Introduction
Gastric cancer is one of the most common human
cancers, with ~988,000 cases/year worldwide. It remains difficult
to treat and ~736,000 patients succumb to the disease each year
(1). In China, gastric cancer is the
leading cause of cancer-related mortality and accounts for ~23% of
all malignant deaths (2). It has been
previously demonstrated that gastric cancer is caused by complex
interactions between genetic and environmental factors. The
dysregulation of potential oncogenic signaling pathways can lead to
increased cell proliferation, evasion of apoptosis and enhanced
invasiveness (3). Furthermore, the
dysregulation of the nuclear factor κB, Wnt/β-catenin and
proliferation/stem cell signaling pathways are identified in 70%
patients with gastric cancer (4).
Frequently rearranged in advanced T cell lymphomas-1 (FRAT1) is a
member of the FRAT family, and is a positive regulator of β-catenin
in the Wnt pathway (5). The
Wnt/β-catenin pathway is closely associated with the pathogenesis
and development of many solid tumors. The overexpression of FRAT1
in ovarian serous adenocarcinomas was significantly associated with
cytoplasmic and nuclear accumulation of β-catenin 6). FRAT1
inhibited GSK-3-mediated phosphorylation of β-catenin and affected
the formation of the destruction complex for β-catenin, leading to
subsequent aberrant nuclear accumulation of β-catenin, which
elevated the transcription activity of β-catenin. The downstream
transcription targets of β-catenin pathway, such as c-myc, were
activated to enhance the cellular growth (7,8).
Additionally, a previous study has demonstrated that FRAT1 is
overexpressed in gastric cancer (9).
The present study aimed to investigate the effects of FRAT1
silencing on the proliferation, apoptosis and the cell cycle of the
human gastric cancer cell line, SGC7901.
Materials and methods
Cell culture
Human gastric adenocarcinoma SGC7901 cells were
obtained from the Institute of Basic Medical Sciences (Chinese
Academy of Medical Science, Beijing, China). Cells were maintained
in RPMI-1640 (GE Healthcare Life Sciences, Logan, UT, USA)
supplemented with 10% fetal bovine serum, 100 U/ml penicillin and
100 mg/ml streptomycin (all from GE Healthcare Life Sciences), at
37°C in a humidified atmosphere containing 5% CO2. The
medium was replaced every two days and cells were passaged twice
weekly.
Transfection
The oligonucleotide sequence
5′-GCAGTTACGTGCAAAGCTT-3′ (Takara Biotechnology Co., Ltd., Dalian,
China), specific to FRATl mRNA was used for the synthesis of small
interfering RNA (siRNA), which was cloned into pSINsi-hu6 vector
(Takara Biotechnology). SGC7901 cells were transfected with the
siFRAT1 vector using Lipofectamine® 2000 (Invitrogen Life
Technologies, Carlsbad, CA, USA), performed according to the
manufacturer's instructions. The transfected SCG7901 cells were
screened using 800 µg/ml G418 (Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) and were defined as group A (10). The non-targeting oligonucleotide
sequence 5′-TCTTAATCGCGTATAAGGC-3′ (Takara Biotechnology) was
transfected as a control group B. The untreated SGC7901 cells were
defined as group C.
FRAT1 mRNA expression analysis
FRAT1 mRNA expression was assessed by reverse
transcription-quantitative polymerase chain reaction (RT-qPCR).
TRIzol reagent RNA kit (Invitrogen; Thermo Fisher Scientific) was
used to extract total RNA, following the protocol provided by the
manufacturer. Takara RNA PCR kit (Takara Biotechnology) was used to
perform the reverse transcriptase polymerase chain reaction. The
following primers were used. FRAT1: Forward,
5′-GGCAGAACCTGGCTACTCTG-3′ and reverse
5′-CACGAGCTTGATTGCAAGTTCAGG-3′; GAPDH: Forward
5′-CCACGCCCTGTCTAAAGTGT-3′ and reverse 5′-GGGGTCATTGATGGCAACAAT
A-3′ (Takara Biotechnology). The complementary DNA mixed with
forward and reverse primers was reacted in Exicycler™ 96 (Bioneer
Co., Daejeon, Korea). An initial denaturation/activation step at
95°C for 10 min was followed by 35 cycles at 95°C for 10 sec, 60°C
for 20 sec and 72°C for 30 sec, finally held at 4°C for 5 min. The
FRAT1 mRNA relative expression ratio was calculated using the
2−ΔΔcq method, and the result of group C was considered
the unit value 1.
FRAT1 and β-catenin protein expression
analysis
Western blot analysis was used to measure the
protein expression levels of FRAT1 and β-catenin. Anti-FRAT1 (cat.
no. ab108405)and anti-β-actin (cat. no. ab95437) antibodies were
purchased from Abcam, Cambridge, UK. The anti-β-catenin antibody
was purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX,
USA). The optical density of bands was measured by Image-Pro Plus
software, version 6.0 (Media Cybernetics, Inc., Rockville, MD,
USA). Protein expression was reported as relative level with
respect to the β-actin in the same sample, and the value of group C
was considered the unit value 1.
Cells proliferation assay
The SGC7901 cells were seeded into a 96-well cell
culture cluster plate at a concentration of 1×104
cells/well in a volume of 100 µl culture medium and cultured for 24
h. A 200 µl RPMI-1640 culture medium supplemented with 10% fetal
bovine serum was then added into every well and the cells were
harvested at 24, 48 and 72 h. The harvested cells were incubated in
200 µl RPMI-1640 culture medium supplemented with 0.5%
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
(KeyGen Biotech. Co., Ltd., Nanjing, China) for 4 h and then 150 µl
of dimethyl sulphoxide was added to dissolve the formazan crystals
for 10 min immediately prior to the assay. Absorbency was measured
at a wavelength of 490 nm using a microplate reader (Bio-Rad,
Berkeley, CA, USA). The assay was repeated ≥3 times for cell
proliferation analysis.
Cells cycle analysis
The SGC7901 cells were cultured in 25 ml culture
bottle to a confluence of 80% and then treated with trypsogen (GE
Healthcare Life Sciences) to harvest the cells. The cells were then
fixed in 75% ethanol overnight at 4°C and incubated with 50 mg/L
RNase A (GE Healthcare Life Sciences) at 37°C for 30 min. Flow
cytometry was performed after the cells were stained with a 50 mg/L
propidium iodide solution (BD FACScan; BD Biosciences, San Diego,
CA, USA) for 20 min in the dark.
Apoptosis assay
The SGC7901 cells were seeded into a 96-well cell
culture cluster plate and incubated for 48 h. The cells were then
harvested by trypsinization and washed twice with 4°C phosphate
buffered saline. Cells were then suspended in 1 ml binding buffer
(0.01 M HEPES/NaOH, 0.14 M NaCl, 2.5 mM CaCl2, pH 7.4)
at a concentration of 1–5×105 cells/ml. Annexin
V-fluorescein isothiocyanate (10 µl) (BD Biosciences) and propidium
iodide (5 µl) (BD Biosciences) were added to the cells, followed by
incubation with gentle mixing for 15 min at room temperature in the
dark. The Annexin V stained cells were analyzed using a BD Model
FACScan (BD Biosciences).
Statistical analysis
Data are presented as the mean ± standard deviation.
SPSS 13.0 (SPSS, Inc., Chicago, IL, USA) was used for data
analysis. P<0.05 was considered to indicate a statistically
significant difference.
Results
FRAT1 expression
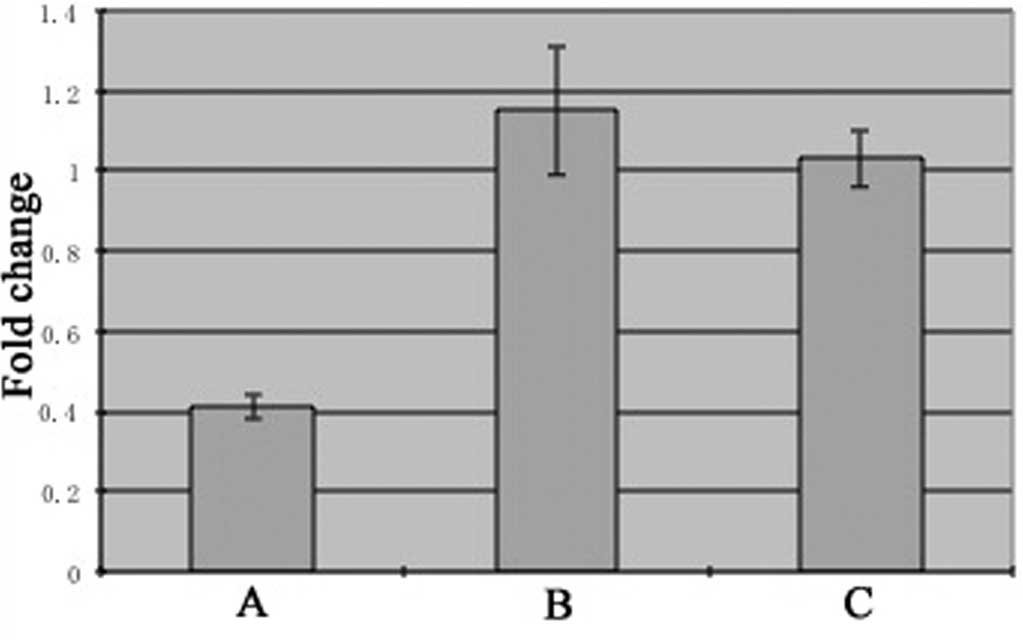
The relative expression of FRAT1 mRNA in SGC7901
cells treated with siFRAT1 (group A; 0.41±0.03) was decreased
significantly compared with control groups B (1.15±0.16, P<0.05)
and C (1.03±0.07, P<0.05) (Fig. 1).
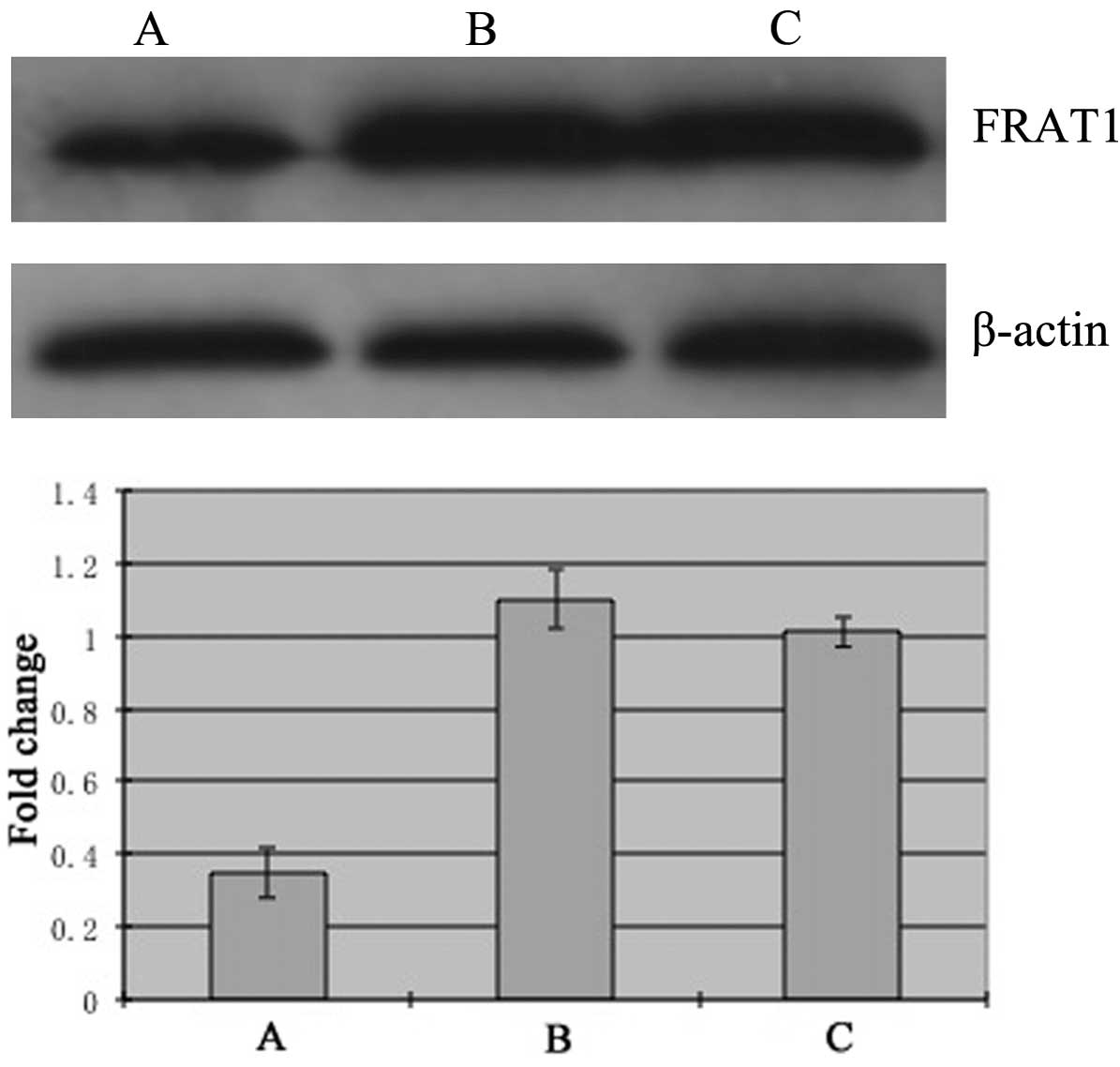
The expression of FRAT1 protein in group A (0.35±0.07) was
decreased significantly compared with control groups B (1.10±0.08,
P<0.05) and C (1.01±0.04, P<0.05) (Fig. 2). These results indicate that the
silencing of FRAT1 by siRNA targeting FRAT1 in SGC7901 cells was
successful.
β-catenin expression
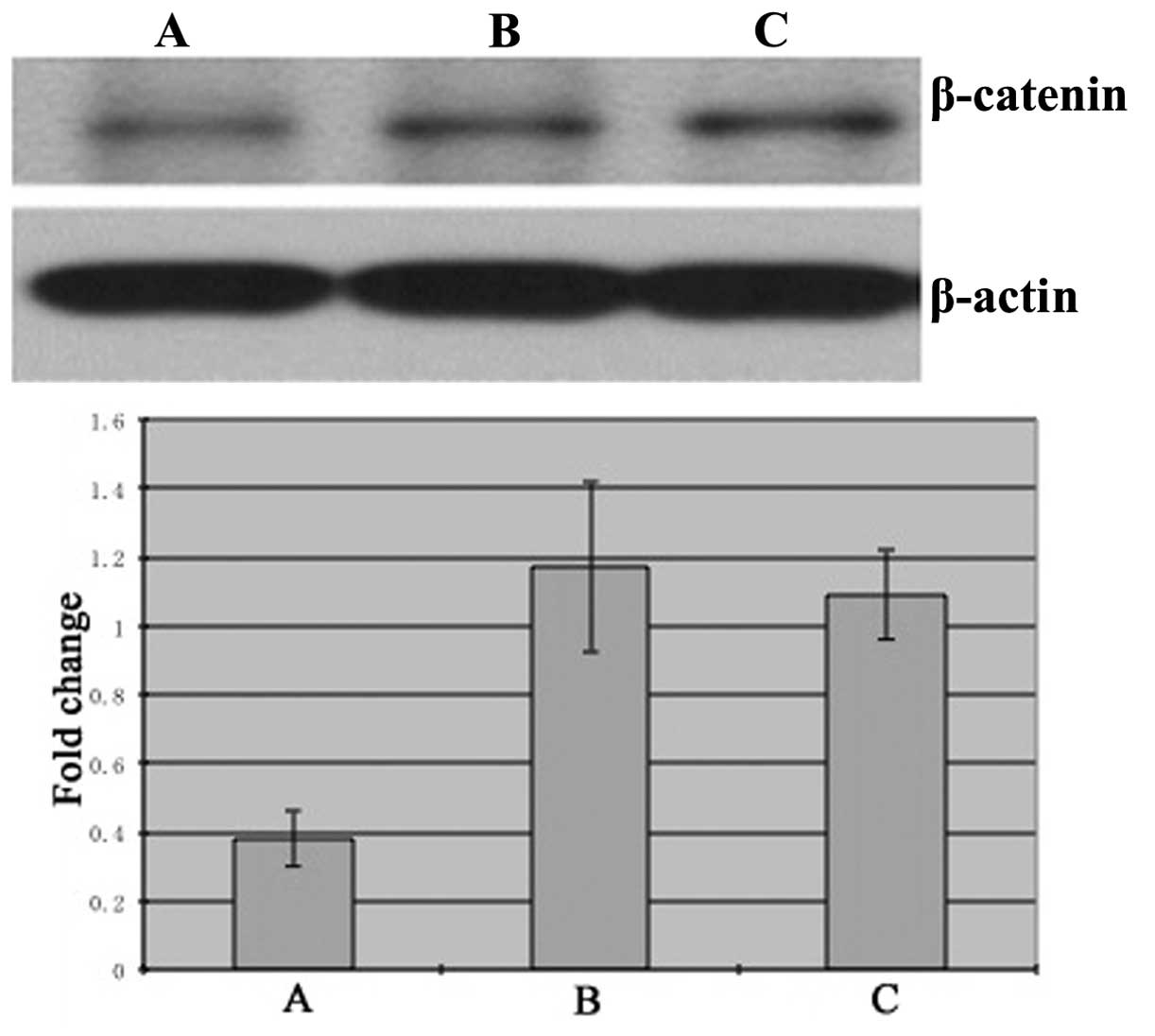
The expression of β-catenin protein in SGC7901 cells
treated with siFRAT1 (group A; 0.38±0.08) was decreased
significantly compared with the control group B (1.17±0.25,
P<0.05) and untreated group C (1.09±0.13, P<0.05) (Fig. 3).
Cells proliferation
The cells proliferation in siFRAT1-treated group A
was decreased significantly compared with control groups B and C at
48 h (P<0.05) and 72 h (P<0.05) (Table I). These results demonstrate that the
silencing of FRAT1 inhibited the proliferation of SGC7901
cells.
 | Table I.Proliferation of SGC7901 cells,
n=3. |
Table I.
Proliferation of SGC7901 cells,
n=3.
|
| Time, h |
|---|
|
|
|
|---|
| Group | 0 | 24 | 48 | 72 |
|---|
| A | 0.26±0.02 | 0.33±0.01 |
0.36±0.01a |
0.39±0.01a |
| B | 0.27±0.03 | 0.35±0.01 | 0.54±0.01 | 0.69±0.00 |
| C | 0.26±0.01 | 0.35±0.02 | 0.57±0.03 | 0.68±0.02 |
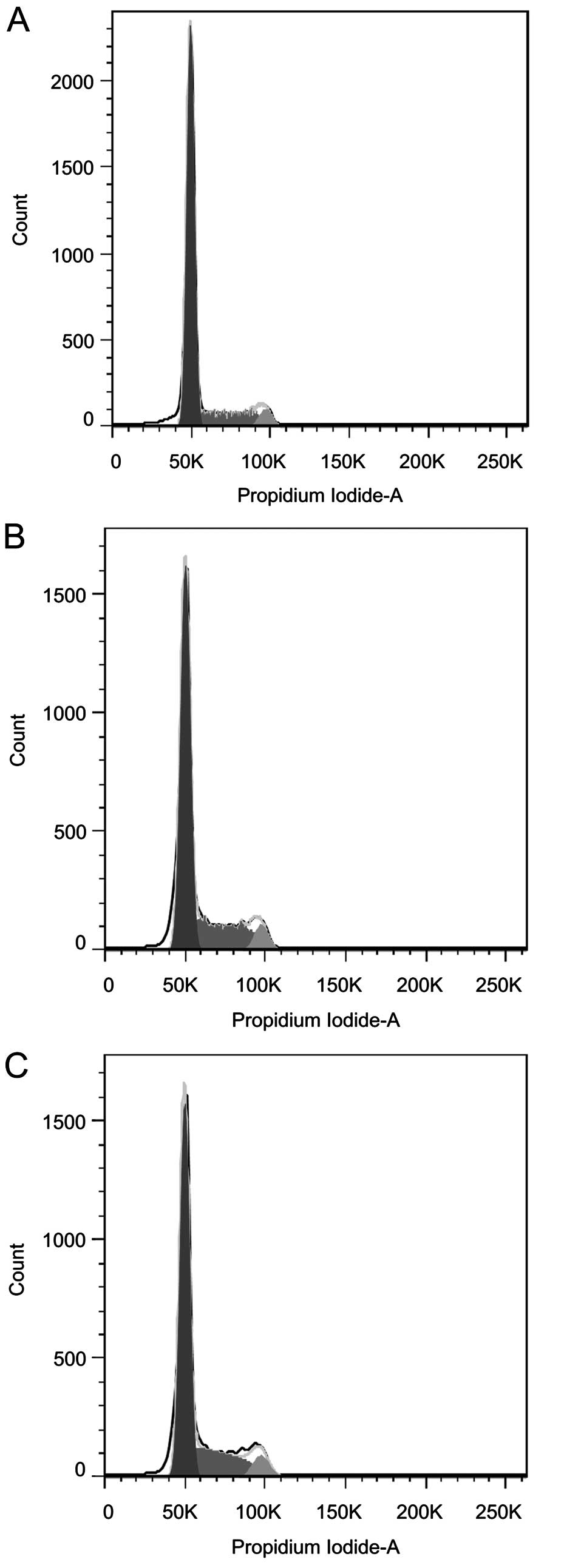
Cells cycle
The cell cycle distribution in siFRAT1-treated group
A was significantly different to that of the control groups B and
C. The G0/G1 stage cells in group A was increased significantly
compared with groups B and C (P<0.05). The S and G2/M stage
cells in group A was decreased significantly compare with groups B
and C (P<0.05) (Table II and
Fig. 4). These results demonstrate
that the silencing of FRAT1 in SGC7901 cells led to increased
arrest at the G0/G1 stage.
 | Table II.Cell cycle distribution of SGC7901
cells, n=3. |
Table II.
Cell cycle distribution of SGC7901
cells, n=3.
|
| Cell cycle phase |
|---|
|
|
|
|---|
| Group | G0/G1 | S | G2/M |
|---|
| A |
73.02±0.52a |
18.75±0.39a |
2.70±0.17a |
| B | 61.77±0.08 | 26.45±0.12 | 6.44±0.11 |
| C | 63.93±0.64 | 25.99±0.62 | 6.45±0.09 |
Cells apoptosis
The fraction of apoptosis in siFRAT1-treated group A
(3.87±0.08) was increased significantly compared with control
groups B (1.62±0.02, P<0.05) and C (1.22±0.02, P<0.05). These
results demonstrate that the silencing of FRAT1 in SGC7901 cells
led to an increase in apoptosis.
Discussion
The FRAT1 gene is located on human chromosome
10q24.1 and encodes a protein comprising 279 amino acids that is
overexpressed in gastric cancer (9).
FRAT1 can inhibit glycogen synthase kinase-3 mediated
phosphorylation of β-catenin and act as a positive regulator of the
Wnt/β-catenin pathway (11,12). The Wnt/β-catenin signaling cascade
modulates the expression of genes that govern cell proliferation,
cell survival, migration, neural development and angiogenesis
during morphogenesis (13–16). It has been previously demonstrated that
FRAT1 plays a important role in tumor progression (17,18). FRAT1
inhibits the phosphorylation of β-catenin, causing it to accumulate
in the nucleus. β-catenin then binds with T cell transcriptional
factor/lymphoid enhancer factor to form a complex, which can drive
c-myc, Cox-2 and cyclin D1 to alter the cell cycle or express
abnormal protein leading to the tumorigenesis (19). Furthermore, overexpression of FRAT1 in
transgenic mice leads to lymphoma progression (20) and knockdown of FRAT1 by RNA
interference inhibits glioblastoma cell growth, migration and
invasion (21). Additionally, the
overexpression of FRAT1 is associated with a malignant phenotype
and poor prognosis in human gliomas (22).
The present study demonstrates that FRAT1 mRNA and
protein expression in SGC7901 cells was inhibited by RNA
interference. The expression of FRAT1 mRNA and FRAT1 protein were
reduced significantly in comparison to untreated cells. The results
also demonstrate that the expression of β-catenin protein was
significantly decreased. Furthermore, the proliferation of FRAT1
silenced SGC7901 cells decreased significantly and the cell cycle
distribution was significantly different from the untreated cells,
with more cells arrested at G0/G1 stage. Apoptosis of FRAT1
silenced SGC7901 cells was increased significantly.
In conclusion, the results of the present study
indicate that FRAT1 is important for the proliferation of SGC7901
cells and silencing of FRAT1 by siRNA can inhibit the proliferation
of the SGC7901 cells. A reduction in the expression of β-catenin
may be a potential mechanism for the effects of FRAT1 silencing on
cell proliferation, apoptosis and cell cycle distribution. FRAT1
may be a potential prognostic biomarker and therapeutic target for
gastric cancer.
Acknowledgements
The study was supported by a fund from the Science
and Technology of Dalian Public Health Bureau, Liaoning, China
(grant no. 2014.142).
Glossary
Abbreviations
Abbreviations:
|
FRAT1
|
frequently rearranged in advanced T
cell lymphomas-1
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
MTT
|
methyl thiazolyl tetrazolium
|
References
|
1
|
Kamangar F, Dores GM and Anderson WF:
Patterns of cancer incidence, mortality, and prevalence across five
continents: defining priorities to reduce cancer disparities in
different geographic regions of the world. J Clin Oncol.
24:2137–2150. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zou XN, Duan JJ, Huangfu XM, Chen WQ and
Zhao P: Analysis of stomach cancer mortality in the national
retrospective sampling survey of death causes in China, 2004-2005.
Zhonghua Yu Fang Yi Xue Za Zhi. 44:390–397. 2010.(In Chinese).
PubMed/NCBI
|
|
3
|
Wu WK, Cho CH, Lee CW, Fan D, Wu K, Yu J
and Sung JJ: Dysregulation of cellular signaling in gastric cancer.
Cancer Lett. 295:144–153. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ooi CH, Ivanova T, Wu J, Lee M, Tan IB,
Tao J, Ward L, Koo JH, Gopalakrishnan V, Zhu Y, et al: Oncogenic
pathway combinations predict clinical prognosis in gastric cancer.
PLoS Genet. 5:e10006762009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jonkers J, Korswagen HC, Acton D, Breuer M
and Berns A: Activation of a novel proto-oncogene, Frat1,
contributes to progression of mouse T-cell lymphomas. EMBO J.
16:441–450. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang Y, Hewitt SM, Liu S, Zhou X, Zhu H,
Zhou C, et al: Tissue microarray analysis of human FRAT1 expression
and its correlation with the subcellular localisation of
beta-catenin in ovarian tumours. Br J Cancer. 94:686–691.
2006.PubMed/NCBI
|
|
7
|
Wang Y, Liu S, Zhu H, Zhang W, Zhang G,
Zhou X, Zhou C, Quan L, Bai J, Xue L, et al: FRAT1 overexpression
leads to aberrant activation of beta-catenin/TCF pathway in
esophageal squamous cell carcinoma. Int J Cancer. 123:561–568.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Guo G, Mao X, Wang P, Liu B, Zhang X,
Jiang X, Zhong C, Huo J, Jin J and Zhuo Y: The expression profile
of FRAT1 in human gliomas. Brain Res. 1320:152–158. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Saitoh T and Katoh M: FRAT1 and FRAT2,
clustered in human chromosome 10q24.1 region, are up-regulated in
gastric cancer. Int J Oncol. 19:311–315. 2001.PubMed/NCBI
|
|
10
|
Yu QG, Gu W, Wang SY, Zhang XX, Yang ZR
and Li WS: Establishment of FRAT1 gene silence HT29 cells model by
RNA interference. China Mod Med. 21:12–15. 2014.
|
|
11
|
Jonkers J, van Amerongen R, van der Valk
M, Robanus-Maandag E, Molenaar M, Destrée O and Berns A: In vivo
analysis of Frat1 deficiency suggests compensatory activity of
Frat3. Mech Dev. 88:183–194. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yost C, Farr GH III, Pierce SB, Ferkey DM,
Chen MM and Kimelman D: GBP, an inhibitor of GSK-3, is implicated
in Xenopus development and oncogenesis. Cell. 93:1031–1041. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Logan CY and Nusse R: The Wnt signaling
pathway in development and disease. Annu Rev Cell Dev Biol.
20:781–810. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liebner S and Plate KH: Differentiation of
the brain vasculature: the answer came blowing by the Wnt. J
Angiogenes Res. 2:12010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ille F and Sommer L: Wnt signaling:
multiple functions in neural development. Cell Mol Life Sci.
62:1100–1108. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Reis M and Liebner S: Wnt signaling in the
vasculature. Exp Cell Res. 319:1317–1323. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Saitoh T, Mine T and Katoh M: Molecular
cloning and expression of proto-oncogene FRAT1 in human cancer. Int
J Oncol. 20:785–789. 2002.PubMed/NCBI
|
|
18
|
Zhang Y, Han Y, Zheng R, Yu JH, Miao Y,
Wang L and Wang EH: Expression of Frat1 correlates with expression
of β-catenin and is associated with a poor clinical outcome in
human SCC and AC. Tumour Biol. 33:1437–1444. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chiurillo MA: Role of the Wnt/β-catenin
pathway in gastric cancer: An in-depth literature review. World J
Exp Med. 5:84–102. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jonkers J, Weening JJ, van der Valk M,
Bobeldijk R and Berns A: Overexpression of Frat1 in transgenic mice
leads to glomerulosclerosis and nephrotic syndrome, and provides
direct evidence for the involvement of Frat1 in lymphoma
progression. Oncogene. 18:5982–5990. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Guo G, Kuai D, Cai S, Xue N, Liu Y, Hao J,
Fan Y, Jin J, Mao X, Liu B, et al: Knockdown of FRAT1 expression by
RNA interference inhibits human glioblastoma cell growth, migration
and invasion. PLoS One. 8:e612062013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Guo G, Zhong CL, Liu Y, Mao XG, Zhang Z,
Jin J, et al: Overexpression of FRAT1 is associated with malignant
phenotype and poor prognosis in human gliomas. Dis Markers.
2015:2897502015. View Article : Google Scholar : PubMed/NCBI
|