Introduction
Sirtuins have attracted significant interest in
aging studies over the last decade. Sirtuins have been shown to
regulate important biological pathways in bacteria, archaea and
eukaryotes by possessing NAD+-dependent histone
deacetylase, mono-ribosyltransferase and tumor suppressor activity
and by regulating gene expression or DNA repair (1–4). Of
the 7 human sirtuins identified, the human SIRT6 gene is
located on the minus strand of chromosome 19p13.3 and is available
in a single copy encoding a 355-amino acid protein (39.1 kDa)
(5). The SIRT6 protein localizes in
the nucleus (6) and is required for
DNA repair and maintenance of genomic stability in mammalian cells,
integrating stress signaling to activate the DNA repair machinery
in response to oxidative stress (7,8).
Knockdown of SIRT6 in human cells renders them prone to
chemically induced double-strand breaks (2,9). SIRT6
deficiency in mice has been shown to lead to the development of an
acute degenerative aging-like phenotype (10). Although studies on SIRT6
knockout mice reported a strong correlation between premature aging
and the absence of the SIRT6 protein (10), the reports on the overexpression of
sirtuin homologues on various model organisms have been
controversial, from no effect (11)
to expansion of lifespan only in male mice (12). Analyses at the mRNA and protein
level revealed SIRT6 expression in the majority of mouse and human
tissues, with particularly high protein levels in the thymus,
skeletal muscle and brain (13,14).
However, there is no sufficient data on SIRT6 promoter
methylation levels and their correlation with human longevity. To
address this issue, we investigated the SIRT6 promoter
methylation levels in 55 individuals of a wide age range by
bisulfite-quantitative polymerase chain reaction (qPCR). The aim of
this study was to elucidate the correlation between the percentage
of SIRT6 promoter methylation and age and investigate
whether it exhibits any association with longevity.
Materials and methods
Subjects and sample collection
This study included 55 individuals (34 females and
21 males), aged 9–95 years, divided into 8 age groups with 10-year
intervals, with the exception of the 1st (9–19) and
the 8th groups (80–95 years). Blood samples were collected at the
Istanbul University Medico-Social Center, Turkey, with the written
consent of the individuals who completed a form indicating their
background regarding gender, age, inherited diseases, cancer,
exposure to chemical or radioactive agents, permanent use of
medicines, smoking habits and presence in the family of individuals
aged >90 years. Genomic DNA was extracted from the blood samples
using the High Pure PCR Template Preparation kit (Roche Diagnostics
GmbH, Mannheim, Germany).
The procedures followed were in accordance with the
current ethical standards.
Primer selection
The promoter sequence for SIRT6 was extracted
and combined from Transcriptional Regulatory Element Database by
Cold Spring Harbor Laboratory, Eukaryotic Promoter Database by the
Swiss Institute of Bioinformatics and Promoter Controls by
SwitchGear Genomics. The CpG islands were identified with the CpG
Island Searcher (GC=66.2%; http://cpgislands.usc.edu). The primers were designed
specifically for bisulfite-converted DNA with the MethPrimer (v1.1
beta) program (http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi).
The primer sequences were 5′-TTAATAAGGGAAATTTTATTGTTTT-3′ for the
forward primer and 5′-CTAACCTCAATACCC CCTAATATTC-3′ for the reverse
primer targeting a 212-bp region on the SIRT6 promoter.
Bisulfite-qPCR
A total of 500 ng of the extracted genomic DNAs were
bisulfite-converted by the EZ DNA Methylation™ kit (D5002; Zymo
Research, Irvine, CA, USA) according to the manufacturer’s
recommendations. The converted DNA samples were dissolved in 15 μl
elution buffer. The concentration of the eluted DNA was measured by
NanoDrop (2000c; Thermo Scientific, Waltham, MA, USA) as RNA due to
the single-stranded nature and uracil content of the
bisulfite-converted DNA. The extracted DNAs were diluted to a
concentration of 20 ng/μl. The specificity of the primers was
confirmed by agarose gel electrophoresis following bisulfite-PCR
using 20 ng sample DNA. To assess the methylation level of the
region of interest, fully methylated (D5015; Zymo Research) and
demethylated (EpiTect® Control DNA, 59665; Qiagen,
Hilden, Germany) bisulfite-converted control DNAs were mixed
accordingly to obtain 100, 80, 60, 20 and 10% methylated standards.
A total of 10 ng of standards and sample DNAs were used for qPCR
(CFX96; Bio-Rad, Hercules, CA, USA) along with 0.5 μM primers, 0.25
mM dNTP, 1× SYBR-Green I (Bioline, Taunton, MA, USA) and 0.5 unit
of ZymoTaq™ DNA polymerase (E2002; Zymo Research) in a total volume
of 10 μl. The PCR conditions were as follows: initial denaturation
at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C
for 30 sec, annealing at 57°C for 30 sec and extension with a plate
reading step at 72°C for 1 min followed by a melting curve analysis
step from 65°C to 95°C by 0.5°C increments for 5 sec. All the
samples were examined in double replicates and the standard DNAs in
triple replicates. The Ct values for the standard DNAs
were used to generate a standard curve. The methylation levels of
all the subjects were calculated according to the standard curve
formula. The average methylation status of the age groups and
standard deviations were calculated.
Statistical analysis
The significance levels of the probable association
of SIRT6 promoter methylation rates with gender, smoking and
longevity were calculated using non-parametric Kruskal-Wallis
one-way analysis of variance. P<0.05 was considered to indicate
a statistically significant difference.
Results
Bisulfite-qPCR
The concentrations of the extracted
bisulfite-converted DNAs were measured to be 23.5–39.1 ng/μl and
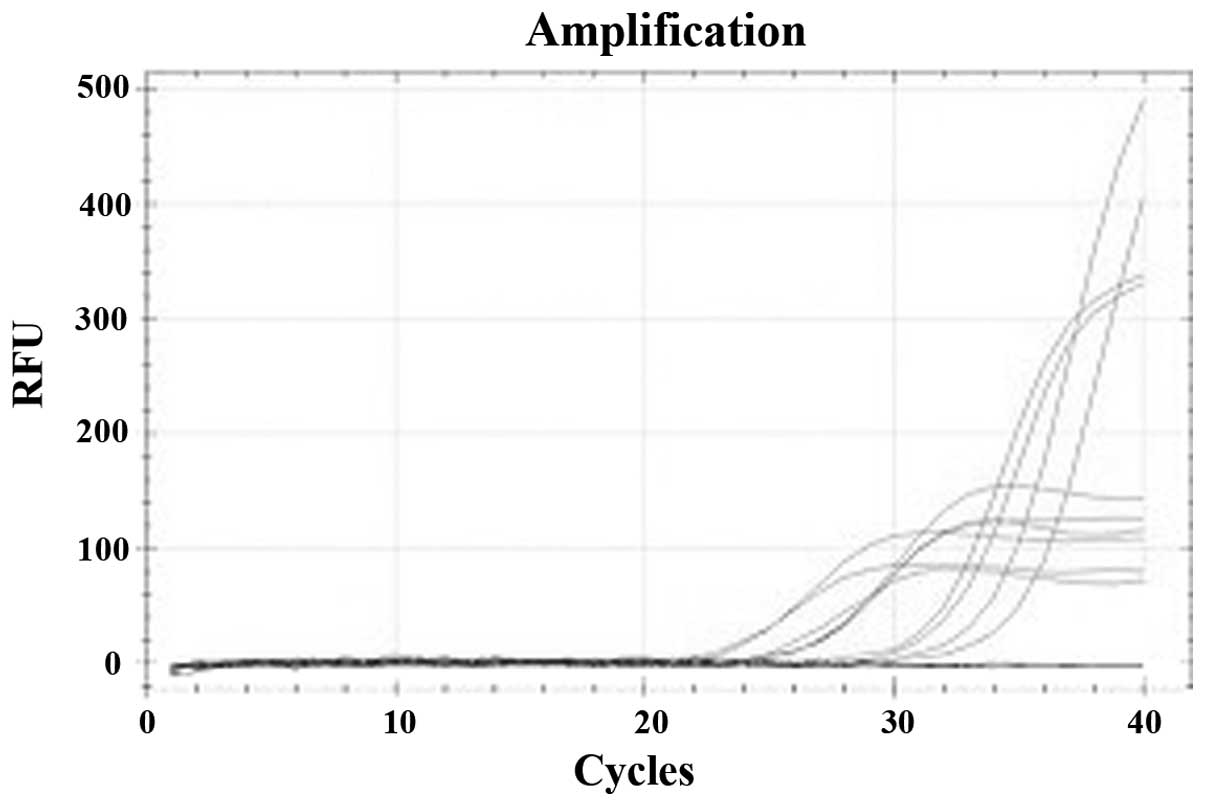
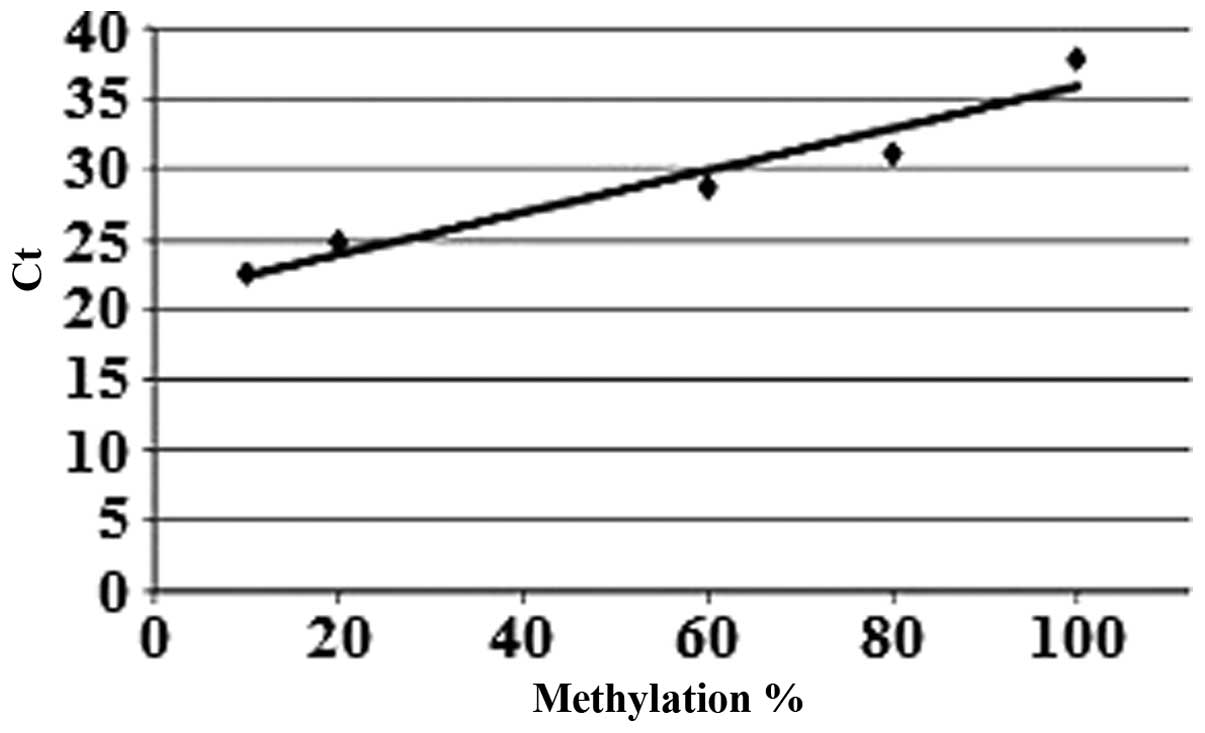
were diluted to 20 ng/μl. Following qPCR analysis, a standard curve
was generated with the mean Ct values of the standards
(Figs. 1 and 2). The SIRT6 promoter methylation
levels of the subjects were calculated using their mean
Ct values according to the standard curve formula: y =
0.1493x + 20.966
The methylation levels and information on the
subjects are presented in Table
I.
 | Table IEvaluation of the subjects. |
Table I
Evaluation of the subjects.
| Group no. | Subject no. | Gender | SIRT6 promoter
methylation (%) | Age (years) | Permanent medical
treatment
+ | Smoking
habit
+ | Extended longevity in
the family
+ |
|---|
| 1 | 1-1 | Male | 66.74 | 9 | | | X |
| 1-2 | Female | 65.25 | 9 | | | |
| 2-1 | Male | 37.18 | 11 | | | X |
| 2-2 | Female | 46.22 | 15 | | | X |
| 2-3 | Male | 15.63 | 15 | | | |
| 2-4 | Female | 15.10 | 17 | X | | X |
| 2-5 | Female | 56.34 | 19 | | | X |
| 2 | 3-1 | Male | 62.76 | 20 | | | |
| 3-2 | Male | 53.97 | 24 | | | |
| 3-3 | Female | 78.08 | 22 | | | X |
| 3-4 | Male | 67.40 | 24 | | X | X |
| 3-5 | Female | 52.57 | 22 | X | | |
| 3-6 | Male | 59.40 | 23 | | X | |
| 3-7 | Female | 68.00 | 20 | | | X |
| 3-8 | Female | 63.75 | 24 | X | | |
| 3-9 | Male | 63.39 | 24 | X | | X |
| 3 | 4-1 | Male | 70.42 | 32 | | | X |
| 4-2 | Female | 59.46 | 32 | X | | X |
| 4-3 | Male | 62.38 | 32 | | X | |
| 4-4 | Female | 67.02 | 32 | X | X | X |
| 4-5 | Male | 71.41 | 32 | X | | |
| 4-6 | Male | 65.43 | 32 | | X | X |
| 4-7 | Female | 65.13 | 33 | X | | X |
| 4-8 | Female | 64.85 | 34 | X | | |
| 4-9 | Female | 61.03 | 34 | | | |
| 4 | 5-1 | Female | 45.98 | 47 | | | X |
| 5-2 | Female | 52.22 | 45 | | | X |
| 5-3 | Male | 65.78 | 44 | | | |
| 5-4 | Female | 78.19 | 42 | X | | |
| 5-5 | Female | 69.98 | 45 | X | | |
| 5-6 | Female | 77.30 | 42 | | | |
| 5-7 | Male | 73.96 | 41 | | X | X |
| 5 | 6-1 | Female | 67.91 | 51 | | | X |
| 6-2 | Female | 59.96 | 51 | X | | |
| 6-3 | Female | 74.64 | 52 | X | X | |
| 6-4 | Female | 68.55 | 52 | X | X | X |
| 6-5 | Male | 76.36 | 53 | X | | |
| 6-6 | Female | 68.26 | 59 | X | | |
| 6 | 7-1 | Female | 65.19 | 63 | X | | |
| 7-2 | Female | 75.60 | 65 | X | | |
| 7-3 | Male | 74.31 | 67 | X | | |
| 7-4 | Male | 72.99 | 65 | X | | X |
| 7-5 | Female | 56.70 | 64 | X | | X |
| 7 | 8-1 | Female | 64.05 | 71 | X | | X |
| 8-2 | Female | 62.30 | 72 | X | | |
| 8-3 | Female | 60.05 | 73 | X | | X |
| 8-4 | Female | 52.22 | 75 | X | | |
| 8-5 | Male | 65.51 | 75 | X | | |
| 8 | 9-1 | Male | 39.87 | 80 | X | | |
| 9-2 | Female | 31.51 | 83 | X | | X |
| 9-3 | Female | 39.11 | 83 | X | | |
| 9-4 | Male | 35.17 | 88 | X | | X |
| 9-5 | Female | 86.39 | 88 | | | X |
| 10-1 | Female | 66.25 | 91 | X | | X |
| 10-2 | Male | 66.74 | 95 | X | | X |
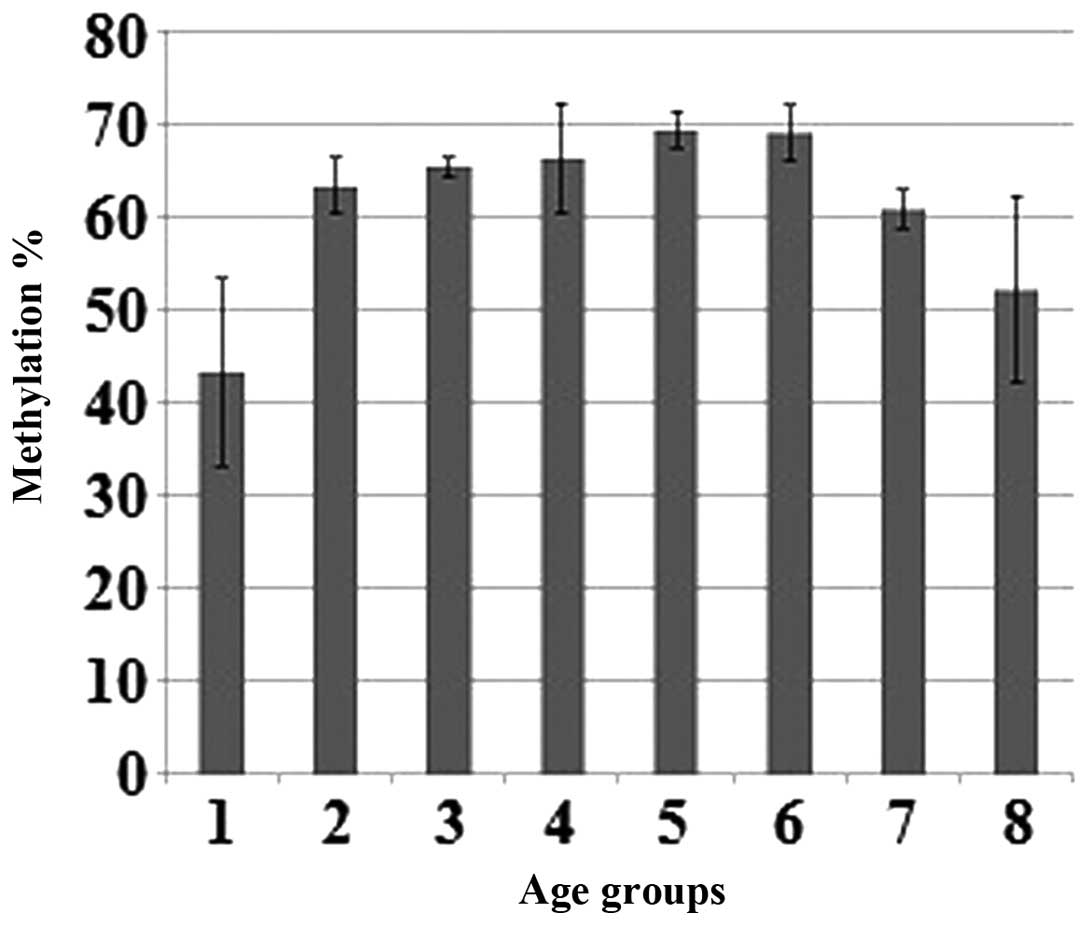
The average methylation percentages in different age
groups were calculated. The methylation of the SIRT6
promoter was found to be 43.21% in the 9–19 age group, an average
of 65.63% in the 20–79 age group and 52.15% in the 80–95 age group
(Fig. 3).
Statistical analysis
The Kruskal-Wallis test confirmed that the
methylation differences between at least two age groups were
significant (P=0.011). However, we were unable to identify a
significant correlation between methylation status and gender
(P=0.806), smoking (P=0.180), family history of cancer (P=0.504),
exposure to chemical or radioactive agents (P=0.085), or extended
longevity in the family (P=0.556).
Discussion
In this study, we determined the promoter
methylation status of the human SIRT6 gene in lymphocytes of
55 subjects aged 9–95 years. Our data indicated that the promoter
was highly methylated between 20 and 79 years. The regulatory
association of methylation of CpG islands in the repression of gene
expression was previously established (15). However, there was no significant
correlation with longevity, gender, permanent medical treatment or
smoking observed in our study. These results are consistent with
those reported by Michishita et al (16), who demonstrated that the
overexpression of SIRT6 in fibroblasts or epithelial cells
was not associated with a lifespan extension compared to normal
cells.
There are currently no available data on the
methylation levels of the human SIRT6 promoter. However, Ren
et al (17) demonstrated
that SIRT6 mRNA levels in porcine brain decreased with age,
which is an indicator of a possible increase in promoter
methylation. By contrast, SIRT6 protein levels were found to be
increased during aging in diabetic mice (18) and following exercise in young and
older mice (19). There is also
accumulating evidence that SIRT6 is regulated in a circadian
fashion. It was proven that NAD+ levels and SIRT1
activity change according to a circadian rhythm (20,21).
SIRT6 is a NAD+-dependent enzyme and is also positively
regulated by SIRT1 (22).
Therefore, it is possible that SIRT6 is also regulated in a
circadian manner. Marquardt et al (23) demonstrated that changing the SIRT6
expression in mice also changed the expression of certain genes
related to the circadian rhythm.
SIRT6 is a chromatin-associated protein expressed in
the majority of tissues. To gain insight into SIRT6 function, a
characterization of the subcellular and tissue distribution
patterns of its expression is required. Apart from age, SIRT6
expression or activity may change during the day, before and after
meals or before and after exercise (19,24,25).
In summary, we demonstrated that promoter methylation of human
SIRT6 is detectable by bisulfite-qPCR, but there was no
correlation with longevity. Further research and clinical studies
are required to fully elucidate the mechanisms underlying the
process of aging.
Acknowledgements
This study was supported by the Istanbul University
Department of Scientific Research Project (grant no. 21953).
References
|
1
|
Kawahara TL, Michishita E, Adler AS, et
al: SIRT6 links histone H3 lysine 9 deacetylation to
NF-kappaB-dependent gene expression and organismal life span. Cell.
136:62–74. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
McCord RA, Michishita E, Hong T, et al:
SIRT6 stabilizes DNA-dependent protein kinase at chromatin for DNA
double-strand break repair. Aging (Albany NY). 1:109–121.
2009.PubMed/NCBI
|
|
3
|
Michishita E, McCord RA, Berber E, et al:
SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric
chromatin. Nature. 452:492–496. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sebastian C, Zwaans BM, Silberman DM, et
al: The histone deacetylase SIRT6 is a tumor suppressor that
controls cancer metabolism. Cell. 151:1185–1199. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mahlknecht U, Ho AD and Voelter-Mahlknecht
S: Chromosomal organization and fluorescence in situ
hybridization of the human sirtuin 6 gene. Int J Oncol. 28:447–456.
2006.PubMed/NCBI
|
|
6
|
Tennen RI, Berber E and Chua KF:
Functional dissection of SIRT6: identification of domains that
regulate histone deacetylase activity and chromatin localization.
Mech Ageing Dev. 131:185–192. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mao Z, Hine C, Tian X, et al: SIRT6
promotes DNA repair under stress by activating PARP1. Science.
332:1443–1446. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cardus A, Uryga AK, Walters G and
Erusalimsky JD: SIRT6 protects human endothelial cells from DNA
damage, telomere dysfunction, and senescence. Cardiovasc Res.
97:571–579. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kaidi A, Weinert BT, Choudhary C and
Jackson SP: Human SIRT6 promotes DNA end resection through CtIP
deacetylation. Science. 329:1348–1353. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mostoslavsky R, Chua KF, Lombard DB, et
al: Genomic instability and aging-like phenotype in the absence of
mammalian SIRT6. Cell. 124:315–329. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Burnett C, Valentini S, Cabreiro F, et al:
Absence of effects of Sir2 overexpression on lifespan in C.
elegans and Drosophila. Nature. 477:482–485. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kanfi Y, Naiman S, Amir G, et al: The
sirtuin SIRT6 regulates lifespan in male mice. Nature. 483:218–221.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Moschen AR, Wieser V, Gerner RR, et al:
Adipose tissue and liver expression of SIRT1, 3, and 6 increase
after extensive weight loss in morbid obesity. J Hepatol.
59:1315–1322. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lee OH, Kim J, Kim JM, et al: Decreased
expression of sirtuin 6 is associated with release of high mobility
group box-1 after cerebral ischemia. Biochem Biophys Res Commun.
438:388–394. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kundu TK and Rao MR: CpG islands in
chromatin organization and gene expression. J Biochem. 125:217–222.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Michishita E, Park JY, Burneskis JM,
Barrett JC and Horikawa I: Evolutionarily conserved and
nonconserved cellular localizations and functions of human SIRT
proteins. Mol Biol Cell. 16:4623–4635. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ren Y, Shan TZ, Zhu LN, Wu T, Guo J and
Wang YZ: Effect of breed on the expression of sirtuins (Sirt1–7 and
antioxidant capacity in porcine brain. Animal. 7:1994–1998.
2013.
|
|
18
|
Ghiraldini FG, Crispim AC and Mello ML:
Effects of hyperglycemia and aging on nuclear sirtuins and DNA
damage of mouse hepatocytes. Mol Biol Cell. 24:2467–2476. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Koltai E, Szabo Z, Atalay M, et al:
Exercise alters SIRT1, SIRT6, NAD and NAMPT levels in skeletal
muscle of aged rats. Mech Ageing Dev. 131:21–28. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nakahata Y, Sahar S, Astarita G, Kaluzova
M and Sassone-Corsi P: Circadian control of the
NAD+salvage pathway by CLOCK-SIRT1. Science.
324:654–657. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bellet MM and Sassone-Corsi P: Mammalian
circadian clock and metabolism - the epigenetic link. J Cell Sci.
123:3837–3848. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim HS, Xiao C, Wang RH, et al:
Hepatic-specific disruption of SIRT6 in mice results in fatty liver
formation due to enhanced glycolysis and triglyceride synthesis.
Cell Metab. 12:224–236. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Marquardt JU, Fischer K, Baus K, et al:
Sirtuin-6-dependent genetic and epigenetic alterations are
associated with poor clinical outcome in hepatocellular carcinoma
patients. Hepatology. 58:1054–1064. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ban N, Ozawa Y, Inaba T, Miyake S,
Watanabe M, Shinmura K and Tsubota K: Light-dark condition
regulates sirtuin mRNA levels in the retina. Exp Gerontol.
48:1212–1217. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kanfi Y, Shalman R, Peshti V, et al:
Regulation of SIRT6 protein levels by nutrient availability. FEBS
Lett. 582:543–548. 2008. View Article : Google Scholar : PubMed/NCBI
|