the importance of LOC102553417 in the
activation of HSCs, the mRNA expression levels of LOC102553417
after TGF-β1-triggered activation of HSC-T6 cells were determined
using RT-qPCR. LOC102553417 mRNA expression levels in the
TGF-β1-induced group were significantly higher compared with those
in the control group (P<0.001; Fig. 1A). Post-transduction of HSC-T6
cells with LOC102553417 silencing vectors and NC vector,
LOC102553417 mRNA expression levels were assessed via RT-qPCR. The
mRNA expression levels of LOC102553417 in the KD1 group
(0.20±0.02), KD2 group (0.12±0.01) and KD3 group (0.16±0.01) were
all significantly downregulated compared with those in the NC
vector group (1.00±0.03; P<0.001). The silencing efficiency of
LOC102553417 KD2 was the greatest (Fig. 1B); therefore, KD2 was selected for
use in the subsequent experiments. CCK-8 assay demonstrated
significantly reduced proliferation of HSCs in
LOC102553417-silenced cells compared with in the NC group
(P<0.001; Fig. 1C).
Furthermore, flow cytometry data demonstrated increased cell
apoptosis in LOC102553417-silenced cells compared with in the NC
group (P<0.001; Fig. 1D and
E).
LOC102553417 competitively binds to
miR-30e, which upregulates MTDH expression
Using the RegRNA2 database predicted that wild-type
(WT) LOC102553417 contained a binding site with miR-30e. MUT
LOC102553417 was designed (Fig.
2A. Using the TargetScan predicted that WT MTDH 3′UTR contained
a binding site with miR-30e. MUT MTDH 3′UTR was designed (Fig. 2B).
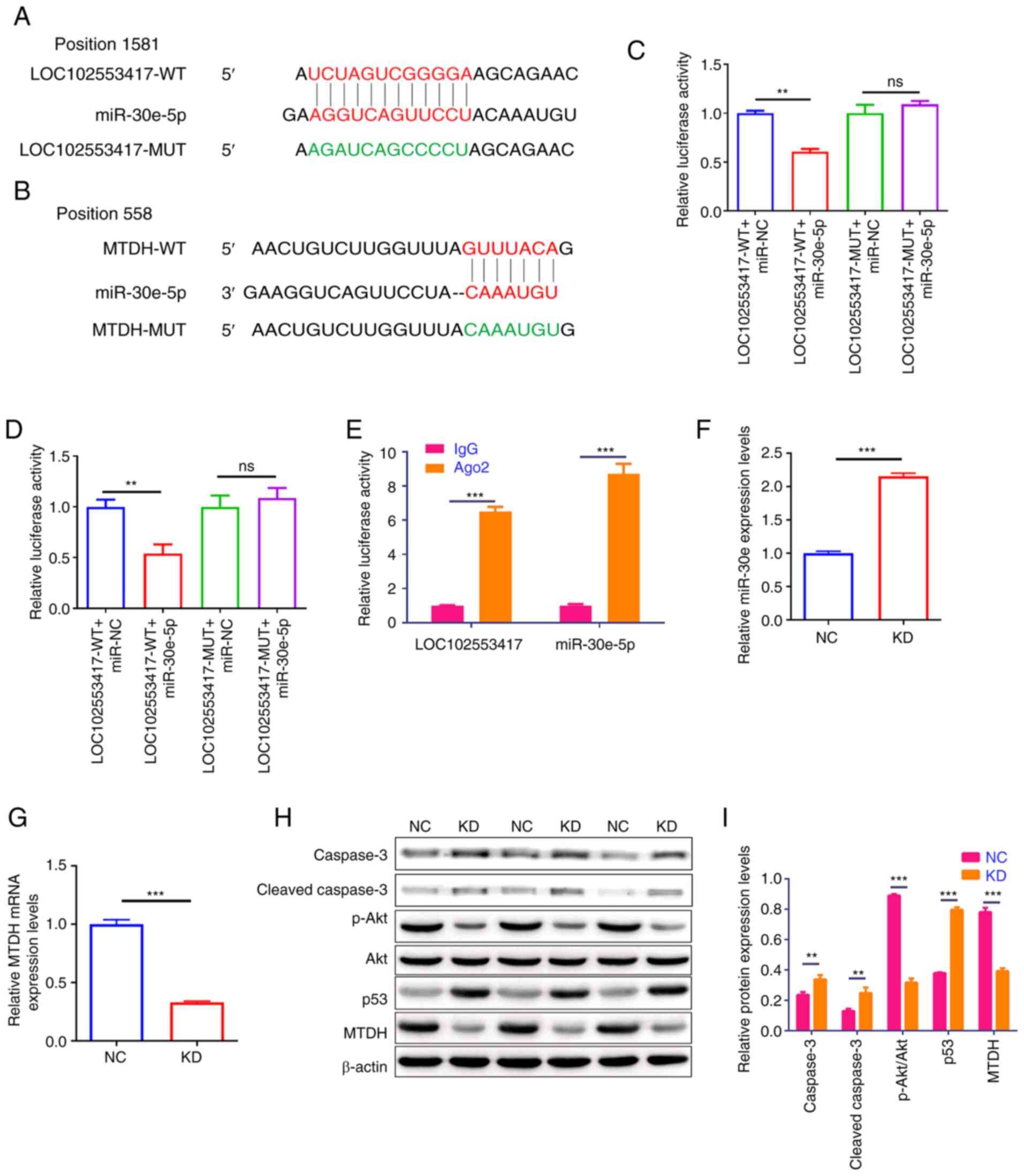 | Figure 2.LOC102553417 silencing suppresses
MTDH expression via competitively binding to miR-30e. Alignment of
the binding site between (A) LOC102553417, miR-30e and the designed
MUT sequence, and (B) miR-30e, MTDH and the designed MUT sequence.
Binding of (C) miR-30e to LOC102553417 and (D) miR-30e to MTDH were
assessed via dual-luciferase reporter assay. (E) Binding of miR-30e
to LOC102553417 was assessed via RNA binding protein
immunoprecipitation. mRNA expression levels of (F) miR-30e and (G)
MTDH were assessed via RT-qPCR. (H) Representative western blotting
images of MTDH, Akt, p-Akt, p53, caspase-3 and cleaved caspase-3
protein bands. (I) Statistical analysis of MTDH, Akt, p-Akt, p53,
caspase-3 and cleaved caspase-3 protein expression levels assessed
via western blotting. Results are expressed as the mean ± SD (n=3).
**P<0.01; ***P<0.001; ns, no significant difference; WT,
wild-type; MUT, mutant; NC, negative control; Ago2, Argonaute
RNA-induced silencing complex catalytic component 2; p,
phosphorylated; miR, microRNA; MTDH, metadherin; RT-qPCR, reverse
transcription-quantitative PCR; NC, negative control; KD,
knockdown. |
The binding of LOC102553417 to miR-30e was
demonstrated by the dual-luciferase reporter assay. Luciferase
activity was significantly reduced in the LOC102553417-WT + miR-30e
mimic compared with that in the LOC102553417-WT + miR-NC group
(P<0.01; Fig. 2C). However,
the LOC102553417-MUT + miR-30e mimic demonstrated no significant
difference in luciferase activity compared with the
LOC102553417-MUT + miR-NC group (P>0.05), which demonstrated the
binding of LOC102553417 to miR-30e at the aforementioned binding
site. The binding of MTDH to miR-30e was also demonstrated using
the luciferase assay. Compared with the MTDH-WT + miR-NC group, the
MTDH-WT + miR-30e mimic group demonstrated significantly reduced
luciferase activity (P<0.01); however the MTDH-MUT + miR-30e
mimic group demonstrated no significant difference in luciferase
activity compared with the MTDH-MUT + miR-NC group (P>0.05;
Fig. 2D), which demonstrated the
binding of miR-30e to MTDH at the binding site predicted using
TargetScan database.
Antibodies to Ago2, a miRNA precursor cleavage
protein, were used to conduct RIP experiments to verify the binding
of miR-30e to LOC102553417. Compared with in the IgG antibody group
(NC antibody), significantly higher expression levels of miR-30e
and LOC102553417 were detected in the Ago2 antibody group
(P<0.001; Fig. 2E).
Subsequently, LOC102553417 was silenced in HSC-T6 cells, the
miR-30e and MTDH expression levels were determined via RT-qPCR, and
the MTDH protein expression levels were assessed via western
blotting. miR-30e relative expression levels in the KD group were
significantly elevated compared with those in the NC group
(P<0.001; Fig. 2F). MTDH mRNA
expression levels (P<0.001; Fig.
2G) and protein expression levels (P<0.001; Fig. 2H and I) were significantly reduced
in the LOC102553417 KD group compared with those in the NC group.
Western blot analysis of the protein expression levels of p-Akt,
Akt, caspase-3, cleaved caspase-3 and p53 was used to assess the
downstream mechanism of the LOC102553417/miR-30e/MTDH axis. Western
blotting demonstrated significant reductions in the p-Akt/Akt ratio
(P<0.001), and significant elevations in p53, caspase-3 and
cleaved caspase-3 protein expression levels (P<0.001) in the
LOC102553417 KD group compared with those in the NC group.
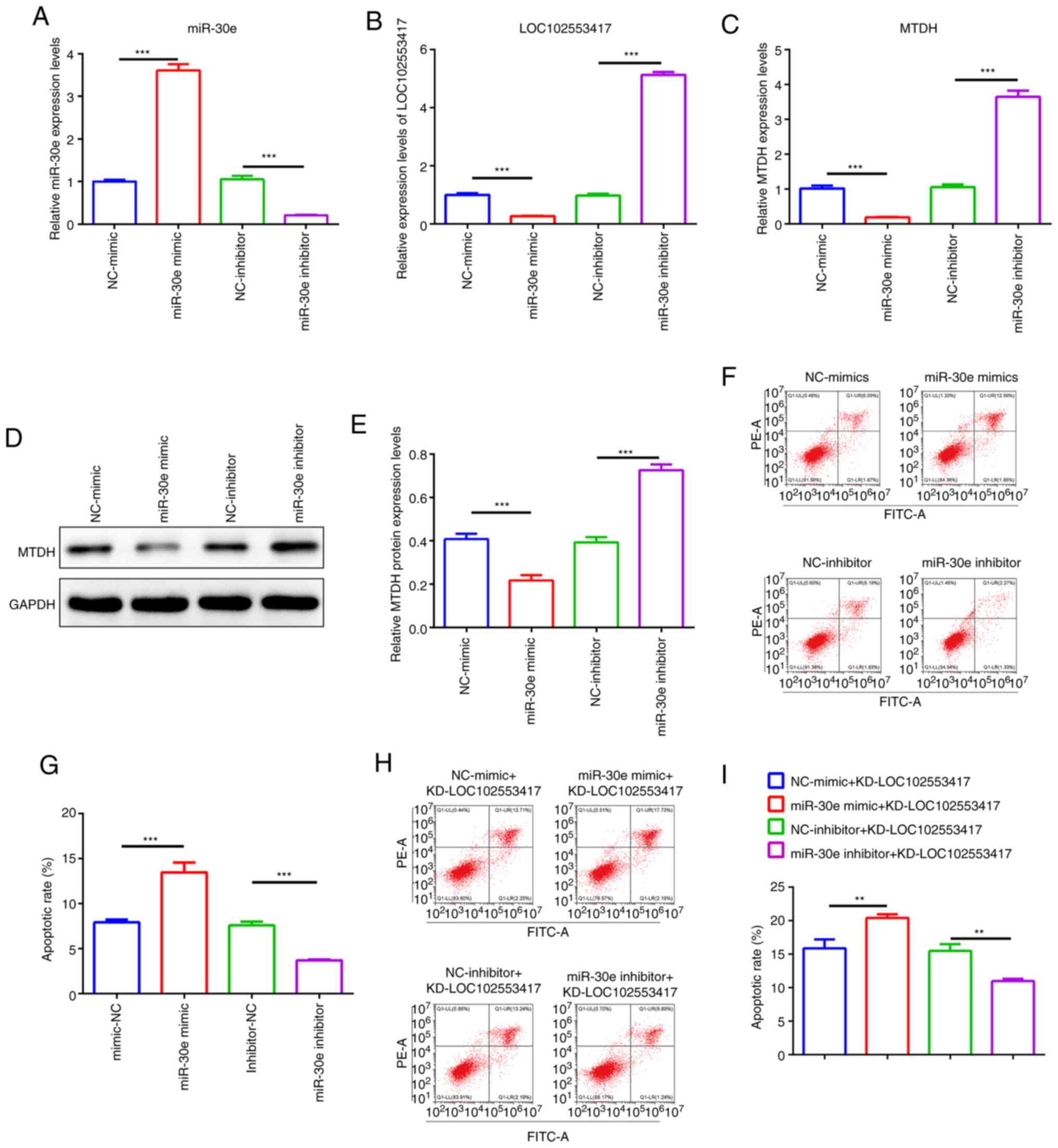
miR-30e inhibits LOC102553417 and MTDH
expression levels and expedites the apoptosis of HSCs in which
LOC102553417 is knocked down
The present study subsequently focused on whether
LOC102553417 could function via the miR-30e/MTDH axis. miR-30e
mimic, miR-30e inhibitor, NC-mimic and NC-inhibitor were
constructed and transfected into HSC-T6 cells, and the mRNA
expression levels of miR-30e, LOC102553417 and MTDH were quantified
via RT-qPCR. miR-30e expression levels in the miR-30e inhibitor
group were significantly reduced compared with those in the
NC-inhibitor group (P<0.001; Fig.
3A); however, LOC102553417 (Fig.
3B) and MTDH (Fig. 3C)
expression levels were significantly increased in the miR-30e
inhibitor group compared with in the NC-inhibitor group
(P<0.001). miR-30e mRNA expression levels were significantly
raised in the miR-30e mimic group compared with in the NC-mimic
group (P<0.001). Furthermore, the miR-30e mimic resulted in
significant reductions in LOC102553417 and MTDH mRNA expression
levels compared with in the NC-mimic group (P<0.001).
Western blotting demonstrated that MTDH protein
expression levels were significantly elevated in the miR-30e
inhibitor group compared with in the NC-inhibitor group (P<0.
001; Fig.3D and E); however, they
were significantly reduced in the miR-30e mimic group compared with
in the NC-mimic group (P<0.001) (Fig. 3D and E). Apoptosis analysis
demonstrated that the apoptotic rate was significantly suppressed
following miR-30e inhibitor transfection compared with that in the
NC-inhibitor group (P<0.001; Fig.
3F and G). Furthermore, a significant increase in apoptotic
rate was demonstrated after miR-30e mimic transfection compared
with the NC-mimic (P<0.001). Compared with LOC102553417
silencing (NC-inhibitor + KD-LOC102553417), simultaneous miR-30e
inhibitor and LOC102553417 silencing (miR-30e inhibitor +
KD-LOC102553417) demonstrated a significantly reduced apoptotic
rate (P<0.001), whereas simultaneous miR-30e mimic and
LOC102553417 silencing (miR-30e mimic + KD-LOC102553417)
demonstrated a significant enhancement in the apoptotic rate
compared with NC-mimic + KD-LOC102553417 (P<0.001) (Fig. 3H and I). These findings indicate
miR-30e inhibits LOC102553417 and MTDH expression levels and
expedites the apoptosis of HSCs in which LOC102553417 is knocked
down.
Discussion
HF is a dynamic process that manifests through
extracellular matrix accumulation triggered by chronic liver injury
of any etiology, including viral infection, alcoholic liver disease
and nonalcoholic steatohepatitis (12). Activation of HSCs is a crucial
event in HF, and their proliferation and apoptosis are highly
relevant to the initiation and development of HF (20). Previous studies have reported that
the proliferation of HSCs can drive the initiation of HF and
stimulating HSC apoptosis could ameliorate HF (21,22). The present study demonstrated that
silencing LOC102553417 contributed to the significant suppression
of HSC-T6 cell proliferation and the significant enhancement of
their apoptotic rate. Therefore, it could be hypothesized that
LOC102553417 may be a driver of HF, which agrees with a previous
finding that LOC102553417 was highly expressed in a rat model of HF
(17), and that targeting
LOC102553417 could be a potential strategy to treat HF.
The gene encoding MTDH is located on the long arm of
human chromosome 8 (8q22) in region 22, with a molecular weight of
~64 kDa and has previously been reported as an miR-30e-5p target
gene (23). MTDH can facilitate
hepatocarcinogenesis and cancer progression (24); in addition, abnormal expression of
miR-30a-5p can impede the proliferative function of liver cancer
cells and enhance their apoptosis via targeting of the
MTDH/PTEN/AKT pathway (25). MTDH
knockout has been shown to impair the proliferation and accelerate
the apoptosis of hepatocellular carcinoma cells via the PTEN/AKT
pathway (26); however, the
significance of MTDH in HF is rarely reported. Previous studies
have reported that miR-30e is abnormally expressed in liver injury
and hepatocellular carcinoma (27,28). Furthermore, hepatitis B virus X
protein may promote the development of liver fibrosis and hepatoma
through the downregulation of miR-30e, which targets prolyl
4-hydroxylase subunit α2 (29).
Moreover, human antigen R has been reported to be involved in
sphingosine 1-phosphate (S1P)-induced bone marrow mesenchymal stem
cell migration and can increase the stabilization of S1P receptor 3
mRNA by competing with miR-30e to regulate liver fibrosis (30). The present study demonstrated that
LOC102553417 loss-of-function expedited HSC apoptosis by inhibiting
MTDH expression through competitively binding to miR-30e, which
could be a key mechanism responsible for the action of LOC102553417
on HSCs, which drive HF. The combination of the miR-30e inhibitor
and KD-LOC102553417 significantly inhibited apoptosis. The results
of the present study demonstrated that the miR-30e inhibitor
significantly decreased the apoptotic rate and that silencing of
LOC102553417 significantly promoted apoptosis. Furthermore, miR-30e
could competitively bind to LOC102553417 and when the miR-30e
inhibitor inhibited the expression of miR-30e, thus reducing the
binding of miR-30e to LOC102553417, the expression of LOC102553417
was significantly increased and the cell apoptotic rate was
significantly reduced. Moreover, miR-30e inhibits LOC102553417 and
MTDH expression levels, and expedites the apoptosis of HSCs in
which LOC102553417 is knocked down. These results further indicate
that the three factors have a mutual regulatory relationship in the
apoptosis of hepatic stellate cells.
Akt is a serine/threonine protein kinase. Activation
of the Akt signaling pathway mainly depends on the activity of
PI3K, which can be stimulated by JAK1 and CD19. Akt phosphorylation
is essential for Akt activation and subsequent PI3K/Akt signaling
pathway activation (31–33). Akt phosphorylates downstream
targets to block cell apoptosis (34). A recent study reported that
activation of the PI3K/AKT/MDM2 signaling pathway could degrade
p53, inhibiting apoptosis (35).
p53 is a key tumor suppressor that suppresses cell proliferation
and induces apoptosis. It predominantly maintains body homeostasis
via diverse regulatory mechanisms, including mediating DNA repair,
impeding cell proliferation, stimulating apoptosis and boosting
metabolism (36,37). The results of the present study
demonstrated that LOC102553417 silencing significantly increased
apoptotic rate, significantly reduced Akt protein phosphorylation
and significantly upregulated p53 protein expression levels.
Furthermore, the present study demonstrated that LOC102553417
silencing could enhance Akt protein phosphorylation via the
miR-30e/MTDH signaling pathway. This may be the downstream pathway
of the LOC102553417-mediated miR-30e/MTDH axis affecting the
apoptosis of HSCs.
Notably, the present study has certain limitations.
Firstly, the significantly upregulated LOC102553417 expression in
clinical HF samples and its clinical significance were not
verified. Furthermore, the present study was only performed on rat
HSCs and needs to be further explored and verified in human cells
and clinical samples. Secondly, an HF cell model was not
established to validate the in vivo function and mechanism
of LOC102553417. Thirdly, pathway inhibitors were not utilized to
verify the downstream pathways of the LOC102553417/miR-30e/MTDH
axis; therefore, an in-depth exploration of their interaction is
warranted in the future.
In conclusion, the present study demonstrated that
silencing LOC102553417 reinforced HSC apoptosis via the
miR-30e/MTDH axis, which could be a crucial regulatory mechanism in
HF and may provide a theoretical basis for HF-targeted therapy.
Acknowledgements
Not applicable.
Funding
The present study was supported by The 2021 Young and
Middle-aged Backbone Talents Research Project of the Affiliated
Hospital of Youjiang Medical University for Nationalities (grant
no. Y20212603), the National Natural Science Foundation of China
(grant nos. 81460123 and 81960303), the Natural Science Foundation
of Guangxi (grant no. 2018GXNSFAA281187), the Clinic Medicine
Research Center of Hepatobiliary Diseases of Guangxi (grant no.
AD17129025), the University-level Scientific Research Project of
Youjiang Medical University for Nationalities (grant no.
yy2019ky008), the Self-funded Scientific Research Project of
Guangxi Zhuang Autonomous Region Health Committee (grant no.
20190953) and the Development Program Project of Baise City
Scientific Research and Technology (grant no. Encyclopedia
20213242).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WW wrote the draft and designed the experiments. CL
performed cell transfection. RH performed bioinformatics analysis
and reviewed and edited the manuscript. JH performed vector design
and RT-PCR and reviewed and edited the manuscript. XC performed
western blotting. LZ performed Cell Counting Kit-8 assay. JW
performed flow cytometry. YBD acquired funding, completed RIP and
dual-luciferase assay, edited the manuscript and provided critical
discussion. CFW analyzed data and acquired funding. WW and CFW
confirm the authenticity of all the raw data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Roehlen N, Crouchet E and Baumert TF:
Liver fibrosis: Mechanistic concepts and therapeutic perspectives.
Cells. 9:8752020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lee YA, Wallace MC and Friedman SL:
Pathobiology of liver fibrosis: A translational success story. Gut.
64:830–841. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Konyn P, Ahmed A and Kim D: Current
epidemiology in hepatocellular carcinoma. Expert Rev Gastroenterol
Hepatol. 15:1295–1307. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Harrison SA, Goodman Z, Jabbar A,
Vemulapalli R, Younes ZH, Freilich B, Sheikh MY, Schattenberg JM,
Kayali Z, Zivony A, et al: A randomized, placebo-controlled trial
of emricasan in patients with NASH and F1-F3 fibrosis. J Hepatol.
72:816–827. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kimura K, Ikoma A, Shibakawa M, Shimoda S,
Harada K, Saio M, Imamura J, Osawa Y, Kimura M, Nishikawa K, et al:
Safety, Tolerability, and preliminary efficacy of the anti-fibrotic
small molecule PRI-724, a CBP/β-catenin inhibitor, in patients with
hepatitis C virus-related cirrhosis: A single-center, open-label,
dose escalation phase 1 trial. EBioMedicine. 23:79–87. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Muir AJ, Levy C, Janssen HLA, Montano-Loza
AJ, Shiffman ML, Caldwell S, Luketic V, Ding D, Jia C, McColgan BJ,
et al: Simtuzumab for primary sclerosing cholangitis: Phase 2 study
results with insights on the natural history of the disease.
Hepatology. 69:684–698. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Friedman SL, Ratziu V, Harrison SA,
Abdelmalek MF, Aithal GP, Caballeria J, Francque S, Farrell G,
Kowdley KV, Craxi A, et al: A randomized, placebo-controlled trial
of cenicriviroc for treatment of nonalcoholic steatohepatitis with
fibrosis. Hepatology. 67:1754–1767. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Friedman SL: Hepatic fibrosis: Emerging
therapies. Dig Dis. 33:504–507. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Altamirano-Barrera A, Barranco-Fragoso B
and Méndez-Sánchez N: Management strategies for liver fibrosis. Ann
Hepatol. 16:48–56. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Higashi T, Friedman SL and Hoshida Y:
Hepatic stellate cells as key target in liver fibrosis. Adv Drug
Del Rev. 121:27–42. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Coll M, Perea L, Boon R, Leite SB,
Vallverdú J, Mannaerts I, Smout A, El Taghdouini A, Blaya D,
Rodrigo-Torres D, et al: Generation of hepatic stellate cells from
human pluripotent stem cells enables in vitro modeling of liver
fibrosis. Cell Stem Cell. 23:101–113.e107. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tsuchida T and Friedman SL: Mechanisms of
hepatic stellate cell activation. Nat Rev Gastroenterol Hepatol.
14:397–411. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fathizadeh H, Hayat SMG, Dao S, Ganbarov
K, Tanomand A, Asgharzadeh M and Kafil HS: Long non-coding RNA
molecules in tuberculosis. Int J Biol Macromol. 156:340–346. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Peng H, Wan LY, Liang JJ, Zhang YQ, Ai WB
and Wu JF: The roles of lncRNA in hepatic fibrosis. Cell Biosci.
8:632018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang K, Shi Z, Zhang M, Dong X, Zheng L,
Li G, Han X, Yao Z, Han T and Hong W: Silencing lncRNA Lfar1
alleviates the classical activation and pyoptosis of macrophage in
hepatic fibrosis. Cell Death Dis. 11:1322020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liao J, Zhang Z, Yuan Q, Liu Q, Kuang J,
Fang Y and Hu X: A lncRNA Gpr137b-ps/miR-200a-3p/CXCL14 axis
modulates hepatic stellate cell (HSC) activation. Toxicol Lett.
336:21–31. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gong Z, Tang J, Xiang T, Lin J, Deng C,
Peng Y, Zheng J and Hu G: Genome-wide identification of long
noncoding RNAs in CCl4-induced liver fibrosis via RNA sequencing.
Mol Med Rep. 18:299–307. 2018.PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chang TH, Huang HY, Hsu JB, Weng SL, Horng
JT and Huang HD: An enhanced computational platform for
investigating the roles of regulatory RNA and for identifying
functional RNA motifs. BMC Bioinformatics. 14 (Suppl 2):S42013.
View Article : Google Scholar
|
|
20
|
Wei S, Wang Q, Zhou H, Qiu J, Li C, Shi C,
Zhou S, Liu R and Lu L: miR-455-3p alleviates hepatic stellate cell
activation and liver fibrosis by suppressing HSF1 expression. Mol
Ther Nucleic Acids. 16:758–769. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Seki E and Brenner DA: Recent advancement
of molecular mechanisms of liver fibrosis. J Hepatobiliary Pancreat
Sci. 22:512–518. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shan L, Liu Z, Ci L, Shuai C, Lv X and Li
J: Research progress on the anti-hepatic fibrosis action and
mechanism of natural products. Int Immunopharmacol. 75:1057652019.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang Z, Qin H, Jiang B, Chen W, Cao W,
Zhao X, Yuan H, Qi W, Zhuo D and Guo H: miR-30e-5p suppresses cell
proliferation and migration in bladder cancer through regulating
metadherin. J Cell Biochem. 120:15924–15932. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sarkar D: AEG-1/MTDH/LYRIC in liver
cancer. Adv Cancer Res. 120:193–221. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li WF, Dai H, Ou Q, Zuo GQ and Liu CA:
Overexpression of microRNA-30a-5p inhibits liver cancer cell
proliferation and induces apoptosis by targeting MTDH/PTEN/AKT
pathway. Tumor Biol. 37:5885–5895. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li WF, Ou Q, Dai H and Liu CA:
Lentiviral-Mediated short hairpin RNA knockdown of MTDH inhibits
cell growth and induces apoptosis by regulating the PTEN/AKT
pathway in hepatocellular carcinoma. Int J Mol Sci. 16:19419–19432.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jiang X, Li W, Tan M, Guo P, Liu X, Pan X,
Yu D, Pang Y, Li D, Wang Q, et al: Identification of miRNAs
involved in liver injury induced by chronic exposure to cadmium.
Toxicology. 469:1531332022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ashmawy AM, Elgeshy KM, Abdel Salam ET,
Ghareeb M, Kobaisi MH, Amin HAA, Sharawy SK and Abdel Wahab AHA:
Crosstalk between liver-related microRNAs and Wnt/β-catenin pathway
in hepatocellular carcinoma patients. Arab J Gastroenterol.
18:144–150. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Feng GX, Li J, Yang Z, Zhang SQ, Liu YX,
Zhang WY, Ye LH and Zhang XD: Hepatitis B virus X protein promotes
the development of liver fibrosis and hepatoma through
downregulation of miR-30e targeting P4HA2 mRNA. Oncogene.
36:6895–6905. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chang N, Ge J, Xiu L, Zhao Z, Duan X, Tian
L, Xie J, Yang L and Li L: HuR mediates motility of human bone
marrow-derived mesenchymal stem cells triggered by sphingosine
1-phosphate in liver fibrosis. J Mol Med (Berl). 95:69–82. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yang CW, Hsu HY, Chang HY, Lee YZ and Lee
SJ: ‘Natural cardenolides suppress coronaviral replication by
downregulating JAK1 via a Na+/K+-ATPase
independent proteolysis’. Biochem Pharmacol. 180:1141222020.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Morbach H, Schickel JN, Cunningham-Rundles
C, Conley ME, Reisli I, Franco JL and Meffre E: CD19 controls
Toll-like receptor 9 responses in human B cells. J Allergy Clin
Immunol. 137:889–898.e6. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Molinaro A, Becattini B, Mazzoli A, Bleve
A, Radici L, Maxvall I, Sopasakis VR, Molinaro A, Bäckhed F and
Solinas G: Insulin-Driven PI3K-AKT signaling in the hepatocyte is
mediated by redundant PI3Kα and PI3Kβ activities and is promoted by
RAS. Cell Metab. 29:1400–1409.e5. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shariati M and Meric-Bernstam F: Targeting
AKT for cancer therapy. Expert Opin Investig Drugs. 28:977–988.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jamal SME, Alamodi A, Wahl RU, Grada Z,
Shareef MA, Hassan SY, Murad F, Hassan SL, Santourlidis S, Gomez
CR, et al: Melanoma stem cell maintenance and chemo-resistance are
mediated by CD133 signal to PI3K-dependent pathways. Oncogene.
39:5468–5478. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Patel KR and Patel HD: p53: An attractive
therapeutic target for cancer. Curr Med Chem. 27:3706–3734. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chao CCK: Mechanisms of p53 degradation.
Clin Chim Acta. 438:139–147. 2015. View Article : Google Scholar : PubMed/NCBI
|