Introduction
Cancer is one of the leading causes of mortality and
has become a worldwide public health problem (1). The exact mechanism of carcinogenesis
remains to be fully elucidated (2).
Previous studies have indicated that the contribution of genetic
variation to cancer development and progression has become clearer
(2,3). However, the identification of
susceptibility genes is problematic and the majority of the
associations have not been replicated due to various problems, such
as significant heterogeneity of the disease.
Obesity has been found to be associated with an
increased risk of cancer. Leptin (LEP, also known as OB for obese)
is an adipocyte-derived hormone that is mainly produced by white
adipose tissue to regulate appetite and weight, body metabolism and
reproductive functions, together with the leptin receptor (LEPR)
(4). The LEP gene, located
at chromosome 7q31.3, encodes a 16-kDa protein that has been
consistently shown to be associated with endocrinological
metabolism (5). Leptin has been
previously indicated to contribute to serum insulin levels and the
development of type 2 diabetes (6),
and to be involved in the pathophysiology of obesity (7,8) and
carcinogenesis (9–14). In addition to regulating body
weight, leptin also affects reproduction, angiogenesis,
hematopoiesis and immune processes (15). There is evidence indicating that
leptin may play a critical role in the initiation and progression
of human cancers (16).
A number of studies have investigated the possible
association between the LEPR Lys656Asn or Ser343Ser genetic
polymorphism and cancer risk, but the results have been conflicting
(14,17–21).
Thus, the association between the LEPR Lys656Asn or
Ser343Ser genetic polymorphism and cancer requires further
investigation. In an attempt to clarify this inconsistency, all the
published hospital and population-based studies prior to June 2014
were combined in a meta-analysis to provide a comprehensive outlook
of the role of the LEPR Lys656Asn or Ser343Ser gene by
multiple research methods and models.
In the present study, a comprehensive meta-analysis
was performed on previous studies to investigate the association of
LEPR Lys656Asn or Ser343Ser genetic polymorphism with all
types of cancer, different types of cancer, ethnicities,
populations, genotype methods and various types of sample size in
cases.
Materials and methods
Search strategy and data extraction
In the meta-analysis, a comprehensive literature
search of the US National Library of Medicine’s PubMed database,
ISI Web of Knowledge, Medline, Embase and Google Scholar Search
(prior to to June 2014) was conducted using the following search
terms, including ‘leptin receptor,’ ‘leptin gene receptor,’ ‘leptin
receptor gene,’ ‘LEPR,’ ‘Lys656Asn,’ ‘K656N,’ ‘rs8179183’ or
‘Ser343Ser’; ’polymorphisms,’ ‘variation,’ ‘mutation’ or ‘SNP;’
‘tumour,’ ‘tumor,’ ‘cancer,’ ‘neoplasm,’ ‘phyma,’ ‘oncoma,’ ‘knub,’
‘carcinoma’ or ‘malignancy;’ and the combined phrases in order to
obtain all the genetic studies on the association of LEPR
Lys656Asn or Ser343Ser genetic polymorphism and cancers. The
references of the original or reviewed studies were also examined
to identify additional studies. Eligible studies were selected
according to the following explicit inclusion criteria: i) A
case-control study on the association between the LEPR
Lys656Asn or Ser343Ser genetic polymorphism and cancer risk, ii)
detailed number of different genotypes for estimating an odds ratio
(OR) with 95% confidence interval (CI), iii) when several
publications reported on the same population data, the largest or
most complete study was chosen, iv) cases with carcinomas were
diagnosed by histopathology and v) animal, case or review studies,
abstracts, editorials, studies with incomplete data and studies
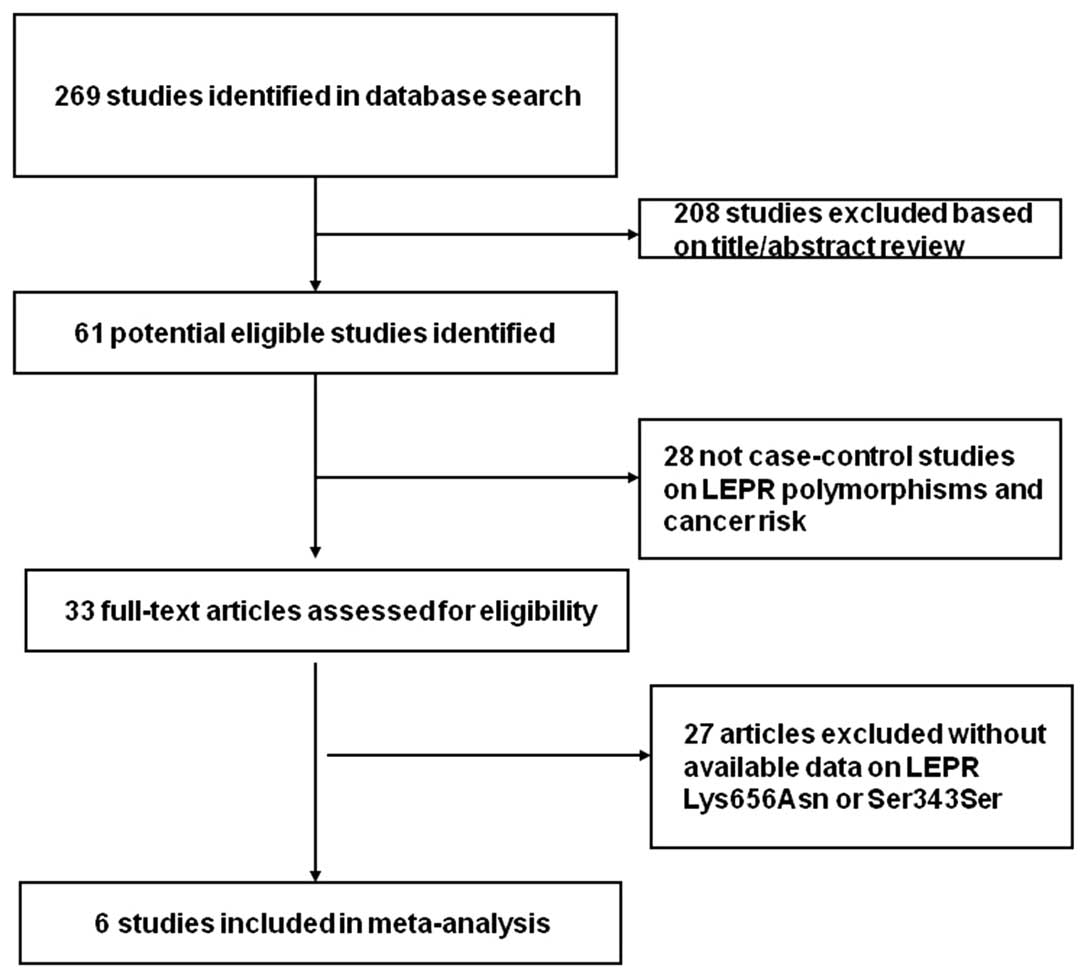
based on pedigree data were excluded (Fig. 1). For each eligible study, the
following information was recorded: First author’s name, year of
publication, country, ethnicity, type of cancer, genotyping
methods, sources of controls, ethnicity of the study population,
genotype and allele distributions, and main results of each
study.
Statistical analysis
The strength of the association between the
LEPR Lys656Asn or Ser343Ser genetic polymorphism and cancer
was assessed by using crude OR with 95% CI. The association between
the LEPR Lys656Asn or Ser343Ser genetic polymorphism and
cancer risk was examined using the following genetic models:
Homozygote co-dominant (CC vs. GG), heterozygote co-dominant (CG
vs. GG), dominant genetic (CC/CG vs. GG), recessive genetic (CC vs.
CG/GG) and additive genetic (C vs. G) models. Firstly, the
Hardy-Weinberg equilibrium (HWE) was checked in the controls for
each study. Subsequently a Q test was performed to evaluate the
heterogeneity (22). Fixed-effects
model was used to pool the data when the P-value of Q test ≥0.05,
otherwise, random-effects model was selected (23). I2 was also used to assess
the heterogeneity in the meta-analysis, and heterogeneity existed
when I2>50% (24).
Sensitivity and subgroup analyses were also performed to explore
the reason of heterogeneity. The funnel plot and Egger’s test were
used to assess the publication bias and P<0.05 was considered to
indicate a statistically significant difference (25). All the statistical analyses were
performed using Stata 12.0 software (StataCorp, College Station,
TX, USA) and Review Manager 5.2 (The Cochrane Collaboration, The
Nordic Cochrane Centre, Copenhagen, Denmark).
Results
Eligible studies
Overall, eight relevant studies comprising 2,480
cases and 3,162 controls were selected in the meta-analysis
(14,17–21).
The main characteristics of these studies are shown in Table I. Genotype and allele distributions
of LEPR Lys656Asn or Ser343Ser genetic polymorphism among
cancer cases and controls and the P-value of HWE in controls are
shown in Table I. All the studies
were case-control studies, including two breast (17,19),
two colorectal (14), two gastric
(21), one esophageal (18) and one lung cancer studies (20). Cancers were histological or
pathological in the majority of the studies. There were four
studies (14,18,19)
investigating the Caucasian population and four studies (17,20,21)
investigating the Asian population. Population-based controls were
carried out in three studies, whereas hospital-based controls were
carried out in five studies. All the studies were reported in
English. The genotyping methods contained the classic polymerase
chain reaction-restriction fragment length polymorphism (PCR-RFLP)
assay, PCR-sequencing, Sequenom iPLEX and SNPstream. The sample
size in the majority of studies was >100 patients. The genotype
distributions of the controls were all in agreement with HWE,
except for four studies not estimable (14,19,20).
 | Table ICharacteristics of the studies
included in the meta-analysis. |
Table I
Characteristics of the studies
included in the meta-analysis.
| Author | Year | Country | Ethnicity | Cancer type | Cases/controls | Source of
controls | Genotype method | Polymorphisms | P-value of HWE in
controls | (Refs.) |
|---|
| Woo et al | 2006 | Korea | Asian | Breast | 45/45 | HB | PCR-sequencing | K656N | 0.632 | (17) |
| Chia et
al | 2007 | USA | Caucasian | Colorectal | 157/191 | HB | PCR-sequencing | K656N, Ser343Ser | - | (14) |
| Doecke et
al | 2008 | Australia | Caucasian | Esophageal | 774/1352 | PB | Sequenom iPLEX | K656N | 0.718 | (18) |
| Teras et
al | 2009 | USA | Caucasian | Breast | 641/650 | PB | SNPstream | K656N | - | (19) |
| Li et
al | 2012 | China | Asian | Lung | 744/832 | PB | PCR-RFLP | K656N | <0.05a | (20) |
| Kim et
al | 2012 | Korea | Asian | Gastric | 48/48 | HB | PCR-RFLP | K656N,
Ser343Ser | 0.703, 0.644 | (21) |
Meta-analysis
Overall, as shown in Table II, the LEPR Lys656Asn or
Ser343Ser genetic polymorphism did not significantly affect the
risk of cancer when all the eligible studies were pooled into the
meta-analysis. When the four studies that the genotype
distributions of the controls were not in agreement with HWE were
excluded, a significant association was not observed in any genetic
model. In all the genetic models, all the P-values of the Q test
were >0.05 and I2 values were <50%. The
sensitivity analysis was performed by deleting one single study
from the overall pooled analysis each time to assess the influence
of the removed data. However, the results revealed that no studies
changed the between-study heterogeneities.
 | Table IIMeta-analysis of the association
between LEPR Lys656Asn and Ser343Ser polymorphism and cancer
risk. |
Table II
Meta-analysis of the association
between LEPR Lys656Asn and Ser343Ser polymorphism and cancer
risk.
| Variables | Studies, n | Homozygous
co-dominant | Heterozygous
co-dominant | Recessive | Dominant | Additive |
|---|
|
|
|
|
|
|---|
| CC vs. GG |
Pheta | CG vs. GG |
Pheta | (CC vs. CG+GG) |
Pheta | (CC+CG vs. GG) |
Pheta | C vs. G |
Pheta |
|---|
| All | 8 | 1.18
(0.92–1.51) | 0.584 | 1.14
(0.99–1.32) | 0.641 | 1.09
(0.86–1.37) | 0.796 | 1.06
(0.95–1.19) | 0.364 | 1.11
(0.99–1.23) | 0.727 |
| HWEb | 4 | 1.04
(0.62–1.74) | - | 1.07
(0.88–1.29) | 0.702 | 1.02
(0.61–1.71) | - | 1.07
(0.89–1.28) | 0.701 | 1.05
(0.90–1.23) | 0.721 |
| Cancer type |
| Breast | 2 | - | - | 0.63
(0.17–2.42) | - | - | - | 0.84
(0.66–1.06) | 0.680 | 0.65
(0.18–2.39) | - |
| Esophageal | 1 | 1.04
(0.62–1.74) | - | 1.06
(0.87–1.29) | - | 1.02
(0.61–1.71) | - | 1.06
(0.87–1.28) | - | 1.04
(0.89–1.23) | - |
| Colorectal | 2 | - | - | - | - | - | - | 1.08
(0.71–1.63) | 0.579 | - | - |
| Lung | 1 | 1.23
(0.92–1.63) | - | 1.25
(1.00–1.55) | - | 1.10
(0.85–1.44) | - | 1.24
(1.02–1.52) | - | 1.15
(1.00–1.34) | - |
| Gastric | 2 | - | - | 1.55
(0.68–3.53) | 0.807 | - | - | 1.55
(0.68–3.53) | 0.807 | 1.50
(0.68–3.31) | 0.812 |
| Ethnicity |
| Caucasian | 4 | 1.04
(0.62–1.74) | - | 1.06
(0.87–1.29) | - | 1.02
(0.61–1.71) | - | 0.98
(0.85–1.13) | 0.453 | 1.04
(0.89–1.23) | - |
| Asian | 4 | 1.23
(0.92–1.63) | - | 1.24
(1.01–1.53)c | 0.730 | 1.10
(0.85–1.44) | - | 1.24
(1.02–1.50)c | 0.731 | 1.16
(1.00–1.33) | 0.752 |
| Source of
controls |
| Population | 3 | 1.18
(0.92–1.51) | 0.584 | 1.14
(0.98–1.32) | 0.272 | 1.09
(0.86–1.37) | 0.796 | 1.06
(0.94–1.19) | 0.052 | 1.11
(0.99–1.23) | 0.370 |
| Hospital | 5 | - | - | 1.21
(0.60–2.41) | 0.525 | - | - | 1.11
(0.78–1.58) | 0.796 | 1.19
(0.61–2.32) | 0.549 |
| Genotype
method |
| PCR-RFLP | 3 | 1.23
(0.92–1.63) | - | 1.26
(1.02–1.56) | 0.861 | 1.10
(0.85–1.44) | - | 1.26
(1.03–1.53)c | 0.855 | 1.16
(1.01–1.34)c | 0.800 |
|
PCR-sequencing | 3 | - | - | 0.63
(0.17–2.42) | - | - | - | 1.03
(0.69–1.53) | 0.651 | 0.65
(0.18–2.39) | - |
| Sequenom
iPLEX | 1 | 1.04
(0.62–1.74) | - | 1.06
(0.87–1.29) | - | 1.02
(0.61–1.71) | - | 1.06
(0.87–1.28) | - | 1.04
(0.89–1.23) | - |
| SNPstream | 1 | - | - | - | - | - | - | 0.84
(0.67–1.07) | - | - | - |
| Sample size in
cases |
| <100 | 3 | - | - | 1.21
(0.60–2.41) | 0.525 | - | - | 1.21
(0.60–2.41) | 0.525 | 1.19
(0.61–2.32) | 0.549 |
| ≥100 | 5 | 1.18
(0.92–1.51) | 0.584 | 1.14
(0.98–1.32) | 0.272 | 1.09
(0.86–1.37) | 0.796 | 1.06
(0.94–1.19) | 0.182 | 1.11
(0.99–1.23) | 0.370 |
The effects of the LEPR Lys656Asn or
Ser343Ser genetic polymorphisms were evaluated according to
specific types of cancer, different ethnicities, different sources
of controls, different detection methods and different sample sizes
in cases. As shown in Table II,
the LEPR Lys656Asn or Ser343Ser genetic polymorphisms were
found to not significantly affect the risk of any type of cancer in
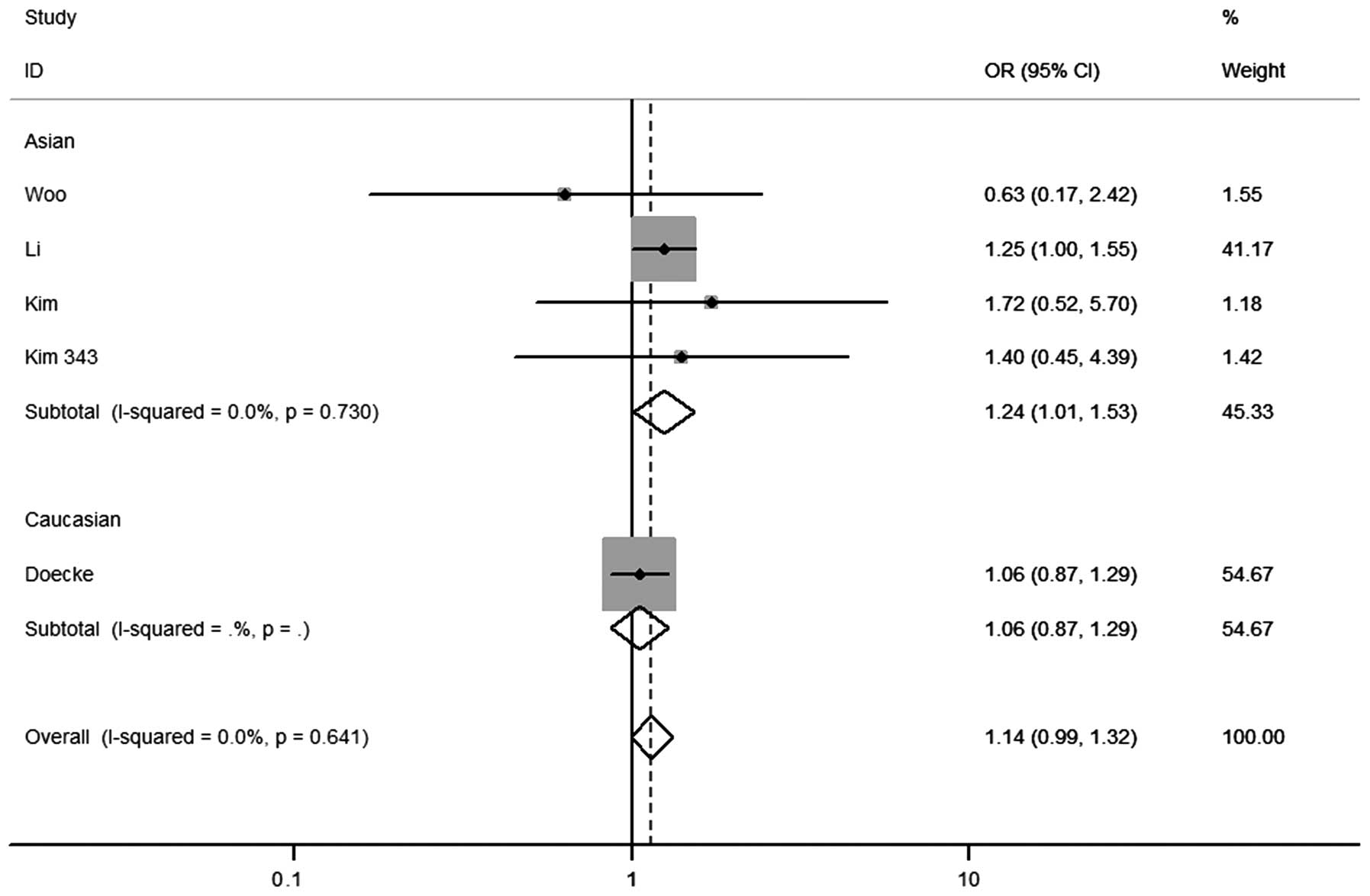
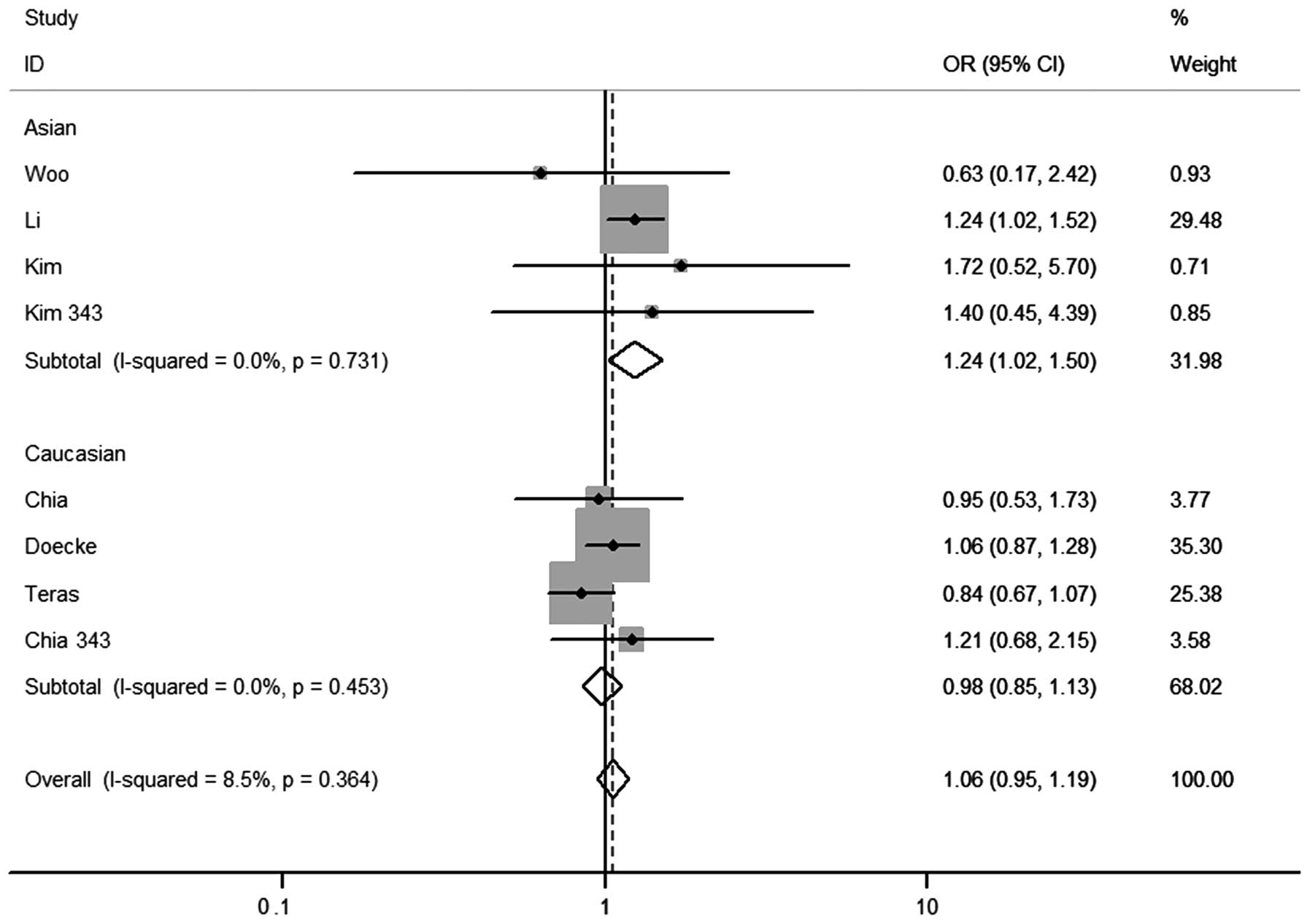
any genetic model tested. In the stratified analysis by ethnicity,
significantly increased risks were found in the Asian population in
the heterozygous co-dominant [OR=1.24 (1.01–1.53)] (Fig. 2) and dominant genetic models
[OR=1.24 (1.02–1.50)] (Fig. 3). For
the Caucasian population, no significant associations were observed
in any genetic model tested. According to the source of controls, a
significant association was not observed in any genetic model in
population- or hospital-based studies. Regarding the detection
method, signification effects in dominant and additive genetic
models were observed in the PCR-RFLP subgroup. According to the
sample size in cases, a significant association was not observed in
any genetic model in small (<100) or big (≥100) sample
studies.
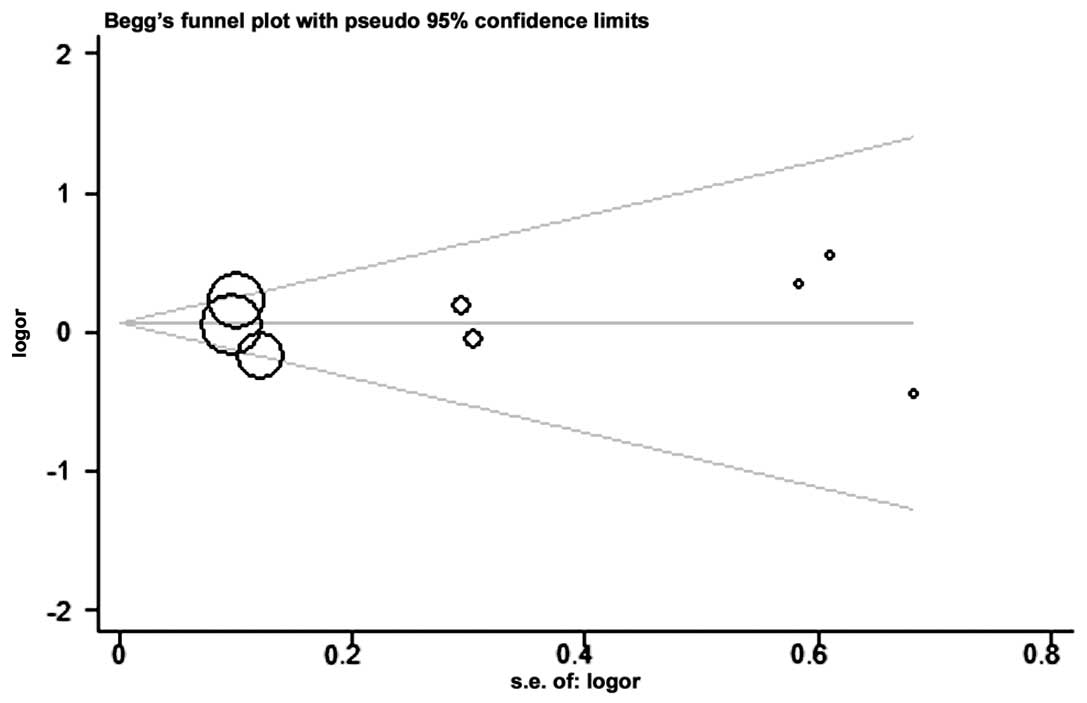
Publication bias
The Begg’s funnel plot and Egger’s test were
performed to assess the publication bias. The shape of the funnel
plots did not reveal any evidence of clear asymmetry in the overall
meta-analysis (Fig. 4). Egger’s
test was used to provide statistical evidence of funnel plot
symmetry. The results did not present any clear evidence of
publication bias (CC vs. GG, P=N/A (not applicable); CG vs. GG,
P=0.891; CC/CG vs. GG, P=0.933; CC vs. CG/GG, P=N/A; and C vs. G,
P=0.926).
Discussion
The present meta-analysis of eight studies involving
2,480 cases and 3,162 controls was conducted in order to yield a
valid conclusion concerning the potential association between the
LEPR Lys656Asn or Ser343Ser genetic polymorphism and cancer
risk. Indications from epidemiological studies have shown that
overweight and obesity may be factors associated with the increased
risk of endometrium, kidney, colon and gallbladder cancers in
females and breast cancer in postmenopausal females (26), and increased mortality rates for
cancers at multiple specific sites (27). Polymorphism-associated low enzyme
activity may cause the reduction of conjugation and thus the
reduced elimination of oxidative intermediates radicals and
electrophiles, resulting in the production of increased
carcinogenic substrates rather than detoxification. Polymorphisms
in LEPR may therefore influence carcinogen levels and
potentially play a role in carcinogenesis. However, studies
focusing on the association of the LEPR Lys656Asn or
Ser343Ser genetic polymorphism with cancer susceptibility have had
controversial conclusions (14,17–21).
This indicates limitations in the studies, including ethnical
differences, small sample sizes and research methodology.
Meta-analysis is known to be a powerful tool for summarizing the
results from various studies by generating a single estimate of the
major effect with an augmented precision.
In the present analysis, the pooled effects for all
the genetic model comparisons indicated no significant association
between the LEPR Lys656Asn or Ser343Ser genetic polymorphism
and any cancer risk. Furthermore, it was found that for the Asian
population, significant associations were observed in heterozygous
co-dominant and dominant genetic models, whereas the Asian
population with the CG genotype had a higher cancer risk compared
to the Caucasian population. Inconsistencies between the two
ethnicities can be explained by the possibility that different
ethnic groups experience multiple lifestyle and environmental
factors. Different genotype and/or allele frequencies of this locus
polymorphism that are present in different populations may result
in various degrees of cancer susceptibility. In the present
meta-analysis, consistent results were observed between hospital-
and population-based studies, but it is still believed that
controls in population-based studies are more representative of the
general population compared to the controls from hospital-based
studies. Several factors, such as environmental factors and genetic
backgrounds, may also contribute to the discrepancy.
There were certain limitations in the meta-analysis.
Firstly, the sample size for any type of cancer investigated was
not sufficiently large, which could increase the probability of
false-positive or false-negative results. Therefore, it may be
difficult to obtain a complete conclusion if the number of included
studies in the subgroup was limited. In addition, the studies
involved in the different ethnicities were warranted to estimate
the effects of this functional polymorphism on cancer risk.
Secondly, as the original data from the eligible studies was not
available, evaluating the roles of specific environmental and
lifestyle factors, such as diet, alcohol consumption and smoking
status, in developing cancer was difficult. Thirdly, the influence
of bias in the present analysis could not be completely excluded as
positive results are published much quicker than studies with
‘negative’ results.
In conclusion, the present meta-analysis indicated
that the LEPR Lys656Asn or Ser343Ser genetic polymorphism
did not significantly affect the risk of cancer, but may increase
the susceptibility of cancers in the Asian population in the
dominant genetic model. This also suggests that the SNP functions
as a dominant mutation, which requires verification or association
with functional studies. Large well-designed epidemiological
studies are also required to validate these findings.
Acknowledgements
The authors gratefully acknowledge the support of
the subjects who participated in the present study. The study was
partly supported by the Natural Science Foundation of Shanghai,
China (grant no. 14ZR1432600).
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar
|
|
2
|
Wright ME, Peters U, Gunter MJ, et al:
Association of variants in two vitamin e transport genes with
circulating vitamin e concentrations and prostate cancer risk.
Cancer Res. 69:1429–1438. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cheung WY and Liu G: Genetic variations in
esophageal cancer risk and prognosis. Gastroenterol Clin North Am.
38:75–91. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Friedman JM and Halaas JL: Leptin and the
regulation of body weight in mammals. Nature. 395:763–770. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Unger RH, Zhou YT and Orci L: Regulation
of fatty acid homeostasis in cells: novel role of leptin. Proc Natl
Acad Sci USA. 96:2327–2332. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lakka HM, Oksanen L, Tuomainen TP, Kontula
K and Salonen JT: The common pentanucleotide polymorphism of the
3′-untranslated region of the leptin receptor gene is associated
with serum insulin levels and the risk of type 2 diabetes in
non-diabetic men: a prospective case-control study. J Intern Med.
248:77–83. 2000.
|
|
7
|
Lönnqvist F, Arner P, Nordfors L and
Schalling M: Overexpression of the obese (ob) gene in adipose
tissue of human obese subjects. Nat Med. 1:950–953. 1995.PubMed/NCBI
|
|
8
|
Yiannakouris N, Yannakoulia M, Melistas L,
Chan JL, Klimis-Zacas D and Mantzoros CS: The Q223R polymorphism of
the leptin receptor gene is significantly associated with obesity
and predicts a small percentage of body weight and body composition
variability. J Clin Endocrinol Metab. 86:4434–4439. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Snoussi K, Strosberg AD, Bouaouina N, Ben
Ahmed S, Helal AN and Chouchane L: Leptin and leptin receptor
polymorphisms are associated with increased risk and poor prognosis
of breast carcinoma. BMC Cancer. 6:382006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu CL, Chang YC, Cheng SP, et al: The
roles of serum leptin concentration and polymorphism in leptin
receptor gene at codon 109 in breast cancer. Oncology. 72:75–81.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Han CZ, Du LL, Jing JX, et al:
Associations among lipids, leptin, and leptin receptor gene
Gin223Arg polymorphisms and breast cancer in China. Biol Trace Elem
Res. 126:38–48. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ribeiro R, Vasconcelos A, Costa S, et al:
Overexpressing leptin genetic polymorphism (-2548 G/A) is
associated with susceptibility to prostate cancer and risk of
advanced disease. Prostate. 59:268–274. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ribeiro R, Araújo AP, Coelho A, et al: A
functional polymorphism in the promoter region of leptin gene
increases susceptibility for non-small cell lung cancer. Eur J
Cancer. 42:1188–1193. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chia VM, Newcomb PA, Lampe JW, et al:
Leptin concentrations, leptin receptor polymorphisms, and
colorectal adenoma risk. Cancer Epidemiol Biomarkers Prev.
16:2697–2703. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Loffreda S, Yang SQ, Lin HZ, et al: Leptin
regulates proinflammatory immune responses. FASEB J. 12:57–65.
1998.PubMed/NCBI
|
|
16
|
Russo VC, Metaxas S, Kobayashi K, Harris M
and Werther GA: Antiapoptotic effects of leptin in human
neuroblastoma cells. Endocrinology. 145:4103–4112. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Woo HY, Park H, Ki CS, Park YL and Bae WG:
Relationships among serum leptin, leptin receptor gene
polymorphisms, and breast cancer in Korea. Cancer Lett.
237:137–142. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Doecke JD, Zhao ZZ, Stark MS, et al;
Australian Cancer Study. Single nucleotide polymorphisms in
obesity-related genes and the risk of esophageal cancers. Cancer
Epidemiol Biomarkers Prev. 17:1007–1012. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Teras LR, Goodman M, Patel AV, et al: No
association between polymorphisms in LEP, LEPR, ADIPOQ, ADIPOR1, or
ADIPOR2 and postmenopausal breast cancer risk. Cancer Epidemiol
Biomarkers Prev. 18:2553–2557. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li Y, Geng J, Wang Y, et al: The role of
leptin receptor gene polymorphisms in determining the
susceptibility and prognosis of NSCLC in Chinese patients. J Cancer
Res Clin Oncol. 138:311–316. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kim EY, Chin HM, Park SM, et al:
Susceptibility of gastric cancer according to leptin and leptin
receptor gene polymorphisms in Korea. J Korean Surg Soc. 83:7–13.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
DerSimonian R and Laird N: Meta-analysis
in clinical trials. Control Clin Trials. 7:177–188. 1986.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mantel N and Haenszel W: Statistical
aspects of the analysis of data from retrospective studies of
disease. J Natl Cancer Inst. 22:719–748. 1959.PubMed/NCBI
|
|
24
|
Higgins JP, Thompson SG, Deeks JJ and
Altman DG: Measuring inconsistency in meta-analyses. BMJ.
327:557–560. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Egger M, Davey Smith G, Schneider M and
Minder C: Bias in meta-analysis detected by a simple, graphical
test. BMJ. 315:629–634. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bianchini F, Kaaks R and Vainio H:
Overweight, obesity, and cancer risk. Lancet Oncol. 3:565–574.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Calle EE, Rodriguez C, Walker-Thurmond K
and Thun MJ: Overweight, obesity, and mortality from cancer in a
prospectively studied cohort of U.S. adults. N Engl J Med.
348:1625–1638. 2003. View Article : Google Scholar : PubMed/NCBI
|