Introduction
Prostate cancer (PC) is one of the most common
cancer types inflicting men, only second to cutaneous cancer, and
the second leading cause of cancer-related mortalities for men
(1). In 2014, an estimated number of
233,000 men were diagnosed with PC and 29,480 men succumbed to the
disease in the USA (2). This indicated
the need to further investigate PC.
Hypertension is the largest attributable risk factor
for mortality worldwide (3), and is
responsible for >50% of stroke and coronary heart disease (CHD).
The risk factors of hypertension include sedentary lifestyle,
stress, visceral obesity, potassium deficiency, obesity, salt
sensitivity, alcohol intake and vitamin D deficiency (4). Previous studies suggested that men with
hypertension are more likely to be diagnosed with PC than those
without hypertension (5). However, the
number of studies exploring the correlation between hypertension
and PC risk are rather limited. Han et al and Takeshita
et al considered that high blood pressure is positively
associated with concurrent serum PSA levels (6,7).
Previous findings showed that the microbial
population in EPS, urine and seminal fluid between the subjects
with PC and benign prostatic hyperplasia (BPH) are significantly
different, indicating a correlation between PC with urinary
microbiota (8).
A small number of investigations regarding the
relationship between hypertension and intestinal bacteria have been
conducted whereas few studies focus on the effect of hypertension
on prostate. A hypothesis was posited in the present study that
hypertension can affect the intestinal bacteria of PC.
Use of traditional culture methods does not allow
for detection of many anaerobic bacteria present in various human
body fluids and tissues (9,10). The 16S rDNA-based polymerase chain
reaction (PCR) is more sensitive than the traditional PCR,
depending on microbial culture techniques (11,12).
Bacterial species are identified by generating clone libraries of
the 16S rDNA followed by sequencing and comparison with databases
containing thousands of ribosomal sequences (13,14). This
method has previously utilized by researchers to evaluate bacterial
16S rDNA sequences in prostatic tissue from patients with PC
(15–17).
The aim of the present study was to compare the
bacterial composition in the biopsy of PC patients in PSA grey-zone
with hypertension with that of the patients without hypertension by
PCR-denaturing gradient gel electrophoresis (DGGE) with 16S rDNA
methods.
Materials and methods
Sample collection
Four biopsy samples were collected from male
patients diagnosed with PC in the First Affiliated Hospital of
Medical School of Zhejiang University (Zhejiang, China). An
ultrasound-guided instrument was used to obtain transperineal
prostate biopsies. All the patients included in the study were
under the age of 65 and the tPSA levels were 4–10 ng/ml. Four
samples were selected from 37 patients and divided into two groups:
i) patients with PC (with and without hypertension); and ii)
patients with BPH (with and without hypertension). The prostate
biopsy samples were placed in sterile centrifuge tubes and stored
at −80°C prior to use.
Procedures performed in studies involving human
participants were in accordance with the Ethical Standards of the
Institutional and/or National Research Committee and with the 1964
Helsinki Declaration and its later amendments or comparable ethical
standards. Informed consent was obtained from all individual
participants included in the study.
DNA extraction
Total genomic DNA was isolated from the biopsy
samples according to the instructions of the QIAamp® DNA
mini kit (Qiagen, Hilden, Germany). The extracted DNA was packed
into three tubes to avoid multi-gelation and stored at −20°C.
PCR amplification
Each DNA sample used in this study was first
amplified with universal bacterial primers. The forward primer 341
(5′-GTATTACCGCGGCTGCTGG-3′) containing a 40-bp GC clamp
(5′-CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGG-3′) and the reverse
primer 534 (5′-ACTCCTACGGGAGGCAGCAG-3′) resulted in fragments of
approximately 200 bp to test the quality of the template and to
exlude the presence of PCR inhibitors. The GC clamp increased the
sensitivity of the DGGE analysis (18). The total PCR reaction volume was 50 µl.
The PCR mixture comprised 1 µl of Bestar Taq DNA polymerase (2.5
U/µl), 5 µl of deoxynucleoside triphosphates (dNTPs, 2 mM each), 5
µl of 10X Bestar Taq buffer (all from DBI Bioscience, Shanghai,
China), 1 µl of each primer (10 µM; Sangon, Shanghai, China) and 2
µl of extracted bacterial DNA (~60 ng). The thermal cycling program
was set at 94°C for 5 min, with 35 cycles of touchdown PCR
denaturation at 94°C for 30 sec, annealing at 60°C for 30 sec, and
72°C extension for 30 sec and a final extension at 72°C for 5 min,
prior to incubation at 4°C.
PCR products were tested by electrophoresis on 1.0%
(w/v) agarose gel. Electrophoresis was performed at 100 V for 20
min with 1X TAE buffer, and visualized by ethidium bromide staining
using a gel imaging system (JS-780; Pei Qing Technology Co., Ltd.,
Shanghai, China). PCR products were stored at −20°C prior to DGGE
electrophoresis.
DGGE electrophoresis
DNA fragments with different sequences were
separated in 8% polyacrylamide (acrylamide: bisacrylamide = 37.5:1;
w/v) gels in 1X TAE buffer with 200 ng of each PCR product. A
denaturing gradient of 40–60% was applied in the DGGE
electrophoresis, formed with deionized formamide and urea. Gels
were electrophorised in 1X TAE buffer at 60°C and 200 V for 3.5 h.
Subsequently, the gels were washed with ultrapure water and stained
with 5% GoldView™ dye for 30 min and photographed. DGGE graphs were
digitized by Quantity One Analysis software (Gene Genius; Syngene,
Frederick, MD, USA).
DGGE band sequencing
Selected DGGE bands (2 and 4) were cut with a
sterile surgical blade under UV and purified with DNA Gel
Extraction kit (SK1135; Sangon). Purified DNA was then amplified
again with the primers described earlier. PCR products were ligated
into the pUCm-T vector and transformed into competent E.
coli DH5α cells (both from Sangon). Recombinant cells were
selected and inoculated into the fluid medium that was loaded with
antibiotic ampicillin and were cultured overnight at 37°C.
Subsequently, bacteria were collected and DNA with the plasmid was
extracted using a UNIQ-10 column kit (Sangon) as per the
manufacturer's protocol. Clones that migrated to the same position
as the original DGGE bands were sequenced (Sangon).
Obtained sequences were searched online based on the
NCBI GenBank database (http://www.ncbi.nlm.nih.gov) BLAST to identify the
closest relative for the partial 16S rRNA gene. The same sequences
were identified when the similarity of the sequence was >97%.
Based on the BLAST results, reference sequences of phylogenetic
neighbor species (up to 97% similarity) were included for
construction of the phylogenetic tree using the MEGA 5 software
package, ver. 5.05, according to the method of neighbor-joining
based on evolutionary distances. The consistency of the tree was
validated by bootstrapping (n=1,000).
Results
PCR efficiency detection
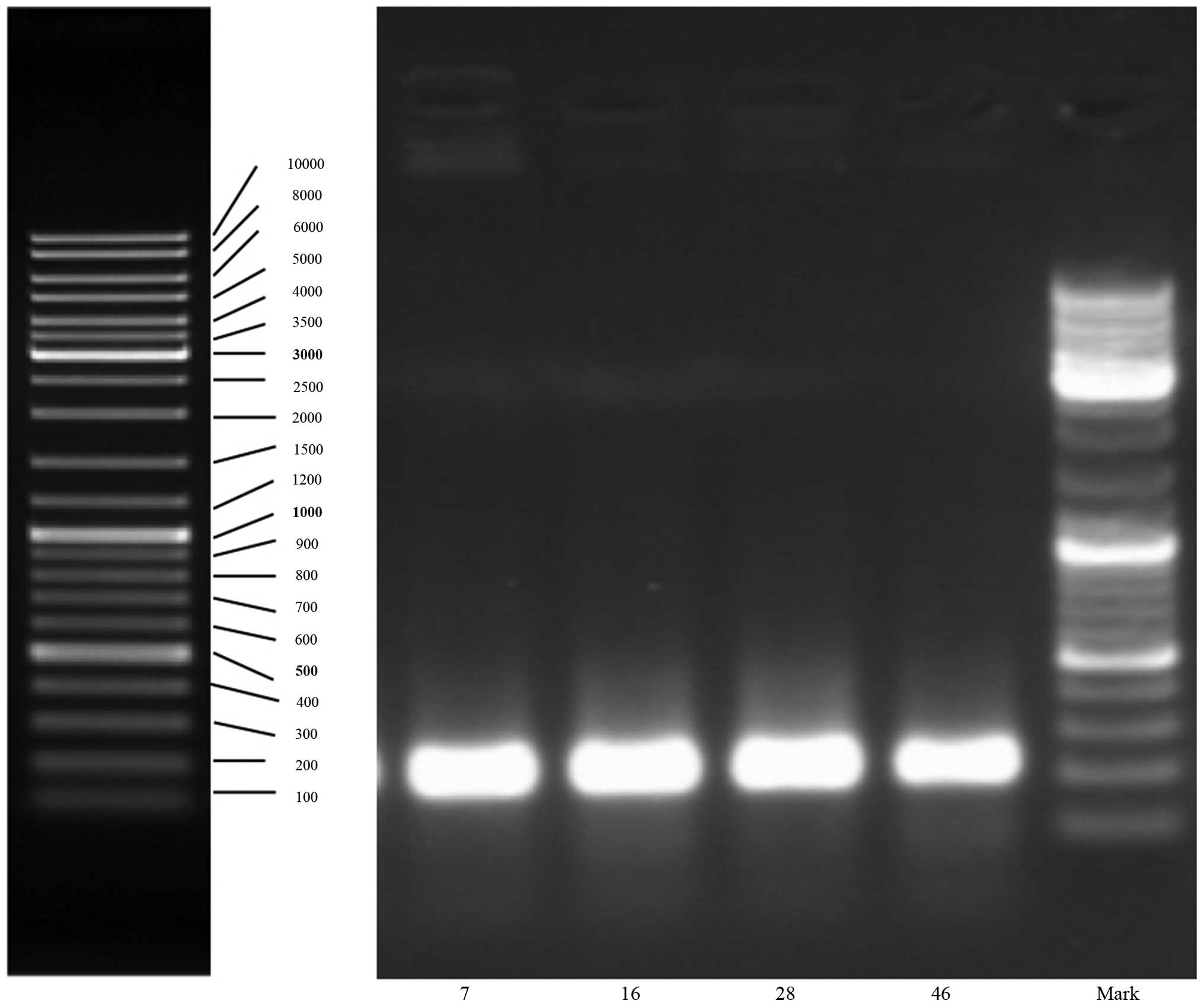
Patient characteristics are provided in Table I. PSA analysis was performed before
prostate biopsy. The total DNA of each biopsy tissue was
successfully extracted and the amplified fragments of PCR were ~230
bp (Fig. 1). Based on the results
obtained, it is evident that 16s RNA gene fragments of bacteria in
the specimens had good amplification efficiency.
 | Table I.Characteristics of patients with PC
groups. |
Table I.
Characteristics of patients with PC
groups.
| Group | Number | PSA (T/F) | Diagnosis | Remarks |
|---|
| PC | 7 |
9.47/1.425 | PC | None |
|
| 16 | 7.035/0.309 | PC | Hypertension |
| BPH | 28 | 5.14/0.69 | BPH | Hypertension |
|
| 46 | 7.29/0.63 | BPH | None |
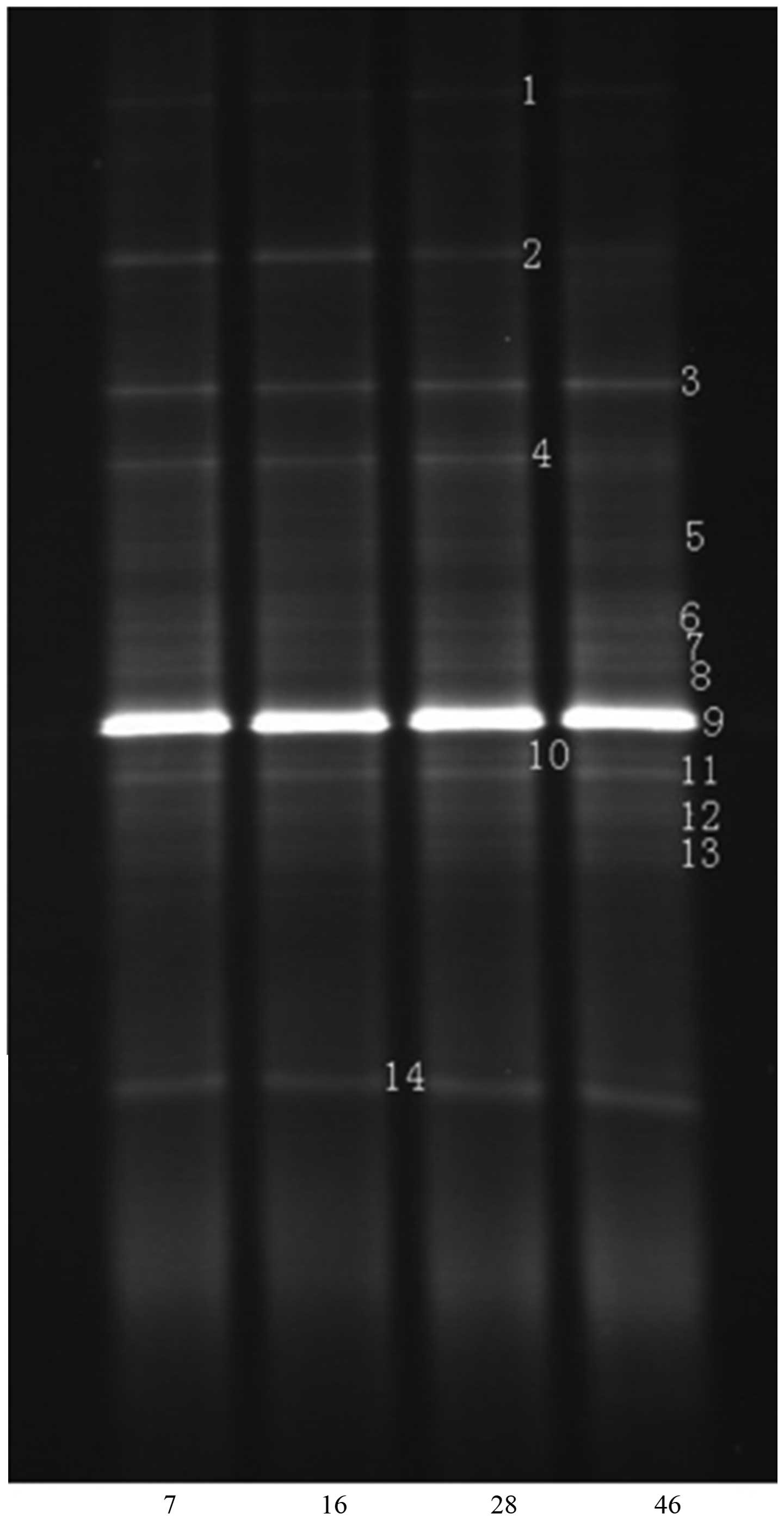
DGGE analysis and sequencing
results
DGGE fingerprinting with primer pair F341-GC and
R534, amplified the total microbial community. The results were
presented with separated bands of different DNA sequences (Fig. 2) and there were 14 discernible bands.
According to the DGGE profile, band 2 and 4 were selected for
sequencing since there was obvious difference between the
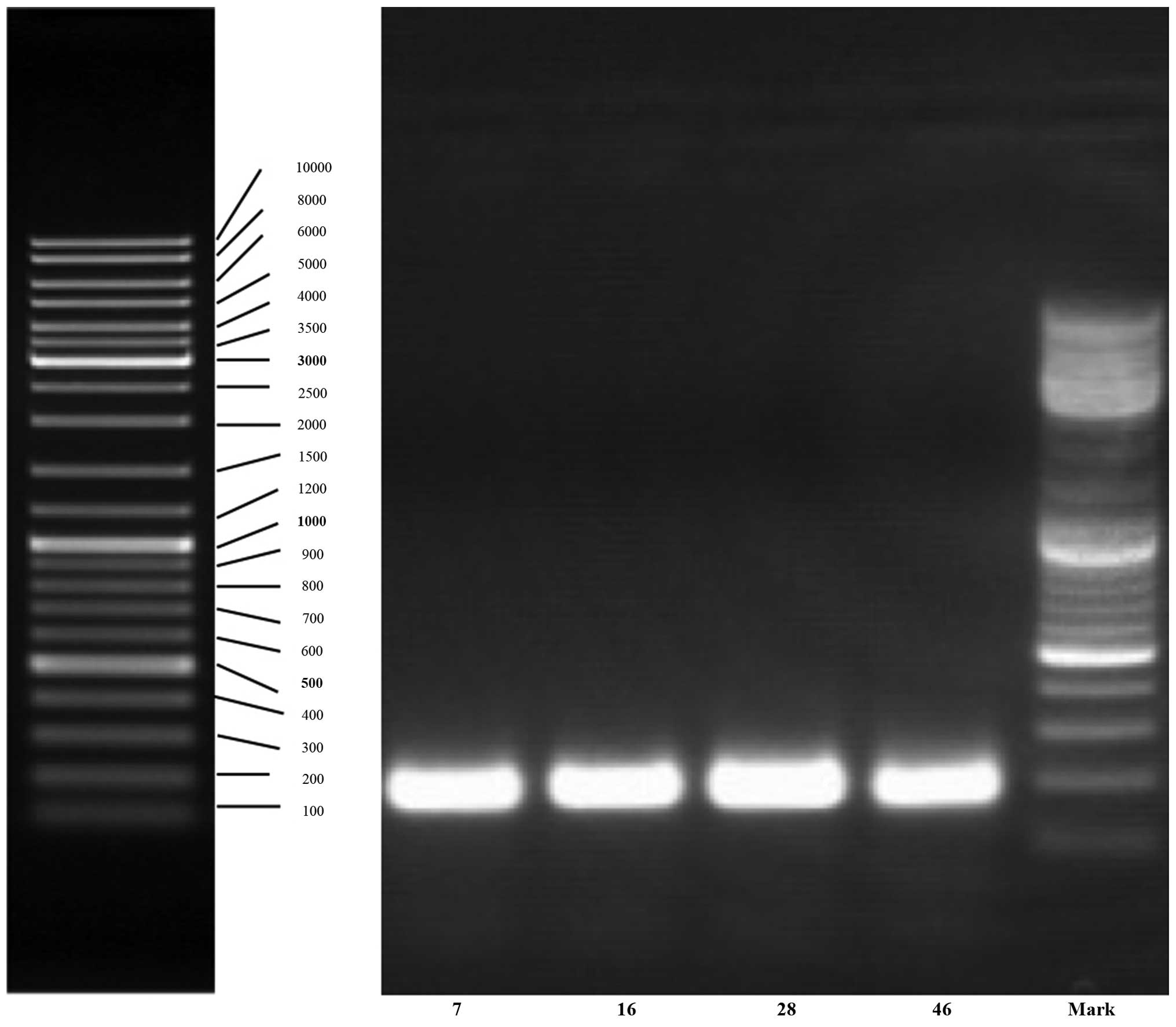
hypertension and non-hypertension groups. The two bands were
amplified again and the lengths of fragments were ~180 bp. The
amplification results are shown in Fig.
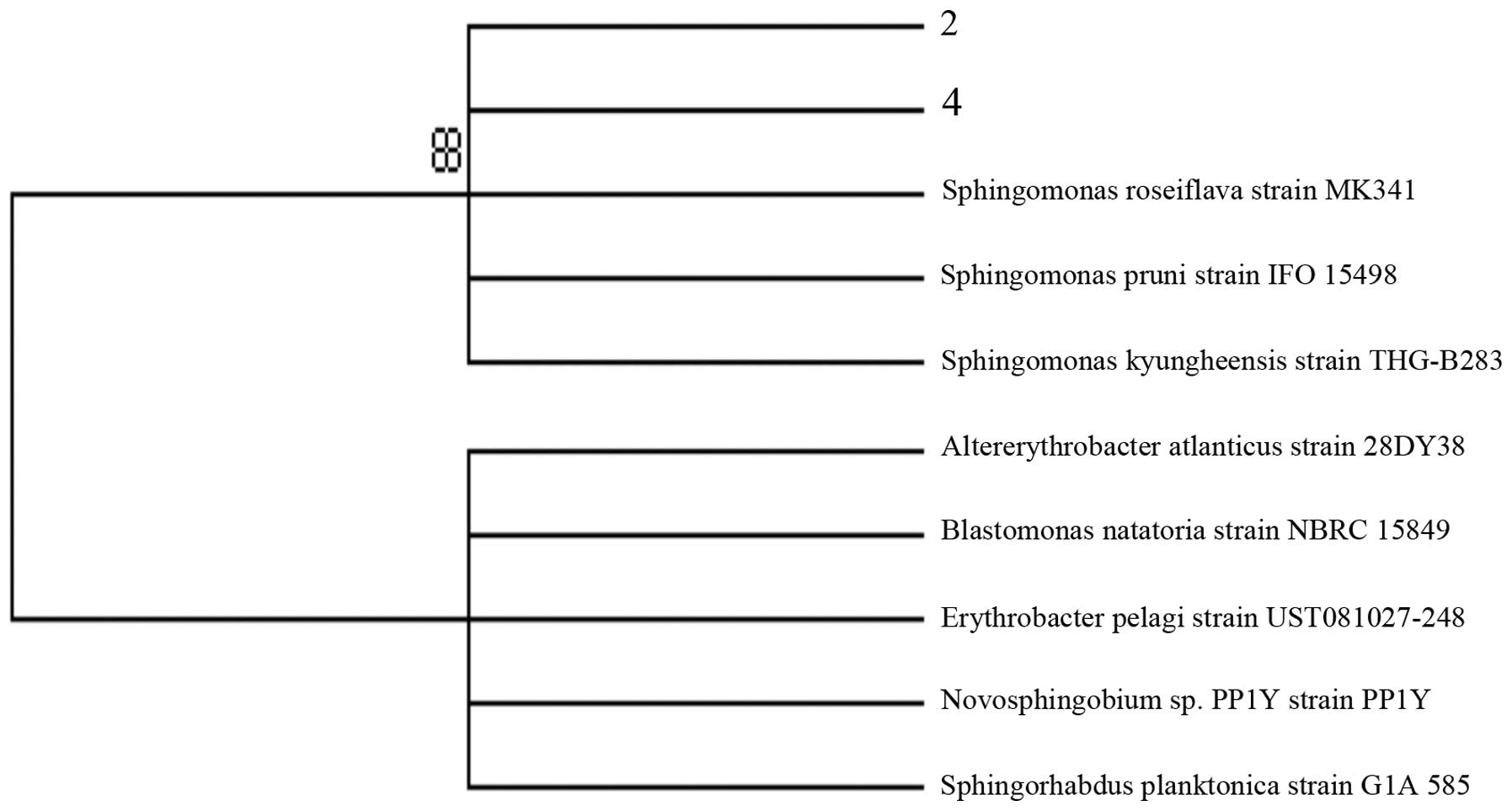
3. Sequences of selected bands 2 and 4 both matched that of
Sphingomonas (Fig. 4). The
result showed that presence of Sphingomonas was less in PC
patients without hypertension compared with PC patients with
hypertension.
Discussion
At present, many microorganisms cannot be cultivated
under the most refined conditions (19), and most bacteria in existence do not
multiply on conventional media according to environmental studies
(20–22). Due to difficult cloning or the lack of
a cloning technique and sequencing of 16S products, very few 16S
sequences from prostate tissue of PC patients have been reported.
Species belonging to the bacterial family Enterobacteriaceae (and
specifically sequences related to E. coli) appear to be the
most commonly detected organism at present (23).
The advent of molecular-based methods for
identifying and characterizing microorganisms has led to a new era
of microbial discovery. The 16S rDNA-based techniques have been
proven to be more substantially sensitive and accurate than
traditional techniques that depend on microbial culture (11,12). Several
studies have evaluated the presence of multiple and diverse
bacterial 16S rDNA sequences in prostate biopsy tissue from BPH and
PC patients (15,24–26). In the
present study, we used PCR-DGGE with 16S rDNA finger printing
analysis to investigate bacterial composition in the biopsy of PC
patients in the PSA grey-zone with or without hypertension.
Comprehending the composition and richness of the
microbial ecosystem in the prostate biopsy tissue which is related
to prostate health is essential for understanding the cause of
prostate diseases, as well as for determining its prevention and
treatment. Exploring the effect of hypertension on the prostate
patients and detection of bacteria in prostate tissue may be useful
to make an important step in determining the etiology of these
syndromes (27).
According to a statistical analysis based on the
clinical data and the PCR-DGGE profiles, the structure of the
bacterial community differed in the PC group. Two bands were
identified as the key variable factors for the discrepancy of BPH
and PC patients. This proved that the Sphingomonas were
greater in BPH than PC. The Sphingomonas is a group of
gram-negative bacteria that belongs to the Alphaproteobacteria
which is one of the most common strains involved in urinary tract
infection. It is likely that bacteria in the male reproductive
tract and urethra may approach prostate tissue, thus its analysis
may more likely reflect shift in the type of bacterial presence in
the prostate with different diseases. We found that
Sphingomonas in PC patients with hypertension was greater
than that in PC patients without hypertension, and there was no
difference between BPH and PC with hypertension. Therefore,
hypertension has become a factor that affects diagnosis of PC in
grey-zone.
The sequencing results showed that particular types
of bacteria exist in the prostate biopsy tissue and these bacteria
were not usually detected in other parts of the human body. We can
predict that the ecological balance of microenvironment in the
prostate biopsy tissue may be important in the manifestation of BPH
and PC. More studies should be performed to detect any
opportunistic, pathogenic bacteria in BPH and PC, to investigate
the potential molecular and cellular mechanisms underpinning the
protective effect of the commensal bacterial and the destructive
mechanism of the pathogenic bacteria.
In addition, common diseases such as hypertension
should be considered in the diagnosis in order to increase accuracy
in the future. Such studies should include many more patients with
BPH and PC with other comorbidity. Bacterial composition from BPH
and PC biopsy tissues may provide further confidence in a
broad-spectrum PCR approach due to its efficiency and less
time-consumption.
In conclusion, Sphingomonas was present in
lower amounts in PC without hypertension when compared to PC with
hypertension. The result may be significant for revealing the
relationship between hypertension with PC.
Acknowledgements
The present study was funded by a grant from the
Medicine and Health Science and Technology Plan Projects of
Zhejiang Province (2012RCA022).
References
|
1
|
Ferlay J, Steliarova-Foucher E,
Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H, Forman D and
Bray F: Cancer incidence and mortality patterns in Europe:
estimates for 40 countries in 2012. Eur J Cancer. 49:1374–1403.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
National Cancer Institute: What you need
to know about TM prostate cancer. http://www.cancer.gov/cancertopicsAccessed.
September 25–2014
|
|
3
|
Lim SS, Vos T, Flaxman AD, Danaei G,
Shibuya K, Adair-Rohani H, Amann M, Anderson HR, Andrews KG, Aryee
M, et al: A comparative risk assessment of burden of disease and
injury attributable to 67 risk factors and risk factor clusters in
21 regions, 1990–2010: a systematic analysis for the Global Burden
of Disease Study 2010. Lancet. 380:2224–2260. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kyrou I, Chrousos GP and Tsigos C: Stress,
visceral obesity, and metabolic complications. Ann N Y Acad Sci.
1083:77–110. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Beebe-Dimmer JL, Dunn RL, Sarma AV, Montie
JE and Cooney KA: Features of the metabolic syndrome and prostate
cancer in African-American men. Cancer. 109:875–881. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Han JH, Choi NY, Bang SH, Kwon OJ, Jin YW,
Myung SC, Chang IH, Kim TH and Ahn SH: Relationship between serum
prostate-specific antigen levels and components of metabolic
syndrome in healthy men. Urology. 72:749–754; discussion 754–755.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Takeshita K, Takahashi S, Tang M, Seeni A,
Asamoto M and Shirai T: Hypertension is positively associated with
prostate cancer development in the TRAP transgenic rat model.
Pathol Int. 61:202–209. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yu H, Meng H, Zhou F, Ni X, Shen S and Das
UN: Urinary microbiota in patients with prostate cancer and benign
prostatic hyperplasia. Arch Med Sci. 11:385–394. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Handschur M, Pinar G, Gallist B, Lubitz W
and Haslberger AG: Culture free DGGE and cloning based monitoring
of changes in bacterial communities of salad due to processing.
Food Chem Toxicol. 43:1595–1605. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Vartoukian SR, Palmer RM and Wade WG:
Strategies for culture of ‘unculturable’ bacteria. FEMS Microbiol
Lett. 309:1–7. 2010.PubMed/NCBI
|
|
11
|
Clarridge JE III: Impact of 16S rRNA gene
sequence analysis for identification of bacteria on clinical
microbiology and infectious diseases. Clin Microbiol Rev.
17:840–862. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Harris KA and Hartley JC: Development of
broad-range 16S rDNA PCR for use in the routine diagnostic clinical
microbiology service. J Med Microbiol. 52:685–691. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Maidak BL, Cole JR, Parker CT Jr, Garrity
GM, Larsen N, Li B, Lilburn TG, McCaughey MJ, Olsen GJ, Overbeek R,
et al: A new version of the RDP (Ribosomal Database Project).
Nucleic Acids Res. 27:171–173. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ranjard L, Poly F and Nazaret S:
Monitoring complex bacterial communities using culture-independent
molecular techniques: Application to soil environment. Res
Microbiol. 151:167–177. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Leskinen MJ, Rantakokko-Jalava K, Manninen
R, Leppilahti M, Marttila T, Kylmälä T and Tammela TL: Negative
bacterial polymerase chain reaction (PCR) findings in prostate
tissue from patients with symptoms of chronic pelvic pain syndrome
(CPPS) and localized prostate cancer. Prostate. 55:105–110. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu SJ, Chen GQ, Ye HY and Wang XF:
Detection and significance of 16S rDNA in the prostatic secretions
of patients with chronic prostatitis. Zhonghua Nan Ke Xue.
12:413–415. 2006.(In Chinese). PubMed/NCBI
|
|
17
|
Shannon BA, Cohen RJ and Garrett KL:
Influence of 16S rDNA primer sequence mismatches on the spectrum of
bacterial genera detected in prostate tissue by universal
eubacterial PCR. Prostate. 68:1487–1491. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mahenthiralingam E, Baldwin A, Drevinek P,
Vanlaere E, Vandamme P, LiPuma JJ and Dowson CG: Multilocus
sequence typing breathes life into a microbial metagenome. PLoS
One. 1:e172006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Amann R, Springer N, Ludwig W, Görtz HD
and Schleifer KH: Identification in situ and phylogeny of
uncultured bacterial endosymbionts. Nature. 351:161–164. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin
ML and Pace NR: Rapid determination of 16S ribosomal RNA sequences
for phylogenetic analyses. Proc Natl Acad Sci USA. 82:6955–6959.
1985. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ward DM, Weller R and Bateson MM: 16S rRNA
sequences reveal uncultured inhabitants of a well-studied thermal
community. FEMS Microbiol Rev. 6:105–115. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wilson KH: Detection of culture-resistant
bacterial pathogens by amplification and sequencing of ribosomal
DNA. Clin Infect Dis. 18:958–962. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sfanos KS, Sauvageot J, Fedor HL, Dick JD,
De Marzo AM and Isaacs WB: A molecular analysis of prokaryotic and
viral DNA sequences in prostate tissue from patients with prostate
cancer indicates the presence of multiple and diverse
microorganisms. Prostate. 68:306–320. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hochreiter WW, Duncan JL and Schaeffer AJ:
Evaluation of the bacterial flora of the prostate using a 16S rRNA
gene based polymerase chain reaction. J Urol. 163:127–130. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Keay S, Zhang CO, Baldwin BR and Alexander
RB: Polymerase chain reaction amplification of bacterial 16s rRNA
genes in prostate biopsies from men without chronic prostatitis.
Urology. 53:487–491. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Krieger JN, Riley DE, Vesella RL, Miner
DC, Ross SO and Lange PH: Bacterial DNA sequences in prostate
tissue from patients with prostate cancer and chronic prostatitis.
J Urol. 164:1221–1228. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Krieger JN, Riley DE, Roberts MC and
Berger RE: Prokaryotic DNA sequences in patients with chronic
idiopathic prostatitis. J Clin Microbiol. 34:3120–3128.
1996.PubMed/NCBI
|