Introduction
PU.1 is a member of the Ezb transformation-specific
sequence family of transcription factors and is expressed mainly in
granulocytic, monocytic and B lymphoid cells (1). The downregulation of PU.1 was reported
to play a role in the pathogenesis of various hematological
malignancies, including acute myeloid leukemia (2), multiple myeloma (3) and acute lymphoblastic leukemia
(4). Furthermore, several studies
indicated that downregulation of PU.1 is required for erythroid
terminal differentiation (5).
We recently demonstrated that the effects of the DNA
methyltransferase inhibitor 5-aza-2′-deoxycytidine (5-azadC) are
correlated with PU.1 expression in PU.1-transgenic chronic myeloid
leukemia-derived K562 cells (6). We
revealed that therapeutic concentrations of 5-azadC induce
erythroid differentiation of these cells. PU.1 expression is
closely associated with the effect of this agent and sufficient
PU.1 levels were shown to accelerate erythroid differentiation and
apoptosis induced by 5-azadC (6).
Several studies indicated that low concentrations of
cytosine arabinoside (Ara-C) induce erythroid differentiation of
K562 cells (7,8). In this study, we investigated whether
the effects of Ara-C are also associated with the expression level
of PU.1.
Materials and methods
Cell culture of PU.1-knockdown K562 (K562
PU.1KD) and PU.1-overexpressing K562 (K562 PU.1OE) cells
K562 PU.1KD and K562 PU.1OE cells were previously
established in our laboratory (9,10) and
were employed in this study. Specifically, we used 2–10 and 3–10 as
K562 PU.1KD cells with vec 5 and vec 6 as their control cells and
H8 and A2 as K562 PU.1OE cells with vec 1 and vec 2 as their
control cells. The K562 PU.1KD cells were maintained in RPMI-1640
medium (Gibco-BRL, Rockville, MD, USA) containing 10%
heat-inactivated fetal bovine serum (HIFBS) and 1 μg/ml puromycin.
The K562 PU.1OE cells were grown in RPMI-1640 containing 10% HIFBS
and 400 μg/ml neomycin. All the cells were cultured under 5%
CO2 at 37°C in a humidified atmosphere.
Assessment of cell viability
The proportion of viable cells was determined by a
dye reduction assay involving
2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium
monosodium salt (WST-8; Dojindo, Kunamoto, Japan), which is a
modification of the
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay.
The effective dose (ED)50 values were calculated from
the data obtained by the cell growth assays. We selected 7
different Ara-C (Sigma, St. Louis, MO, USA) doses, with the group
without Ara-C serving as control, and performed the assays 5 days
after addition of Ara-C. The viable cells (%) were calculated as
the ratio of the absorbance (490 nm) of Ara-C-treated to that of
Ara-C-untreated cells in the cell growth assays. The calculated
ratios were analyzed using the website http://www.vector.co.jp/soft/win95/edu/se248471.html
and the ED50 values were obtained.
mRNA expression analysis
cDNAs were prepared from cells using reverse
transcription (Transcriptor First Strand cDNA synthesis kit; Roche
Diagnostics, Mannheim, Germany). Quantitative polymerase chain
reaction (qPCR) was performed using QuantiTect SYBR Green PCR
reagent (Qiagen, Miami, FL, USA) according to the manufacturer’s
protocol and using an Opticon Mini Real-Time PCR instrument
(Bio-Rad, Hercules, CA, USA) as previously described (11). The primer sequences were as follows:
α-globin: forward, 5′-AAGGTCGGCGCGCACGCT-3′ and reverse,
5′-CTCAGGTCGAAGTGCGGG-3′; human β-globin: forward,
5′-CTCATGGCAAGAAAGTGCTCG-3′ and reverse,
5′-AATTCTTTGCCAAAGTGATGGG-3′; and GAPDH: forward,
5′-GAAGGTGAAGGTCGGAGT-3′ and reverse; 5′-GAAGATGGTGATGGGATTTC-3′.
The thermal cycling conditions for α-globin were incubation at 95°C
for 15 min, followed by 35 cycles at 95°C for 15 sec, at 55°C for
15 sec and at 72°C for 15 sec. The thermal cycling conditions for
β-globin and GAPDH were incubation at 95°C for 15 min, followed by
35 cycles at 95°C for 30 sec, at 55°C for 30 sec and at 72°C for 45
sec. The copy number of each sample was calculated as previously
described (12).
Statistical analysis
Data are expressed as means ± standard error of the
mean and *P<0.05 was considered to indicate a
statistically significant difference, whereas
**P<0.01. Comparison of the means was performed with
the Student’s t-test (http://www.physics.csbsju.edu) between all the
controls and each group of K562 PU.1KD or K562 PU.1OE cells.
Results
ED50 of Ara-C is increased in
K562 PU.1KD cells
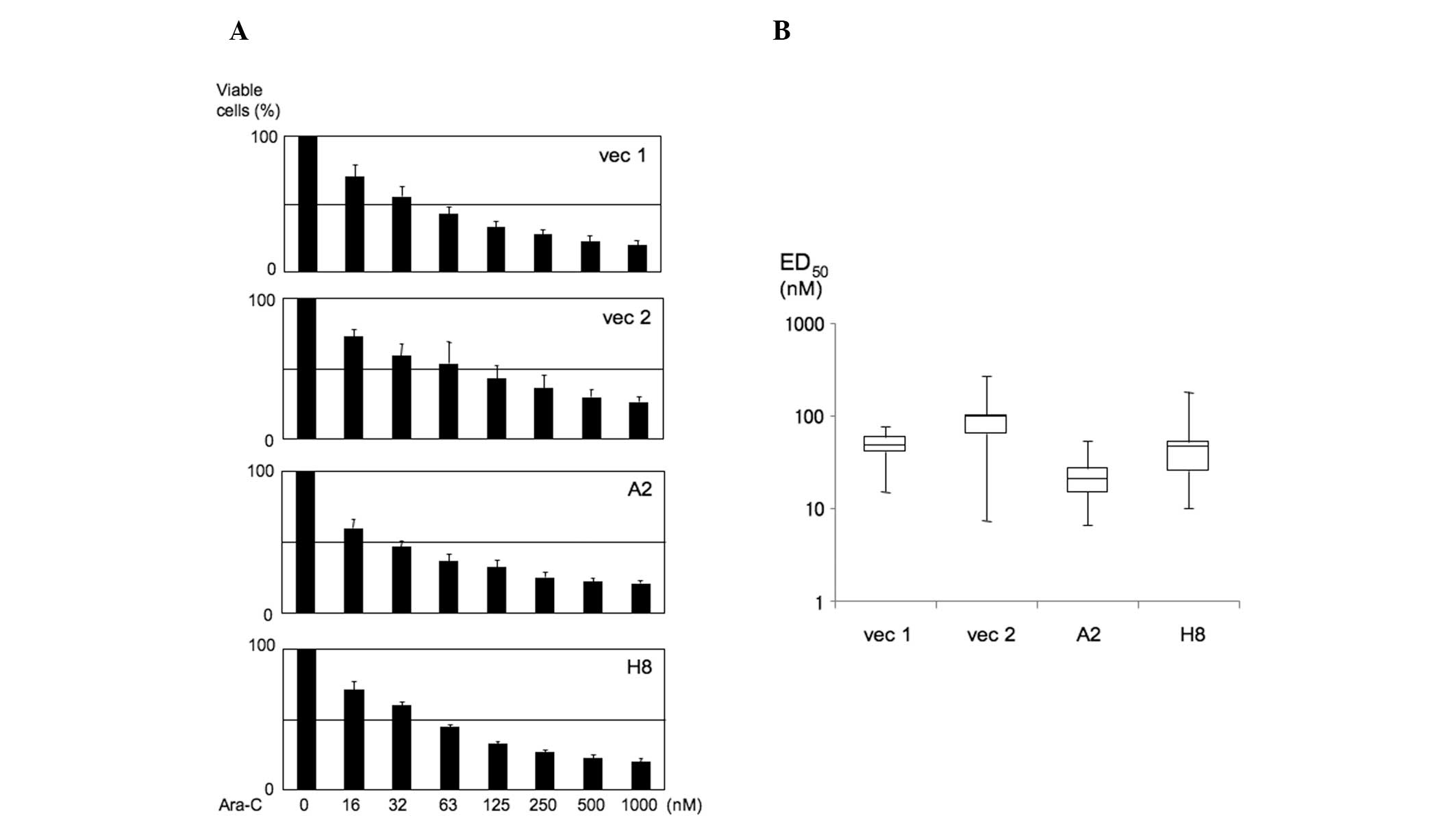
We first assessed the viability of K562 PU.1OE cells
in the presence of various concentrations of Ara-C, as depicted in
Fig. 1A. The range of the Ara-C
dose and time of incubation for erythroid differentiation was based
on the results published by Sztiller-Sikorska et al
(8). Based on the data presented in
Fig. 1A, we calculated
ED50 from the Ara-C effect on cell viability. As a
result, in K562 PU.1OE A2 cells, the ED50 value tended
to be decreased (6.55–53.2 nM, median 20.38 nM; P=0.099). However,
there was no difference between K562 PU.1OE H8 cells and their
controls [H8: 10.1–179.26 nM, median 45.7 nM; vec 1 (control):
14.9–76.24 nM, median 47.80 nM; vec 2 (control): 7.28–267.17 nM,
median 95.3 nM] (Fig. 1B).
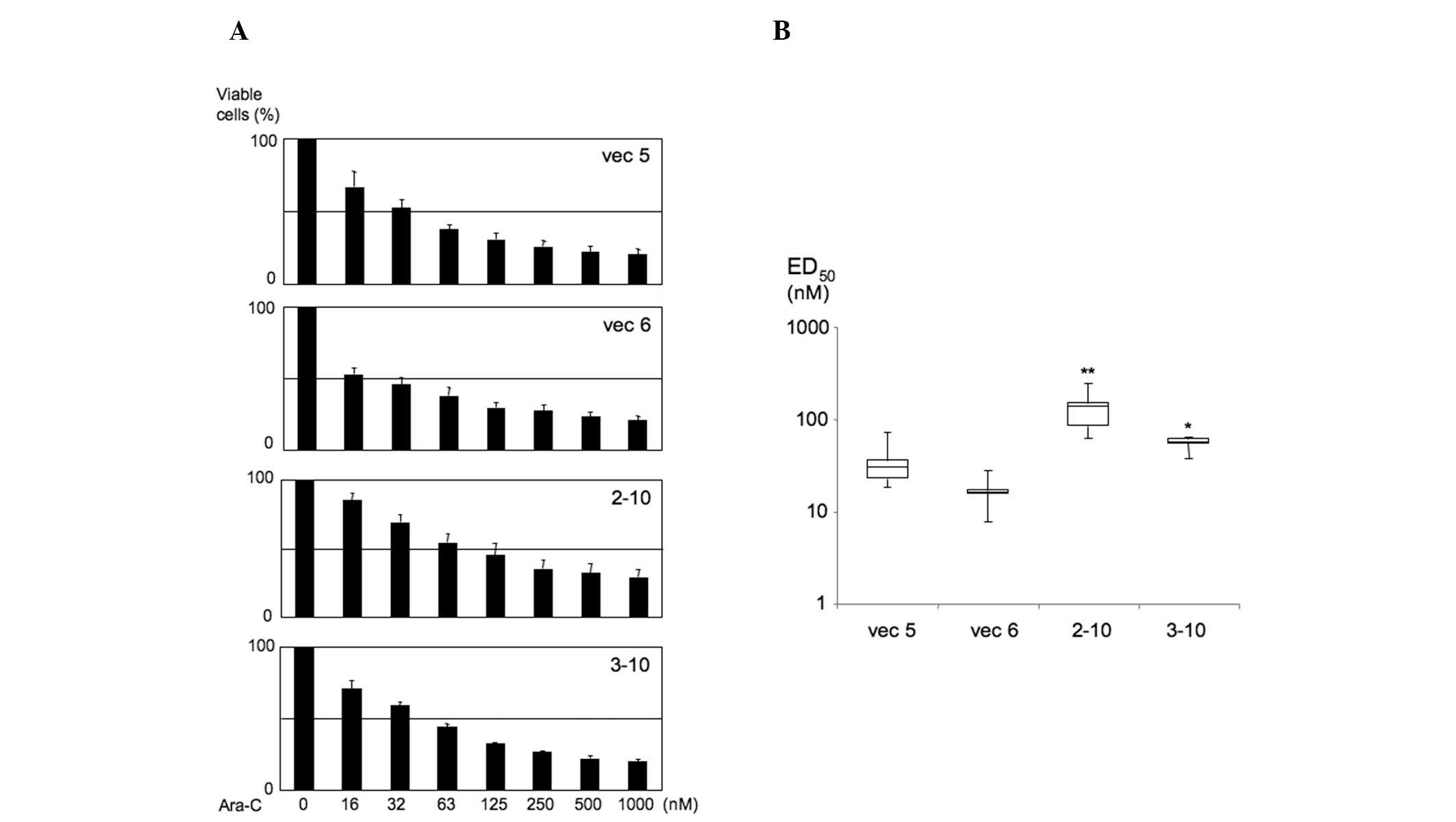
We then assessed cell viability (Fig. 2A) and ED50 value
(Fig. 2B) in K562 PU.1KD cells. In
contrast to the results of the K562 PU.1OE cells, the
ED50 value was distinctly increased in 2–10 cells
(62.75–250.59 nM, median 137.16 nM; P=0.0042), which exhibited the
lowest PU.1 expression among all cell lines (9,10),
compared to their controls (vec 5: 18.53–72.06 nM, median 30.47 nM;
vec 6: 7.88–28.46 nM, median 16.17 nM). Relatively high
ED50 values were also observed in K562 PU.1KD 3–10 cells
(37.56–65.02 nM, median 56.93 nM; P=0.025), which exhibited the
second lowest expression of PU.1 next to 2–10 cells. Collectively,
these findings demonstrated that the Ara-C effect on cell viability
was suppressed in K562 PU.1KD cells.
β-globin expression is significantly
disturbed in K562 PU.1KD cells following treatment with Ara-C
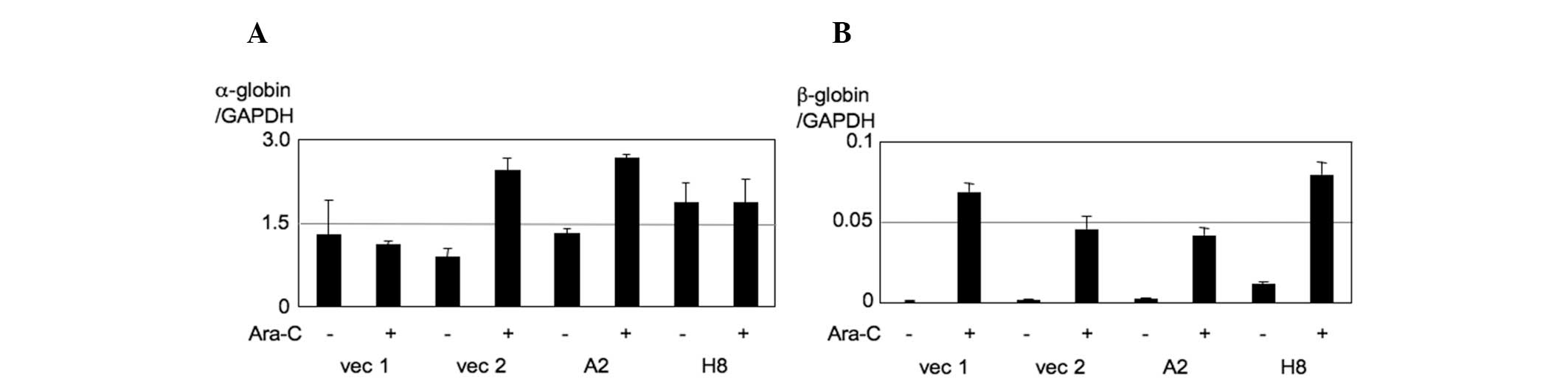
We then investigated whether the indicated dose of
Ara-C leads to the differentiation of K562 cells and whether the
effects of ED50 values are associated with the induction
of erythroid differentiation genes. For this purpose, we assessed
the expression of α-globin and β-globin by qPCR. In K562 PU.1OE
cells, the baseline expression of α-globin is extremely high
(α-globin copy number/GAPDH copy number: 1.1–1.8) and moderately
affected by the addition of Ara-C (Fig.
3A). By contrast, Ara-C significantly induces β-globin
expression, suggesting that measuring β-globin expression may be a
good marker to evaluate Ara-C-induced K562 cell differentiation.
However, there were no obvious differences between K562 PU.1OE
cells and their controls (Fig. 3B).
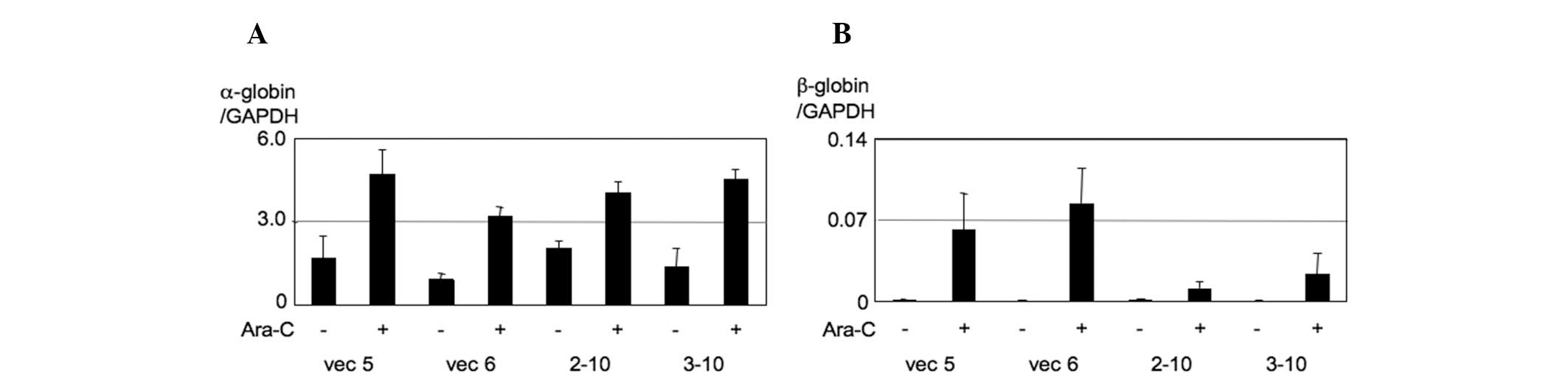
We then investigated these effects using K562 PU.1KD cells. As a
result, we observed that β-globin expression was markedly induced
by Ara-C and this induction was significantly disturbed in K562
PU.1KD 2–10 and 3–10 cells (Fig.
4). These findings were consistent with ED50 values,
suggesting that the Ara-C effect on cell viability is associated
with the effect on erythroid differentiation of K562 cells and
these effects are disturbed by the lack of expression of PU.1.
Discussion
It was previously reported that constitutive
upregulation of PU.1 is considered to be the main cause for the
inhibition in the differentiation process of murine erythroleukemia
(MEL) cells and PU.1 downregulation is required for terminal
erythroid differentiation (5,13–15).
However, several studies indicated a requirement for PU.1
expression for erythroid differentiation (6,16,17).
Back et al (16)
demonstrated a requirement of PU.1 expression in erythroid
differentiation. The authors of that study produced a line of
PU.1-deficient mice carrying a green fluorescent protein reporter
at this locus and revealed that PU.1-deficient fetal erythroid
progenitors lose their self-renewal capacity and undergo
proliferation arrest, premature differentiation and apoptosis
(16). Wontakal et al
(17) demonstrated that PU.1
regulates an extensive network of genes that constitute major
pathways for controlling the growth and survival of immature
myeloid cells. That study further revealed that fetal liver
erythroid progenitors, the earliest erythroid-committed cells, are
significantly reduced in mice with low PU.1 expression in
vivo (17). The findings of
these studies, including those of the present study, suggest that
sufficient expression of PU.1 is indispensable for erythroid
differentiation. As previously described (18), one possible explanation for this
discrepancy between studies employing MEL cells, is that K562 cells
express the endogenous ɛ-globin and γ-globin genes, but not the
adult stage-specific β-globin gene and, therefore, have been
considered as a model for embryonic-fetal stages of erythroid
development (19,20). Wontakal’s and Back’s studies
employed fetal liver erythroid progenitors from mice, which are
used for analyzing the embryonic-fetal stages, whereas MEL cells
are considered to be a model for fetal-adult development (20), which are employed in the majority of
the previous studies analyzing the functions of PU.1 during
erythroid differentiation (6,16,17).
The roles of PU.1 may differ during the different
stages of erythroid differentiation. Proper expression of PU.1 is
required for the differentiation of immature erythroid cells. We
hypothesized that, at least in certain hematological malignancies,
measuring the expression of PU.1 may be useful in predicting the
efficacy of low-dose chemotherapies in affecting erythroid
differentiation.
Acknowledgements
This study was supported in part by Grants-in-Aid
for Scientific Research (no. 23590687) from the Ministry of
Education, Science and Culture of Japan, the Takeda Science
Foundation and a foundation from Kitasato University School of
Allied Health Sciences (Grant-in-Aid for Research Project, no.
2013–1001). The authors would like to thank Dr Takashi Satoh for
helpful discussions.
References
|
1
|
Chen HM, Zhang P, Voso MT, et al:
Neutrophils and monocytes express high levels of PU.1 (Spi-1) but
not Spi-B. Blood. 85:2918–2928. 1995.PubMed/NCBI
|
|
2
|
Rosenbauer F, Wagner K, Kutok JL, et al:
Acute myeloid leukemia induced by graded reduction of a
lineage-specific transcription factor, PU.1. Nat Genet. 36:624–630.
2004. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pettersson M, Sundstrom C, Nilsson K and
Larsson LG: The hematopoietic transcription factor PU.1 is
downregulated in human multiple myeloma cell lines. Blood.
86:2747–2753. 1995.PubMed/NCBI
|
|
4
|
Sokalski KM, Li SK, Welch I, Cadieux-Pitre
HA, Gruca MR and DeKoter RP: Deletion of genes encoding PU.1 and
Spi-B in B cells impairs differentiation and induces pre-B cell
acute lymphoblastic leukemia. Blood. 118:2801–2808. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kihara-Negishi F, Suzuki M, Yamada T,
Sakurai T and Oikawa T: Impaired repressor activity and biological
functions of PU.1 in MEL cells induced by mutations in the
acetylation motifs within the ETS domain. Biochem Biophys Res
Commun. 335:477–484. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Aoyama S, Nakano H, Danbara M, Higashihara
M, Harigae H and Takahashi S: The differentiating and apoptotic
effects of 2-aza-5′-deoxycytidine are dependent on the PU.1
expression level in PU.1-transgenic K562 cells. Biochem Biophys Res
Commun. 420:775–781. 2012.PubMed/NCBI
|
|
7
|
Woessmann W, Zwanzger D and Borkhardt A:
ERK signaling pathway is differentially involved in erythroid
differentiation of K562 cells depending on time and the inducing
agent. Cell Biol Int. 28:403–410. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sztiller-Sikorska M, Jakubowska J, Wozniak
M, Stasiak M and Czyz M: A non-apoptotic function of caspase-3 in
pharmacologically-induced differentiation of K562 cells. Br J
Pharmacol. 157:1451–1462. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Iseki Y, Imoto A, Okazaki T, Harigae H and
Takahashi S: Identification of annexin 1 as a PU.1 target gene in
leukemia cells. Leuk Res. 33:1658–1663. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Imoto A, Okada M, Okazaki T, Kitasato H,
Harigae H and Takahashi S: Metallothionein-1 isoforms and vimentin
are direct PU.1 downstream target genes in leukemia cells. J Biol
Chem. 285:10300–10309. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Iseki Y, Nakahara M, Kubo M, Obata F,
Harigae H and Takahashi S: Correlation of PU.1 and signal
regulatory protein α1 expression in PU.1 transgenic K562 cells. Int
J Mol Med. 29:319–323. 2012.PubMed/NCBI
|
|
12
|
Takahashi S, Harigae H, Kameoka J, Sasaki
T and Kaku M: AML1B transcriptional repressor function is impaired
by the Flt3 internal tandem duplication. Br J Haematol.
130:428–436. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rao G, Rekhtman N, Cheng G, Krasikov T and
Skoultchi AI: Deregulated expression of the PU.1 transcription
factor blocks murine erythroleukemia cell terminal differentiation.
Oncogene. 14:123–131. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yamada T, Kondoh N, Matsumoto M, Yoshida
M, Maekawa A and Oikawa T: Overexpression of PU.1 induces growth
and differentiation inhibition and apoptotic cell death in murine
erythroleukemia cells. Blood. 89:1383–1393. 1997.PubMed/NCBI
|
|
15
|
Yamada T, Abe M, Higashi T, et al: Lineage
switch induced by overexpression of Ets family transcription factor
PU.1 in murine erythroleukemia cells. Blood. 97:2300–2307. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Back J, Dierich A, Bronn C, Kastner P and
Chan S: PU.1 determines the self-renewal capacity of erythroid
progenitor cells. Blood. 103:3615–3623. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wontakal SN, Guo X, Will B, et al: A large
gene network in immature erythroid cells is controlled by the
myeloid and B cell transcriptional regulator PU.1. PLoS Genet.
7:e10013922011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Takahashi S: Opposing role, depending on
the stage, of PU.1 during erythroid differentiation. J Blood Lymph.
2:e1082012. View Article : Google Scholar
|
|
19
|
Donze D, Townes TM and Bieker JJ: Role of
erythroid Kruppel-like factor in human gamma- to beta-globin gene
switching. J Biol Chem. 270:1955–1959. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wall L, Destroismaisons N, Delvoye N and
Guy LG: CAAT/enhancer-binding proteins are involved in beta-globin
gene expression and are differentially expressed in murine
erythroleukemia and K562 cells. J Biol Chem. 271:16477–16484. 1996.
View Article : Google Scholar : PubMed/NCBI
|