Introduction
The GATA binding protein 3 (GATA3) is a member of
the GATA family of zinc finger transcription factors that bind to
the consensus 5′-(A/T)GATA(A/G)-3′ motif (1). Human GATA3 exhibits 85% amino acid
homology with human GATA1 in the DNA-binding domain, with no
homology elsewhere in the protein, located on the 10p15 band of the
human genome (2). GATA3, similar to
other GATA family members, plays important roles in vertebrate
embryonic organogenesis, including the mammary gland, sympathetic
nervous system, parathyroid gland, kidney, inner ear, skin and
T-cell lineages (3–9). Each GATA family member exhibits a
distinctive pattern of expression in tissues and cell lines. The
GATA3 protein is highly expressed in T-lymphoid cells and was
suggested to be involved in the regulation of T-cell receptor α-
and β-chain genes (2). GATA3 was
identified in the luminal cells of mammary ducts and the body cells
of terminal end buds, suggesting that GATA3 actively maintains
luminal epithelial differentiation in the adult mammary gland,
which suggests important implications in the pathogenesis of breast
cancer (10). The majority of
breast cancers arise from luminal epithelial cells; therefore,
GATA3 appears to control a set of genes involved in the
differentiation and proliferation of breast cancer cells. The
expression of GATA3 is strongly associated with estrogen receptor
(ER) expression in breast cancer (11) and there is accumulating evidence
that GATA3 may be used as a clinical marker to determine response
to hormonal therapy and refine the prognosis of breast cancer
patients (12,13). The GATA3 gene was recently
identified as a potential tumor marker and putative tumor
suppressor gene in breast cancer, whose expression may be
associated with a more fvorable prognosis and prolonged
disease-free survival in breast cancer patients (14). A meta-analysis reported that
GATA3 was one of the most significant genes exhibiting low
expression in invasive carcinomas of the breast with poor clinical
outcome, whereas low GATA3 expression was associated with a higher
histological grade, positive nodes, larger tumor size, negative ER
and progesterone receptor and HER2-neu overexpression (15).
To the best of our knowledge, microRNAs (miRNAs) may
act as tumor suppressors and oncogenes by genetic variations in the
3′ untranslated region (3′UTR) binding sites, regulating the
target-gene expression post-transcriptionally (16). Chou et al (17) demonstrated that GATA3 increased the
level of expression of miRNA (miR)-29b, which in turn repressed a
network of prometastatic microenvironmental components, including
angiopoietin-like 4, lysyl oxidase, matrix metalloproteinase 9 and
vascular endothelial growth factor A, through binding to specific
sequence motifs in their 3′UTR. The realisation that the
GATA3-miR-29b axis regulates the tumor microenvironment and
inhibits metastasis may open up novel possibilities for therapeutic
intervention in breast cancer. However, the role of genetic
variations in the miRNA binding sites of GATA3 has not been
fully elucidated. Therefore, we tested our hypothesis that the
GATA3 3′UTR variants may be associated with its mRNA
expression by performing a bioinformatics analysis and
genotype-phenotype association analysis based on the HapMap
database.
Materials and methods
Bioinformatics and selection of
polymorphisms
We identified the single-nucleotide polymorphisms
(SNPs) in the GATA3 gene and coding region by searching the
National Center for Biotechnology Information online database
(http://www.ncbi.nlm.nih.gov/SNP/). We
limited the SNPs to those with a minor allele frequency (MAF) of
>0.05 among different populations and used the SNP Function
Prediction bioinformatics tool (http://snpinfo.niehs.nih.gov/snpinfo/snpfunc.htm)
to predict the potential miRNA binding sites. We then calculated
the genotype distributions of all the selected GATA3 3′UTR
SNPs among different populations according to the database. In
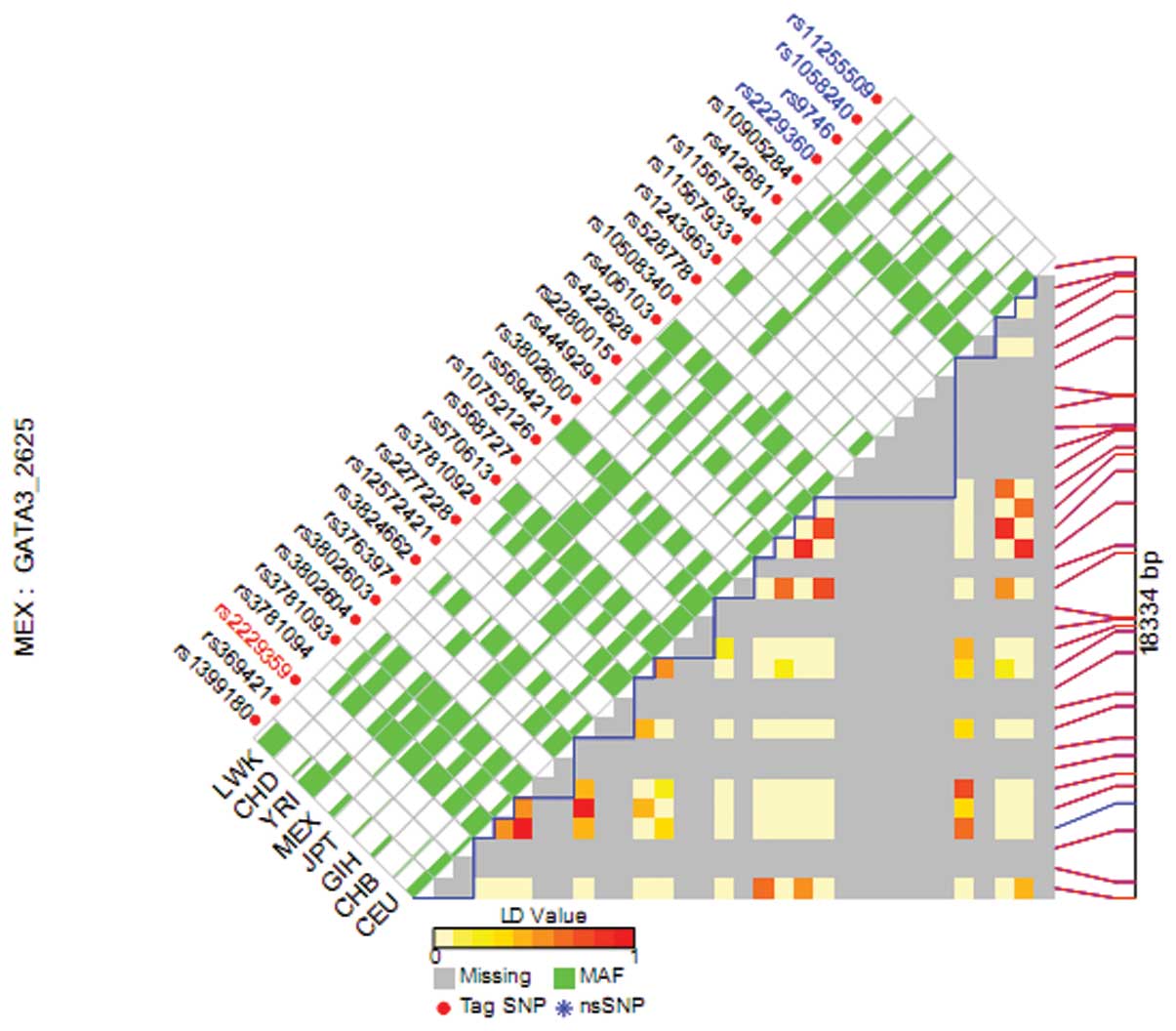
addition, the pairwise linkage disequilibrium (LD) values of all
the SNPs in the same gene were calculated and the SNPs not in LD
(r2<0.8) were selected. Subsequently, we plotted LD
maps of those SNPs in GATA3 gene with the LD TagSNP
Selection online program (http://snpinfo.niehs.nih.gov/snpinfo/snptag.htm).
Genotype and mRNA expression data of
lymphoblastoid cell lines from the HapMap database
We used the data on GATA3 genotypes and mRNA
levels available online (http://app3.titan.uio.no/biotools/tool.php?app=snpexp)
to analyse the genotype-phenotype association (18). The gene expression variation was
analysed by using genome-wide expression arrays (47,294
transcripts) from Epstein-Barr virus-transformed lymphoblastoid
cell lines from 270 HapMap individuals (128 females and 142 males)
(19). The genotyping data from the
HapMap phase II release 23 dataset consisted of 3.96 million SNP
genotypes from 270 individuals belonging to 4 populations (20). The SNPexp v1.2 web tool (Norwegian
PSC Research Center, Clinic for Specialized Surgery and Medicine,
Rikshospitalet, Oslo University Hospital, Oslo, Norway) was used to
analyse and visualize the correlation between HapMap genotypes and
gene expression levels. The probe GI_4503928-S, representing the
gene ‘GATA3’ was found in the file
‘illumina_Human_WG-6_array_content.csv’ and a correlation analysis
was then performed between the SNP genotype and expression levels
for the probe GI_4503928-S (additive model assumed).
Statistical methods
We analysed the SNP genotype and phenotype
correlation with the Chi-square test. The statistics test were
two-sided and P<0.05 was considered to indicate a statistically
significant difference.
Results
GATA3 3′UTR selected variants and
putative miRNA binding sites
In total, we identified 685 SNPs in the GATA3
gene region and 73 in the coding region (http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi).
Among these SNPs, 30 were located in the 3′UTR, of which 4
(rs2229360, rs58582188, rs9746 and rs1058240) exhibited a MAF of
>0.05. The only SNP with putative miRNA binding sites revealed
by SNP Function Prediction was rs1058240 (Table I). As presented in Table I, rs1058240 has three potential mRNA
binding sites, including hsa-miR-1299, hsa-miR-182 and hsa-miR-95.
We listed the genotype frequencies of 3 SNPs among different
populations. rs58582188 was excluded, as it was not found in the
database (Table II). We calculated
the pairwise LD values of all the SNPs in the GATA3 gene
with a MAF of >0.05 and selected the SNPs not in LD
(r2<0.8) to plot LD maps with the online SNP Function
Prediction bioinformatics tool. The color of each SNP spot changing
from red to white reflects the decrease in its D’ value (Fig. 1).
 | Table ISelected SNPs of 3′UTR and putative
miRNA binding sites. |
Table I
Selected SNPs of 3′UTR and putative
miRNA binding sites.
| SNPs | Alleles | MAF | Putative miRNA
binding sites |
|---|
| rs2229360 | C/T | 0.0845 | NA |
| rs58582188 | −/A/T | 0.0964 | NA |
| rs9746 | A/G | 0.1915 | NA |
| rs1058240 | A/G | 0.1488 | hsa-miR-1299,
hsa-miR-182, hsa-miR-95 |
 | Table IIFrequency distributions of selected
variables among different populations. |
Table II
Frequency distributions of selected
variables among different populations.
| Genotypes | European | Asian | African |
|---|
| rs2229360 |
| CC | 0.982 | 0.453 | 0.957 |
| CT | 0.018 | 0.453 | 0.043 |
| TT | 0.000 | 0.093 | 0.000 |
| T alleles | 0.009 | 0.320 | 0.022 |
| rs9746 |
| AA | 0.726 | 0.453 | 0.699 |
| AG | 0.257 | 0.453 | 0.265 |
| GG | 0.018 | 0.093 | 0.035 |
| G alleles | 0.0146 | 0.320 | 0.168 |
| rs1058240 |
| AA | 0.619 | 1.000 | 0.584 |
| AG | 0.336 | 0.000 | 0.363 |
| GG | 0.044 | 0.000 | 0.053 |
| G alleles | 0.212 | 0.000 | 0.235 |
GATA3 mRNA expression by genotype in
lymphoblastoid cell lines
For the mRNA expression of the GATA3 gene, we
used the available HapMap-cDNA expression database for the
correlation analysis of the GATA3 genotype and mRNA
expression level in Epstein-Barr virus-transformed lymphoblastoid
cell lines from 270 HapMap individuals. For rs2229360, 268 cell
lines with available values were collected. A total of 6 (2.2%)
cell lines had the TT genotype, 42 (15.7%) had the TC genotype and
220 (82.1%) had the CC genotype. After excluding one cell line with
unavailable values for rs1058240, 5 (1.9%) cell lines had the GG
genotype, 62 (23.0%) had the GA genotype and 202 (75.1%) had the AA
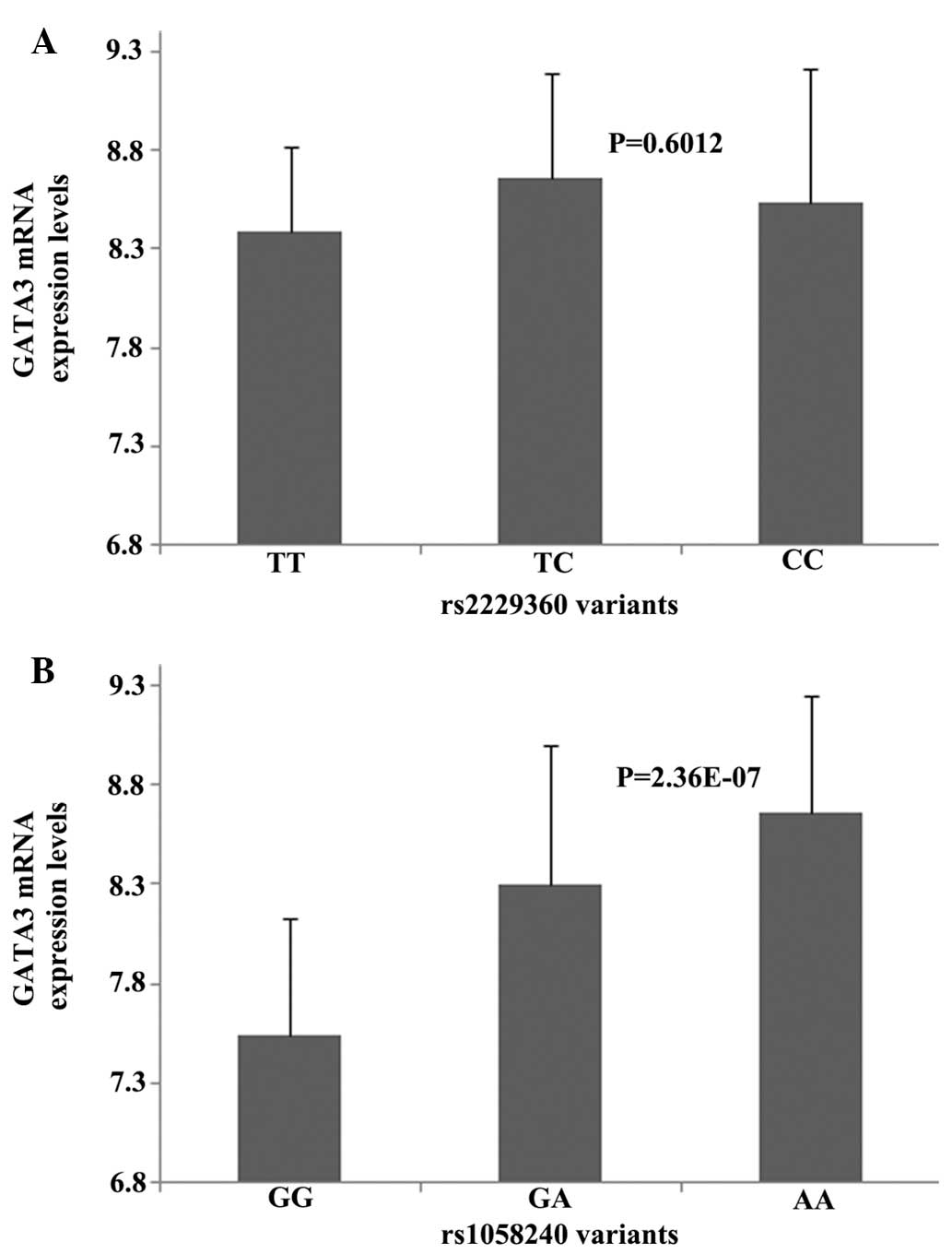
genotype. The GATA3 mRNA expression levels of the cell lines
with rs2229360 and rs1058240 are illustrated in Fig. 2. There was no significant difference
in GATA3 mRNA expression among the cell lines carrying
rs2229360 variants (P=0.6012; Fig.
2A). The AA genotype of rs1058240 exhibited a significantly
higher GATA3 mRNA expression level compared to the GG and GA
genotypes (P=2.36E-07; Fig.
2B).
Discussion
The zinc finger transcription factor GATA3 was first
identified in the early 1990s and is considered to be a marker of
luminal breast cancers. GATA3 was confirmed to be necessary for the
differentiation and maintenance of the luminal epithelium in the
adult mammary gland (10). Usary
et al (21) demonstrated
that mutations in the second zinc finger may affect DNA binding,
indicating its crucial role in the development and progression of
breast cancer. GATA3 was identified as one of the most
significantly mutated genes in breast cancer by whole-exome
sequencing and GATA3 gene mutations were identified in 4
patients with luminal tumors, including 3 previously unknown
frameshift mutations near the 3′-end of the coding sequence
(22). GATA3 mutations in
breast cancer may be associated with loss of DNA binding, aberrant
nuclear localization, decrease in transactivational activity and
alterations in invasiveness, but not cell proliferation (23). It is well known that miRNAs may
function as tumor suppressors and oncogenes through interactions
with the 3′UTR of their mRNA targets and may control the
target-gene expression post-transcriptionally (16). For example, GATA3 may promote
differentiation, suppress metastasis and alter the tumor
microenvironment in breast cancer by inducing miR-29b expression
(17). It was also demonstrated
that miR-30c was transcriptionally regulated by GATA3 in breast
tumors (24). However, the roles of
genetic variants in GATA3 gene 3′UTR and its
post-transcriptional regulation has not been fully elucidated. Our
data demonstrated that rs1058240 located in the GATA3 3′UTR
displays 3 putative miRNA binding sites by using bioinformatics
analysis; this SNP is significantly associated with the mRNA
expression level, suggesting it may be partly involved in
GATA3 post-transcriptional regulation. Our findings may
enable a better understanding of the roles miRNA variants in
GATA3 3′UTR play in its mRNA expression and open up novel
possibilities for therapeutic intervention in breast cancer.
In conclusion, our results indicated the vital role
of GATA3 variants in 3′UTR in the post-transcriptional
regulation of mRNA expression. However, the association of the
regulation of GATA3 transcription with variations in the
3′UTR requires further validation to facilitate the development of
novel therapeutic strategies.
Acknowledgements
This study was supported by grants from the National
Natural Science Foundation of China (no. 81272252) and the Natural
Science Foundation of Jiangsu Province (no. BK2011656).
References
|
1
|
Merika M and Orkin SH: DNA-binding
specificity of GATA family transcription factors. Mol Cell Biol.
13:3999–4010. 1993.PubMed/NCBI
|
|
2
|
Joulin V, Bories D, Eleouet JF, Labastie
MC, Chretien S, Mattei MG and Romeo PH: A T-cell specific TCR delta
DNA binding protein is a member of the human GATA family. EMBO J.
10:1809–1816. 1991.PubMed/NCBI
|
|
3
|
Asselin-Labat ML, Sutherland KD, Barker H,
et al: Gata-3 is an essential regulator of mammary-gland
morphogenesis and luminal-cell differentiation. Nat Cell Biol.
9:201–209. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lim KC, Lakshmanan G, Crawford SE, Gu Y,
Grosveld F and Engel JD: Gata3 loss leads to embryonic lethality
due to noradrenaline deficiency of the sympathetic nervous system.
Nat Genet. 25:209–212. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Grigorieva IV, Mirczuk S, Gaynor KU, et
al: Gata3-deficient mice develop parathyroid abnormalities due to
dysregulation of the parathyroid-specific transcription factor
Gcm2. J Clin Invest. 120:2144–2155. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Grote D, Souabni A, Busslinger M and
Bouchard M: Pax 2/8-regulated Gata 3 expression is necessary for
morphogenesis and guidance of the nephric duct in the developing
kidney. Development. 133:53–61. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
van der Wees J, van Looij MA, de Ruiter
MM, et al: Hearing loss following Gata3 haploinsufficiency is
caused by cochlear disorder. Neurobiol Dis. 16:169–178.
2004.PubMed/NCBI
|
|
8
|
Kaufman CK, Zhou P, Pasolli HA, et al:
GATA-3: an unexpected regulator of cell lineage determination in
skin. Genes Dev. 17:2108–2122. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ting CN, Olson MC, Barton KP and Leiden
JM: Transcription factor GATA-3 is required for development of the
T-cell lineage. Nature. 384:474–478. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kouros-Mehr H, Slorach EM, Sternlicht MD
and Werb Z: GATA-3 maintains the differentiation of the luminal
cell fate in the mammary gland. Cell. 127:1041–1055. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Voduc D, Cheang M and Nielsen T: GATA-3
expression in breast cancer has a strong association with estrogen
receptor but lacks independent prognostic value. Cancer Epidemiol
Biomarkers Prev. 17:365–373. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Parikh P, Palazzo JP, Rose LJ, Daskalakis
C and Weigel RJ: GATA-3 expression as a predictor of hormone
response in breast cancer. J Am Coll Surg. 200:705–710. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fang SH, Chen Y and Weigel RJ: GATA-3 as a
marker of hormone response in breast cancer. J Surg Res.
157:290–295. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bong PN, Zakaria Z, Muhammad R, et al:
Expression and mutational analysis of GATA3 in Malaysian breast
carcinomas. Malays J Pathol. 32:117–122. 2010.PubMed/NCBI
|
|
15
|
Mehra R, Varambally S, Ding L, et al:
Identification of GATA3 as a breast cancer prognostic marker by
global gene expression meta-analysis. Cancer Res. 65:11259–11264.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar
|
|
17
|
Chou J, Lin JH, Brenot A, Kim JW, Provot S
and Werb Z: GATA3 suppresses metastasis and modulates the tumour
microenvironment by regulating microRNA-29b expression. Nat Cell
Biol. 15:201–213. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Holm K, Melum E, Franke A and Karlsen TH:
SNPexp - A web tool for calculating and visualizing correlation
between HapMap genotypes and gene expression levels. BMC
Bioinformatics. 11:6002010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Stranger BE, Forrest MS, Dunning M, et al:
Relative impact of nucleotide and copy number variation on gene
expression phenotypes. Science. 315:848–853. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
International HapMap Consortium. The
International HapMap Project. Nature. 426:789–796. 2003. View Article : Google Scholar
|
|
21
|
Usary J, Llaca V, Karaca G, et al:
Mutation of GATA3 in human breast tumors. Oncogene. 23:7669–7678.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Banerji S, Cibulskis K, Rangel-Escareno C,
et al: Sequence analysis of mutations and translocations across
breast cancer subtypes. Nature. 486:405–409. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gaynor KU, Grigorieva IV, Allen MD, et al:
GATA3 mutations found in breast cancers may be associated with
aberrant nuclear localization, reduced transactivation and cell
invasiveness. Horm Cancer. 4:123–139. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bockhorn J, Dalton R, Nwachukwu C, et al:
MicroRNA-30c inhibits human breast tumour chemotherapy resistance
by regulating TWF1 and IL-11. Nat Commun. 4:13932013. View Article : Google Scholar : PubMed/NCBI
|