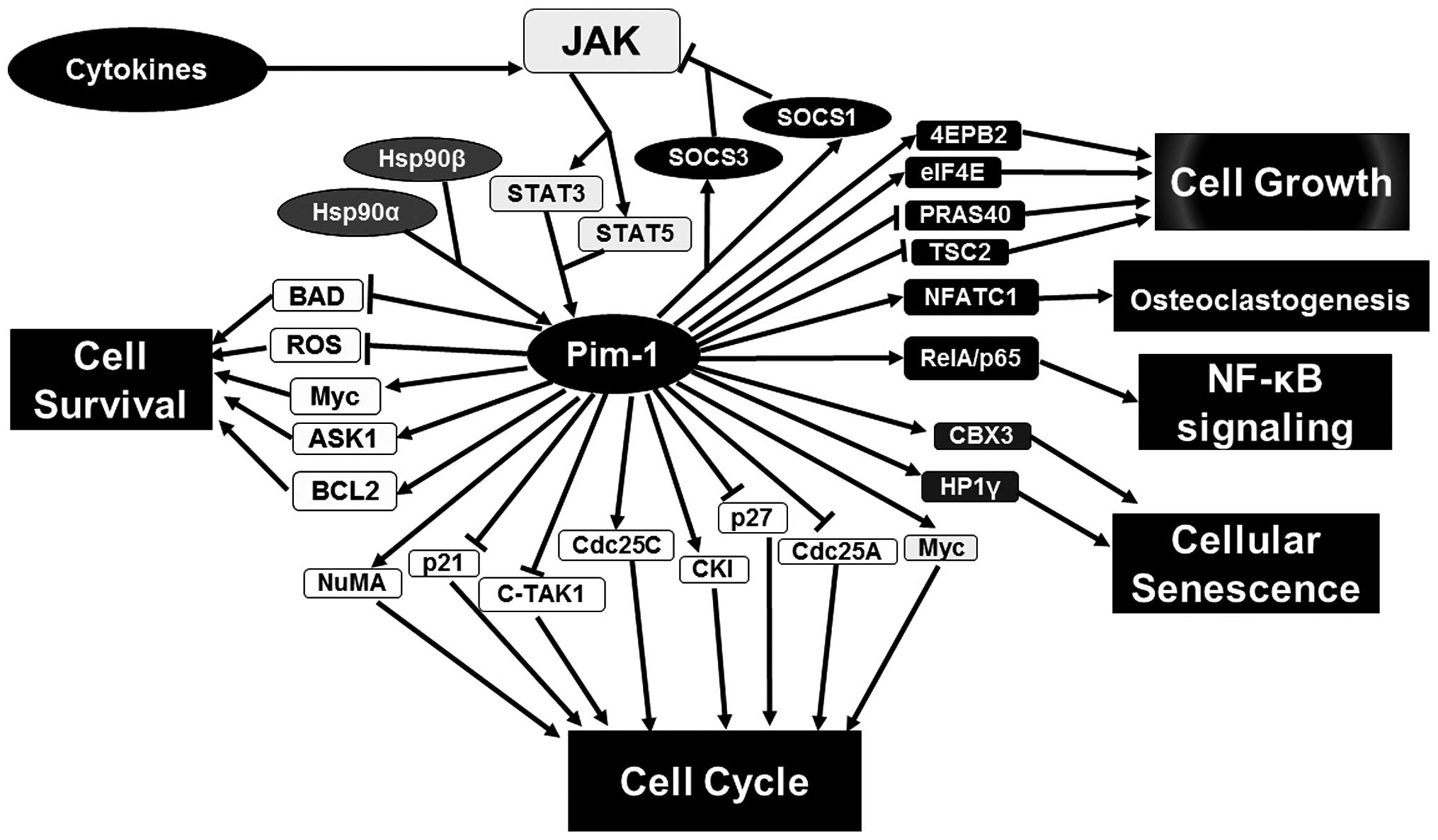
Proviral integration site for Moloney murine
leukemia virus-1 (Pim-1) kinase is observed to interact with
numerous proteins participating in various signaling pathways
(Fig. 1) (1,2). The
Pim-1 gene was originally identified as a proviral
integration site for Moloney murine leukemia virus-1. Pim-1 is a
proto-oncogene that encodes a serine/threonine kinase, which has a
crucial role in oncogenesis (3). This
proto-oncogene was originally found in hematopoietic cells as a
member of the Pim family (Pim-1, Pim-2 and
Pim-3). Transcription of Pim-1 can be activated by
several interleukins, such as interleukin-2 (IL-2), IL-3 and IL-6.
It has been shown that the Pim-1 kinase has an essential role in
cytokine-induced signal transduction by controlling transcription
factors (4). Upregulation of Pim-1 is
correlated with cell proliferation induced by mitogens or
cytokines, while downregulation of Pim-1 is correlated with growth
retention due to the absence of cytokines (3). Additionally, deficiency of Pim-1 kinase
leads to failure in cell survival and growth (1–3). Recent
studies have shown that Pim-1 is required in drug resistance and
has important roles in prostate cancer. In addition, new functions
of Pim-1 have been revealed in immunotherapy, senescence bypass,
epigenetic dynamics and cancer metastasis.
Pim-1 transcription can be activated by interleukins
followed by signaling transduction to the nucleus through two
families of proteins, Janus kinase (JAK)/signal transducers and
activators of transcription (STAT) (Fig.
1). STAT proteins can increase the expression of Pim-1 kinase
by binding to the promoter of the Pim-1 gene. This is widely
found in the classical upstream of the Pim-1 signaling pathway.
However, a previous study indicated that Pim-1 can, in turn,
downregulate the JAK/STAT pathway (9).
In detail, Pim-1 expression is induced by STAT3 and STAT5 whereupon
Pim-1 kinase phosphorylates and stabilizes SOCS1 and SOCS3. Upon
phosphorylation, SOCS proteins become more suppressive by
interacting with active JAK proteins and blocking phosphorylation
of STAT proteins (Fig. 1) (10). In addition, phosphorylated STAT3
triggers Pim-1 expression during human pulmonary hypertension,
which in turn contributes to proliferation of smooth muscle cells
(11). Thus, Pim-1 may form a feedback
loop with the JAK/STAT pathway for tight regulation of its own
expression and function. Numerous Pim-1 phosphorylation substrates
have been identified, which are involved in cell cycle, cell growth
and cell survival (Fig. 1). For
example, Pim-1 phosphorylates cell cycle regulator p21, which
thereby dissociates p21 with proliferating cell nuclear antigen
binding to regulate cell cycle and proliferation (12) and phosphorylates p27 to promote cell
cycle progression (13). Cell survival
depends on signals that inhibit apoptosis. One of the main
regulators of cell survival is the B-cell lymphoma-2 (Bcl-2) family
comprising Bcl-2, Bcl-xL and myeloid cell leukemia-1 (MCL-1)
(14). According to studies by Kumar
et al (15), the inactivation
of Bcl-2-associated death promoter (BAD) can occur due to
phosphorylation at Ser-75 by Pim-1, thus strengthening the
dissociation of Bak with Bcl-xL. Therefore, Pim-1 is important for
Bcl-xL pro-survival effect. This statement was further supported by
the fact that inhibition of Pim-1 kinase suppresses phosphorylation
of Bad, which in turn increases LY294002-induced apoptosis in
prostate cancer LNCaP cells (15). In
addition, multiple signaling networks are regulated by Pim-1 and
have been reviewed by numerous researchers suggesting that Pim-1
may be a master regulator of cell function (1) (Fig. 1). The
following text will focus on the essential new functions of Pim-1
in cancer.
Increasing evidence has shown that Pim-1 would be a
novel and essential drug target in numerous types of cancer
(16), in particular prostate cancer
(17). Pim-1 transcription is
regulated by interleukins (ILs), which implicates that Pim-1 can be
a potential target for immunotherapy. Our previous study showed
that IL-6 can induce Pim-1L and Pim-1S expression (5). Treatment with neutralized IL-6 antibody,
results in the decrease of Pim-1L and Pim-1S expression in prostate
cancer cells (5). This suggests that
targeting Pim-1 has a great potential in immunotherapy.
Directly targeting Pim-1 using a specific monoclonal
antibody to Pim-1 has been tested in preclinical studies. Treatment
with the specific antibody to Pim-1 (mAb P9) in SCID mice
inoculated with DU145 cells subcutaneously decreased the tumor
growth. Additionally, the growth rate of tumors generated from
C57BL/6 mice inoculated with TRAMP-C1 cells also was decreased by
this antibody (18). Antibody P9
induces apoptotic pathway by specific interaction with Pim-1. They
also found that the treatment with Pim-1 antibody P9 significantly
inhibited the level of Pim-1 kinase in prostate cancer cell lines,
such as PC-3, DU145 and TRAMP-C1 with changes in protein kinase B
(or AKT), heat-shock protein 90 and caspase pathways (18). A previous study also showed that the
treatment with P9 decreased the growth of human leukemia cell lines
(19). Our previous studies identified
that Pim-1L (44 kDa) expresses on cell membrane to mediate drug
resistance in prostate cancer cells (5,20). Combined
treatment with the Pim-1 antibody P9 and chemotherapy drugs
decreased prostate cancer cell growth (18).
Human cluster of differentiation 4 [CD4(+)]
CD25(high)FOXP3(+) T regulatory cells (T regs), which have
functional plasticity, can differentiate into effector T cells
induced by inflammation (21). FOXP3
is a specific transcription factor that determines development of T
regs and is critical for obtaining the inhibitory abilities of T
regs (22). According to recent
studies FOXP3 is regulated through phosphorylation, which affects
its DNA binding ability and stability (22,23). Pim-1
expression can be regulated through T cell receptor signaling and
IL-6 in in vitro-expanded T regs (21). Recent studies have shown that human
FOXP3 is phosphorylated by Pim-1 kinase at Ser-422, which blocks
FOXP3 chromatin-binding activity for expression of target genes.
When in vitro-expanded T reg cells were treated with a
Pim-1-specific inhibitor,
3-cyano-4-phenyl-6-(3-bromo-6-hydroxy)phenyl-2(1H)-pyridone, they
exhibited increased suppressive activity. This means that Pim-1
kinase diminishes the suppressive activity of in
vitro-expanded T regs by inhibiting human FOXP3 (21). From several studies, it has been
revealed that T regs may be one of the reasons for unsuccessful
cancer immunotherapies due to inhibiting tumor elimination and
activity of tumor-associated antigen-specific lymphocytes (24). Therefore, the efficiency of antitumor
immunotherapies can be improved through suppression of T regs via
inhibition of FOXP3 by targeting Pim-1 kinase.
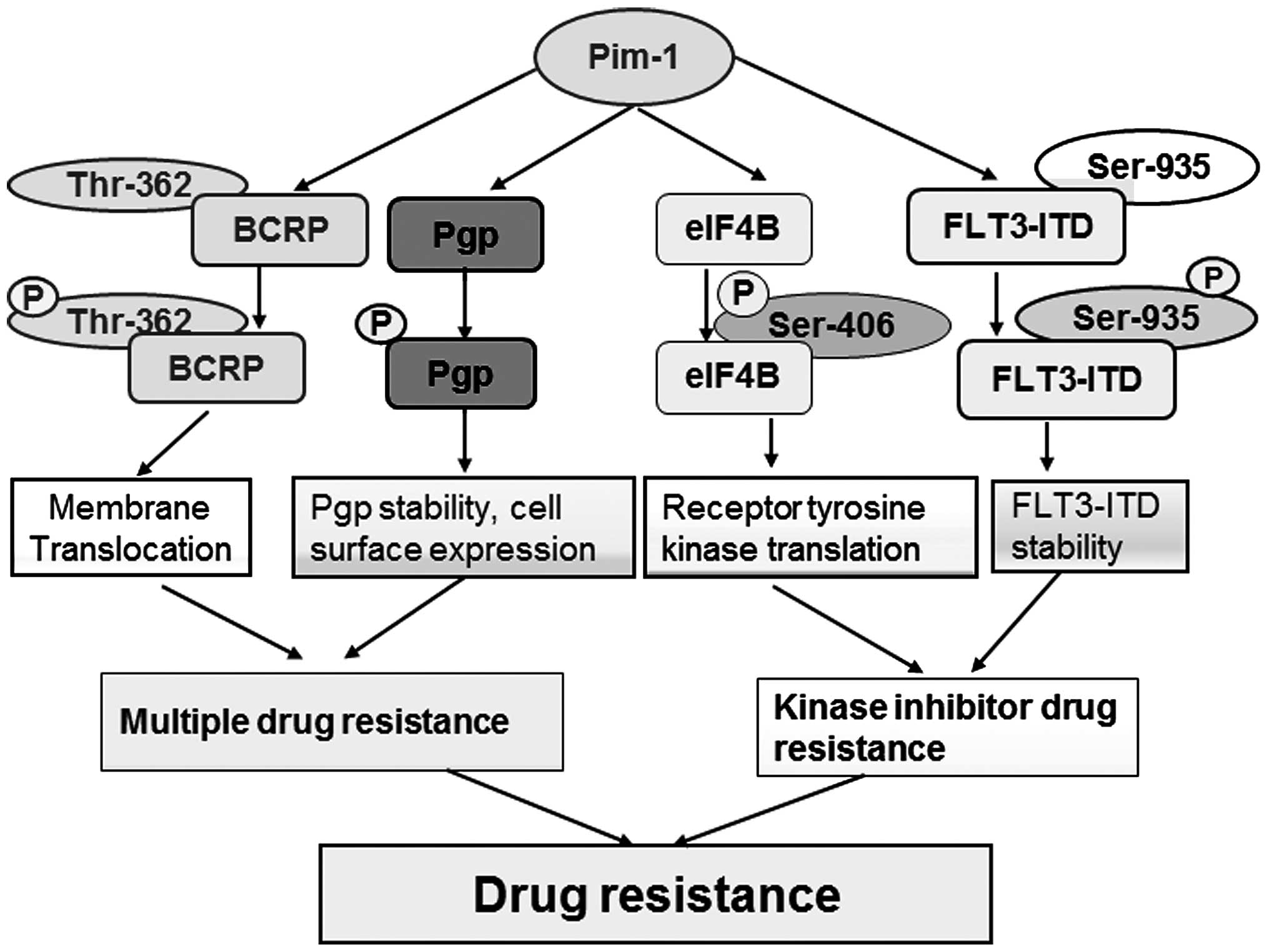
Emerging evidence has shown that Pim-1 kinase has
been associated with the drug-resistant abilities of cancer cells
(25). Pim-1 mediates drug resistance
through interaction with and phosphorylation of Etk (5), P-glycoprotein (Pgp) (26), breast cancer resistant protein (BCRP)
(20) and fms-like tyrosine kinase 3
(FLT3) (27,28) (Fig. 2).
The original findings on Pim-1-mediated drug resistance come from
the early study that Pim-1 overexpression allows cells to undergo
prolonged survival upon withdrawal of IL-3 (29). Following this, Pim-1-mediated drug
resistance in prostate cancer was identified as a mechanism of
inhibiting p53-induced apoptosis (5).
Mechanistically, Pim-1L competes with p53 to bind non-receptor
tyrosine kinase Etk. Etk signaling has an important role in this
drug resistance as Pim-1L, but not Pim-1S, directly interacts with
Etk at the plasma membrane while Etk signaling can promote cell
survival by inhibiting p53 (30).
Thus, Pim-1L showed a higher ability to protect the prostate cancer
cells to undergo apoptosis induced by chemotherapy drugs. At that
time, it was unclear whether Pim-1 mediated drug resistance was
only due to reduced cell apoptosis or through a mechanism of
multiple drug resistance mediated by adenosine triphosphate-binding
cassette (ABC) drug transporters. Subsequent discoveries benefited
from a yeast two-hybrid system using full-length Pim-1L to screen
novel Pim-1L-binding proteins. Our first study regarding
Pim-1L-mediated multiple drug resistance explained the molecular
mechanism that Pim-1L phosphorylates BCRP at Thr-362 resulting in
BCRP dimerization and its translocation to the plasma membrane
(20). This suggests that
translocation of the phosphorylated ABC transporter by Pim-1
promotes drug resistance via efflux drugs outside of the cells
(10,20). Pgp is another member of the ABC family
(31). It is known that Pgp must
translocate to the plasma membrane to enhance drug efflux activity
(32). Pim-1 kinase also
phosphorylates Pgp and protects it from proteasomal degradation
through stabilizing Pgp and enhancing its cell surface expression
(26). Combined inhibitors of Pgp and
Pim-1 enhance drug efficiency by increased apoptosis of drug
resistant cancer cells (26).
In addition to Etk, another tyrosine kinase that
interacts with Pim-1 for drug resistance is FLT3. FLT3 is a
receptor tyrosine kinase that can be found in normal cells, as well
as in cancerous cells (33). However,
FLT3 tends to mutate by internal tandem duplication (ITD), which
accounts for 30% of acute myeloid leukemia (AML). AML patients with
FLT3-ITD frequently develop resistance to FLT3 inhibitors. It has
been shown that STAT5 can be activated by mislocalized and
phosphorylated FLT3-ITD, which in turn promotes expression of Pim-1
kinase (34–36). Recently, our previous study revealed
that Pim-1 participates in a positive-feedback loop regulating
FLT3-ITD expression and stability through phosphorylation at
Ser-935. Therefore, Pim-1 kinase facilitates abnormal signaling of
FLT3-ITD in cancer cells, and enhancing their drug resistance
(37). Combined inhibition of Pim-1
and FLT3 increases cancer cell sensitivity to either drug alone
(37).
Cellular senescence can be described as an arrest of
the proliferative abilities of the cell, so that the cell loses the
ability to divide (38). Certain
characteristics of senescent cells are heterochromatin formation
and telomere shortening. It has long been believed that cellular
senescence serves as a protective mechanism against cancer
(39). The exact role of
oncogene-induced senescence in cancer is largely unknown. Recently,
it was revealed that Pim-1 expression is elevated upon aging in
human fibroblast cells and Pim-1 inhibition reduces replicative and
oncogene-induced senescence (40). In
addition, it has been shown that expression of Pim-1 is activated
through IL-6/STAT3 signaling, thus Pim-1 mediates cytokine-induced
cellular senescence (40). Notably, it
was found that Pim-1 kinase has the potential to rejuvenate human
cardiac progenitor cells (hCPCs). According to obtained results,
hCPC cell lines transduced with lentivirus for overexpressing Pim-1
were less susceptible to replicative senescence, had longer
telomeres and increased abilities to proliferate (41). These two contradictive findings provide
a foundation to further study in this area. Results that state
participation of Pim-1 in premature aging via heterochromatin
formation make the kinase a potential target in activating cellular
senescence through cytokines in cancer therapy. By contrast, the
observations showing that Pim-1 kinase has the capacity to
rejuvenate hCPC also makes Pim-1 a potential target in deactivating
the rejuvenation process in tumor cells. Furthermore, recent new
findings show that nuclear localized Pim-1 (refer to Pim-1S) can
promote senescence bypass of hCPC stem cells through downregulation
of p16 and p53 (42). The new finding
that mitochondrial localization of Pim-1 (mito-Pim1) increases hCPC
cell survival and decreases apoptosis further supports that
distinct cellular localization of Pim-1 fine-tunes the signaling
networks for differential functions, such as maintaining
mitochondrial integrity, energy and survival for senescence bypass.
However, which factors determine the functional switch of Pim-1 in
differential genetic context should be further investigated.
Elevation of Pim-1 kinase has been found in numerous
types of cancer, in particular male hormone-related prostate cancer
(2). Tissue microarray analysis using
Pim-1S (17) and Pim-1L antibodies
(5) showed that Pim-1S and Pim-1L are
largely upregulated only in the advanced, but not in the early,
stage of prostate cancer. Thus, Pim-1S and Pim-1L can be used as a
biomarker for prostate cancer. However, these two isoforms of Pim-1
show distinct roles in hormone-regulated signaling. Androgen
receptor (AR) has a central role in prostate cancer progression.
Pim-1S and Pim-1L phosphorylate AR at different sites. Pim-1S and
Pim-1L can interact with and phosphorylate AR at Ser-213, but only
Pim-1L can phosphorylate AR at Thr-850 (43). Pim-1S and Pim-1L mediated
phosphorylation results in recruiting the distinct ubiquitin E3
ligase. Our previous study showed that Pim-1S-induced Ser-213
phosphorylation of AR promotes AR degradation through ubiquitin E3
ligase Mdm2 depending on cell cycle (43). However, Pim-1L-induced Thr-850
phosphorylation stabilizes AR through ubiquitin E3 ligase RNF6 and
enhances AR target gene transcription under low-androgen conditions
(43). More data showed that Pim-1S
and Pim-1L can promote prostate cancer cell growth even in
low-androgen conditions (43). These
data suggest that Pim-1 has pivotal roles in hormone refractory
prostate cancer. Similar findings were reported that Pim-1S
phosphorylates AR at Ser-213 and inhibits AR target genes, such as
tumor suppressor genes NKX3.1 (44,45).
However, Pim-1S-mediated phosphorylation at Ser-213 also inhibits
AR target gene PSA. This paradox between oncogenic Pim-1S
and PSA most likely is caused by ubiquitination and degradation of
AR following phosphorylation at Ser-213. Pim-1L may switch AR
target genes by RNF6, as RNF6 regulates AR target genes specificity
(46).
Phosphorylation of heterochromatin protein 1γ at
Ser-93 by Pim-1 promotes its binding with histone H3K9me3, which
leads to heterochromatin formation and suppression of gene
transcriptions responsible for proliferation (40). Another epigenetic regulation of Pim-1
involves oncogenic transcription factor c-Myc (Myc). One study
showed that Pim-1 can directly regulate Myc transcriptional
activity (47). Pim-1 overexpression
alone is not enough to transform benign prostate RWPE1 cell line to
malignantly form (48). However, Pim-1
overexpression combined with Myc leads to development of the
advanced form of prostate carcinoma (49). Recent studies have shown that Pim-1
associates with Myc and can thereby regulate the epigenetic
dynamics of oncogene expression. Pim-1-mediated co-regulation
consists of ~20% of the Myc-regulated genes. Pim-1 phosphorylates
histone H3 at Ser-10 (H3S10) on the nucleosome at the MYC-binding
sites. This suggests that Pim-1 regulates transcriptional
activation, which contributes to Myc-transforming activity
(50). Thus, Pim-1 is a
transcriptional cofactor of Myc that phosphorylates the chromatin
at Myc-binding sites and regulates epigenetic dynamics for cellular
transformation.
Given the elevation of Pim-1 in highly advanced
stages of cancers, in addition to the important role of Pim-1 in
cell survival and proliferation, whether Pim-1 directly regulates
cancer cell invasion to induce metastasis remains to be elucidated.
Recently, one study showed Pim-1 overexpressing prostate cancer PC3
cells induced tumor invasion to prostate-draining lymph nodes, but
also into the lungs to form metastases in a Xenograft model
(51). Mechanistically, Pim-1
phosphorylates CXCR4 at Ser-339 for cell migration and invasion
(51). The epithelial-mesenchymal
transition (EMT) is known to be one of the mechanisms of
metastasis. Pim-1 is expressed at high levels in the stroma of
human prostate cancer samples (52).
Inducible overexpression of Pim-1 in immortalized prostate
fibroblast cell lines increased the differentiation of
myofibroblasts and transition of cancer-associated fibroblasts
(52). Pim-1 in fibroblasts
upregulated the expression levels of secreted proteins of
extracellular matrix collagen 1A1, chemokine CCL5, and the
platelet-derived growth factor receptors (52). In addition, Pim-1 upregulated c-MET, a
well-known EMT inducer through translational regulation (53). Pim-1 regulated MET through the control
of the translation of c-MET by regulating the phosphorylation of
eukaryotic initiation factor 4B (eIF4B) at Ser-406 (53). As c-MET kinase is an inducer of cancer
metastasis, these findings suggest Pim-1 may have a significant
potential in cancer metastasis by crosstalk with multiple
signaling.
Targeting Pim-1 in prostate cancer would be
promising for preventing the cancer recurrence caused by kinase
inhibitor drugs in clinical treatment. For example, the PI3K/AKT
pathway is a strong signaling pathway that promotes cell
proliferation and survival in numerous types of cancer including
prostate cancer. However, a single treatment targeting this pathway
has been a significant obstacle for therapy efficiency. One of the
mechanisms is that Akt inhibition can induce upregulation of
numerous receptor tyrosine kinases, such as c-MET, HER2 and insulin
receptor growth factor in prostate cancer cells through Pim-1
mediated regulation of translation in a cap-independent manner, but
internal ribosome entry-dependent manner (53). Furthermore, Pim-1 inhibition by
inhibitor SMI-4a represses the resistance to Akt inhibitor drugs
(54).
Pim-1 kinase is a critical enzyme that is involved
in cell growth, differentiation, survival, apoptosis, senescence
and drug resistance. Interaction of Pim-1 with different proteins
and association with various signaling pathways make it one of the
important antitumor targets. Numerous Pim-1 inhibitors are under
preclinical studies or clinical trials, such as P9 monoclonal
antibodies and AZD1208. An increasing number of new Pim-1
inhibitors are still developing and undergoing preclinical
investigations. These efforts further suggest that Pim-1 is
believed to be a master drug target in numerous types of cancer. In
addition, the fact that Pim-1 kinase inhibits transcriptional
activity of FOXP3 makes it an even more noteworthy antitumor
target, as it is thought that T regs are responsible for decreasing
the efficiency of cancer immunotherapies. Furthermore, Pim-1 can
promote drug resistance, a trait of cancer stem cells, through
interaction with and phosphorylation of Pgp, BCRP and FLT3-ITD,
which links Pim-1 as a promising targeted therapy in cancer stem
cells (69). The new findings of the
role of Pim-1 in cellular senescence in differential cancer
microenvironment (70) allow us to be
cautious for cancer treatment in individual therapy. As Pim-1 is a
potential biomarker of prostate cancer and crosstalk with numerous
signaling pathways, targeting Pim-1 in immunotherapy and
personalized therapy would be of great significance for the next
generation of precision medicine in cancer.
The study was supported in part by the seed grant to
Y.X.
|
1
|
Narlik-Grassow M, Blanco-Aparicio C and
Carnero A: The PIM family of serine/threonine kinases in cancer.
Med Res Rev. 34:136–159. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Warfel NA and Kraft AS: PIM kinase (and
Akt) biology and signaling in tumors. Pharmacol Ther. 151:41–49.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li J, Loveland BE and Xing PX: Anti-Pim-1
mAb inhibits activation and proliferation of T lymphocytes and
prolongs mouse skin allograft survival. Cell Immunol. 272:87–93.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Aho TLT, Sandholm J, Peltola KJ, Mankonen
HP, Lilly M and Koskinen PJ: Pim-1 kinase promotes inactivation of
the pro-apoptotic Bad protein by phosphorylating it on the Ser112
gatekeeper site. FEBS Lett. 571:43–49. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xie Y, Xu K, Dai B, Guo Z, Jiang T, Chen H
and Qiu Y: The 44 kDa Pim-1 kinase directly interacts with tyrosine
kinase Etk/BMX and protects human prostate cancer cells from
apoptosis induced by chemotherapeutic drugs. Oncogene. 25:70–78.
2006.PubMed/NCBI
|
|
6
|
Saris CJM, Domen J and Berns A: The pim-1
oncogene encodes two related protein-serine/threonine kinases by
alternative initiation at AUG and CUG. EMBO J. 10:655–664.
1991.PubMed/NCBI
|
|
7
|
Kumar A, Mandiyan V, Suzuki Y, Zhang C,
Rice J, Tsai J, Artis DR, Ibrahim P and Bremer R: Crystal
structures of proto-oncogene kinase Pim1: A target of aberrant
somatic hypermutations in diffuse large cell lymphoma. J Mol Biol.
348:183–193. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bachmann M and Möröy T: The
serine/threonine kinase Pim-1. Int J Biochem Cell Biol. 37:726–730.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yin J, Shine L, Raycroft F, Deeti S,
Reynolds A, Ackerman KM, Glaviano A, O'Farrell S, O'Leary O, Kilty
C, et al: Inhibition of the Pim1 oncogene results in diminished
visual function. PLoS One. 7:e521772012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Magnuson NS, Wang Z, Ding G and Reeves R:
Why target PIM1 for cancer diagnosis and treatment? Future Oncol.
6:1461–1478. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hofmann AD, Takahashi T, Duess J, Gosemann
JH and Puri P: Increased expression of activated pSTAT3 and PIM-1
in the pulmonary vasculature of experimental congenital
diaphragmatic hernia. J Pediatr Surg. 50:908–911. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang Y, Wang Z and Magnuson NS: Pim-1
kinase-dependent phosphorylation of p21Cip1/WAF1 regulates its
stability and cellular localization in H1299 cells. Mol Cancer Res.
5:909–922. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Morishita D, Katayama R, Sekimizu K,
Tsuruo T and Fujita N: Pim kinases promote cell cycle progression
by phosphorylating and down-regulating p27Kip1 at the
transcriptional and posttranscriptional levels. Cancer Res.
68:5076–5085. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lam LT, Zhang H, Xue J, Hessler P, Tahir
SK, Chen J, Jin S, Souers AJ and Leverson JD: Colorectal cancer
cell lines with high BCL-XL and low MCL-1 expression are sensitive
to a potent and selective BCL-XL inhibitor. Cancer Res. 74(Suppl
19): 27592014. View Article : Google Scholar
|
|
15
|
Kumar JK, Ping RYS, Teong HF, Goh S and
Clément MV: Activation of a non-genomic Pim-1/Bad-Pser75 module is
required for an efficient pro-survival effect of Bcl-xL induced by
androgen in LNCaP cells. Int J Biochem Cell Biol. 43:594–603. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Block KM, Hanke NT, Maine EA and Baker AF:
IL-6 stimulates STAT3 and Pim-1 kinase in pancreatic cancer cell
lines. Pancreas. 41:773–781. 2012.PubMed/NCBI
|
|
17
|
Dhanasekaran SM, Barrette TR, Ghosh D,
Shah R, Varambally S, Kurachi K, Pienta KJ, Rubin MA and Chinnaiyan
AM: Delineation of prognostic biomarkers in prostate cancer.
Nature. 412:822–826. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hu XF, Li J, Vandervalk S, Wang Z,
Magnuson NS and Xing PX: PIM-1-specific mAb suppresses human and
mouse tumor growth by decreasing PIM-1 levels, reducing Akt
phosphorylation, and activating apoptosis. J Clin Invest.
119:362–375. 2009.PubMed/NCBI
|
|
19
|
Li J, Hu XF, Loveland BE and Xing PX:
Pim-1 expression and monoclonal antibody targeting in human
leukemia cell lines. Exp Hematol. 37:1284–1294. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xie Y, Xu K, Linn DE, Yang X, Guo Z,
Shimelis H, Nakanishi T, Ross DD, Chen H, Fazli L, et al: The
44-kDa Pim-1 kinase phosphorylates BCRP/ABCG2 and thereby promotes
its multimerization and drug-resistant activity in human prostate
cancer cells. J Biol Chem. 283:3349–3356. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li Z, Lin F, Zhuo C, Deng G, Chen Z, Yin
S, Gao Z, Piccioni M, Tsun A, Cai S, et al: PIM1 kinase
phosphorylates the human transcription factor FOXP3 at serine 422
to negatively regulate its activity under inflammation. J Biol
Chem. 289:26872–26881. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nie H, Zheng Y, Li R, Guo TB, He D, Fang
L, Liu X, Xiao L, Chen X, Wan B, et al: Phosphorylation of FOXP3
controls regulatory T cell function and is inhibited by TNF-α in
rheumatoid arthritis. Nat Med. 19:322–328. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Morawski PA, Mehra P, Chen C, Bhatti T and
Wells AD: Foxp3 protein stability is regulated by cyclin-dependent
kinase 2. J Biol Chem. 288:24494–24502. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Oleinika K, Nibbs RJ, Graham GJ and Fraser
AR: Suppression, subversion and escape: The role of regulatory T
cells in cancer progression. Clin Exp Immunol. 171:36–45. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Isaac M, Siu A and Jongstra J: The
oncogenic PIM kinase family regulates drug resistance through
multiple mechanisms. Drug Resist Updat. 14:203–211. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xie Y, Burcu M, Linn DE, Qiu Y and Baer
MR: Pim-1 kinase protects P-glycoprotein from degradation and
enables its glycosylation and cell surface expression. Mol
Pharmacol. 78:310–318. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Natarajan K, Bhullar J, Shukla S, Burcu M,
Chen ZS, Ambudkar SV and Baer MR: The Pim kinase inhibitor SGI-1776
decreases cell surface expression of P-glycoprotein (ABCB1) and
breast cancer resistance protein (ABCG2) and drug transport by
Pim-1-dependent and -independent mechanisms. Biochem Pharmacol.
85:514–524. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kim KT, Baird K, Ahn JY, Meltzer P, Lilly
M, Levis M and Small D: Pim-1 is up-regulated by constitutively
activated FLT3 and plays a role in FLT3-mediated cell survival.
Blood. 105:1759–1767. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lilly M, Sandholm J, Cooper JJ, Koskinen
PJ and Kraft A: The PIM-1 serine kinase prolongs survival and
inhibits apoptosis-related mitochondrial dysfunction in part
through a bcl-2-dependent pathway. Oncogene. 18:4022–4031. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jiang T, Guo Z, Dai B, Kang M, Ann DK,
Kung HJ and Qiu Y: Bi-directional regulation between tyrosine
kinase Etk/BMX and tumor suppressor p53 in response to DNA damage.
J Biol Chem. 279:50181–50189. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Riganti C, Gazzano E, Gulino GR, Volante
M, Ghigo D and Kopecka J: Two repeated low doses of doxorubicin are
more effective than a single high dose against tumors
overexpressing P-glycoprotein. Cancer Lett. 360:219–226. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gribar JJ, Ramachandra M, Hrycyna CA, Dey
S and Ambudkar SV: Functional characterization of
glycosylation-deficient human P-glycoprotein using a vaccinia virus
expression system. J Membr Biol. 173:203–214. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Meshinchi S and Appelbaum FR: Structural
and functional alterations of FLT3 in acute myeloid leukemia. Clin
Cancer Res. 15:4263–4269. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Schmidt-Arras D, Böhmer SA, Koch S, Müller
JP, Blei L, Cornils H, Bauer R, Korasikha S, Thiede C and Böhmer
FD: Anchoring of FLT3 in the endoplasmic reticulum alters signaling
quality. Blood. 113:3568–3576. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Stout BA, Bates ME, Liu LY, Farrington NN
and Bertics PJ: IL-5 and granulocyte-macrophage colony-stimulating
factor activate STAT3 and STAT5 and promote Pim-1 and cyclin D3
protein expression in human eosinophils. J Immunol. 173:6409–6417.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Choudhary C, Olsen JV, Brandts C, Cox J,
Reddy PN, Böhmer FD, Gerke V, Schmidt-Arras DE, Berdel WE,
Müller-Tidow C, et al: Mislocalized activation of oncogenic RTKs
switches downstream signaling outcomes. Mol Cell. 36:326–339. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Natarjan K, Xie Y, Burcu M, Linn DE, Qui Y
and Baer MR: Pim-1 kinase phosphorylates and stabilizes 130 kDa
FLT3 and promotes aberrant STAT5 signaling in acute myeloid
leukemia with FLT3 internal tandem duplication. PLoS One.
8:e764532013.PubMed/NCBI
|
|
38
|
Lanigan F, Geraghty JG and Bracken AP:
Transcriptional regulation of cellular senescence. Oncogene.
30:2901–2911. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Vargas J, Feltes BC, Poloni JG and Bonatto
D: Senescence, an endogenous anticancer mechanism. Fronti Biosci
(Landmark Ed.). 17:2616–2643. 2012. View
Article : Google Scholar
|
|
40
|
Jin B, Wang Y, Wu CL, Liu KY, Chen H and
Mao ZB: PIM-1 modulates cellular senescence and links IL-6
signaling to heterochromatin formation. Aging Cell. 13:879–889.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mohsin S, Khan M, Nguyen J, Alkatib M,
Siddiqi S, Hariharan N, Wallach K, Monsanto M, Gude N, Dembitsky W,
et al: Rejuvenation of human cardiac progenitor cells with Pim-1
kinase. Circ Res. 113:1169–1179. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Samse K, Emathinger J, Hariharan N,
Quijada P, Ilves K, Völkers M, Ormachea L, De La Torre A, Orogo AM,
Alvarez R, et al: Functional ffect of Pim1 depends upon
intracellular localization in human cardiac progenitor cells. J
Biol Chem. 290:13935–13947. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Linn DE, Yang X, Xie Y, Alfano A, Deshmukh
D, Wang X, Shimelis H, Chen H, Li W, Xu K, et al: Differential
regulation of androgen receptor by PIM-1 kinases via
phosphorylation-dependent recruitment of distinct ubiquitin E3
ligases. J Biol Chem. 287:22959–22968. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ha S, Iqbal NJ, Mita P, Ruoff R, Gerald
WL, Lepor H, Taneja SS, Lee P, Melamed J, Garabedian MJ, et al:
Phosphorylation of the androgen receptor by PIM1 in hormone
refractory prostate cancer. Oncogene. 32:3992–4000. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Song H, Zhang B, Watson MA, Humphrey PA,
Lim H and Milbrandt J: Loss of Nkx3.1 leads to the activation of
discrete downstream target genes during prostate tumorigenesis.
Oncogene. 28:3307–3319. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Xu K, Shimelis H, Linn DE, Jiang R, Yang
X, Sun F, Guo Z, Chen H, Li W, Chen H, et al: Regulation of
androgen receptor transcriptional activity and specificity by
RNF6-induced ubiquitination. Cancer Cell. 15:270–282. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kim J, Roh M and Abdulkadir SA: Pim1
promotes human prostate cancer cell tumorigenicity and c-MYC
transcriptional activity. BMC Cancer. 10:2482010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kim J, Eltoum IE, Roh M, Wang J and
Abdulkadir SA: Interactions between cells with distinct mutations
in c-MYC and Pten in prostate cancer. PLoS Genet. 5:e10005422009.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang J, Kim J, Roh M, Franco OE, Hayward
SW, Wills ML and Abdulkadir SA: Pim1 kinase synergizes with c-MYC
to induce advanced prostate carcinoma. Oncogene. 29:2477–2487.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zippo A, De Robertis A, Serafini R and
Oliviero S: PIM1-dependent phosphorylation of histone H3 at serine
10 is required for MYC-dependent transcriptional activation and
oncogenic transformation. Nat Cell Biol. 9:932–944. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Santio NM, Eerola SK, Paatero I,
Yli-Kauhaluoma J, Anizon F, Moreau P, Tuomela J, Härkönen P and
Koskinen PJ: Pim kinases promote migration and metastatic growth of
prostate cancer xenografts. PLoS One. 10:e01303402015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zemskova MY, Song JH, Cen B, Cerda-Infante
J, Montecinos VP and Kraft AS: Regulation of prostate stromal
fibroblasts by the PIM1 protein kinase. Cell Signal. 27:135–146.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Cen B, Xiong Y, Song JH, Mahajan S, DuPont
R, McEachern K, DeAngelo DJ, Cortes JE, Minden MD, Ebens A, et al:
The Pim-1 protein kinase is an important regulator of MET receptor
tyrosine kinase levels and signaling. Mol Cell Biol. 34:2517–2532.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Cen B, Mahajan S, Wang W and Kraft AS:
Elevation of receptor tyrosine kinases by small molecule AKT
inhibitors in prostate cancer is mediated by Pim-1. Cancer Res.
73:3402–3411. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Tong Y, Stewart KD, Thomas S, Przytulinska
M, Johnson EF, Klinghofer V, Leverson J, McCall O, Soni NB, Luo Y,
et al: Isoxazolo[3,4-b]quinoline-3,4(1H,9H)-diones as unique,
potent and selective inhibitors for Pim-1 and Pim-2 kinases:
Chemistry, biological activities, and molecular modeling. Bioorg
Med Chem Lett. 18:5206–5208. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Holder S, Lilly M and Brown ML:
Comparative molecular field analysis of flavonoid inhibitors of the
PIM-1 kinase. Bioorg Med Chem. 15:6463–6473. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Blanco-Aparicio C, Collazo AM, Oyarzabal
J, Leal JF, Albarán MI, Lima FR, Pequeño B, Ajenjo N, Becerra M,
Alfonso P, et al: Pim 1 kinase inhibitor ETP-45299 suppresses
cellular proliferation and synergizes with PI3K inhibition. Cancer
Lett. 300:145–153. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Yang Q, Chen LS, Neelapu SS and Gandhi V:
Combination of Pim kinase inhibitor SGI-1776 and bendamustine in
B-cell lymphoma. Clin Lymphoma Myeloma Leuk. 13(Suppl 2):
S355–S362. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Keeton E, McEachern K, Alimzhanov M, Wang
S, Cao Y, Bao L, Palakurthi S, Grondine M, Chen Y, Dillman K, et
al: Efficacy and biomarker modulation by AZD1208, a novel, potent
and selective pan-Pim kinase inhibitor, in models of acute myeloid
leukemia. Cancer Res. 72:27962012. View Article : Google Scholar
|
|
60
|
Mondello P, Cuzzocrea S and Mian M: Pim
kinases in hematological malignancies: Where are we now and where
are we going? J Hematol Oncol. 7:952014. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Hospital MA, Green AS, Lacombe C, Mayeux
P, Bouscary D and Tamburini J: The FLT3 and Pim kinases inhibitor
SGI-1776 preferentially target FLT3-ITD AML cells. Blood.
119:1791–1792. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Foulks JM, Carpenter KJ, Luo B, Xu Y,
Senina A, Nix R, Chan A, Clifford A, Wilkes M, Vollmer D, et al: A
small-molecule inhibitor of PIM kinases as a potential treatment
for urothelial carcinomas. Neoplasia. 16:403–412. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Keeton EK, McEachern K, Dillman KS,
Palakurthi S, Cao Y, Grondine MR, Kaur S, Wang S, Chen Y, Wu A, et
al: AZD1208, a potent and selective pan-Pim kinase inhibitor,
demonstrates efficacy in preclinical models of acute myeloid
leukemia. Blood. 123:905–913. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Kirschner AN, Wang J, van der Meer R,
Anderson PD, Franco-Coronel OE, Kushner MH, Everett JH, Hameed O,
Keeton EK, Ahdesmaki M, et al: PIM kinase inhibitor AZD1208 for
treatment of MYC-driven prostate cancer. J Natl Cancer Inst.
107:dju4072014. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Hogan C, Hutchison C, Marcar L, Milne D,
Saville M, Goodlad J, Kernohan N and Meek D: Elevated levels of
oncogenic protein kinase Pim-1 induce the p53 pathway in cultured
cells and correlate with increased Mdm2 in mantle cell lymphoma. J
Biol Chem. 283:18012–18023. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Turaka A, Buyyounouski MK, Hanlon AL,
Horwitz EM, Greenberg RE and Movsas B: Hypoxic prostate/muscle PO2
ratio predicts for outcome in patients with localized prostate
cancer: Long-term results. Int J Radiat Oncol Biol Phys.
82:e433–e439. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Guo Z, Wang A, Zhang W, Levit M, Gao Q,
Barberis C, Tabart M, Zhang J, Hoffmann D, Wiederschain D, et al:
PIM inhibitors target CD25-positive AML cells through concomitant
suppression of STAT5 activation and degradation of MYC oncogene.
Blood. 124:1777–1789. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Liang C and Li YY: Use of regulators and
inhibitors of Pim-1, a serine/threonine kinase, for tumour therapy
(Review). Mol Med Rep. 9:2051–2060. 2014.PubMed/NCBI
|
|
69
|
Xie Y and Bayakhmetov S: PIM1 kinase as a
promise of targeted therapy in prostate cancer stem cells (Review).
Mol Clin Oncol. 4:13–17. 2016.
|
|
70
|
Xie Y, Lu W, Liu S, Yang Q, Carver BS and
Chen Z: The essential role of ARF in prostate cancer
microenvironment. BJU Int. 116:41. 2015.
|