Introduction
Bladder cancer is one of the most common cancer
ranking fourth in incidence in Western countries and first in China
(1,2).
There are ~380,000 new cases and 150,000 fatalities per year
worldwide (3). Bladder cancer is
staged via the tumor-node-metastasis system, which describes the
extent of invasion (Tis-T4) (4,5).
Approximately 75% of patients present with non-invasive bladder
cancer (NIBC; stage Ta), but a quarter suffer muscle-invasive
bladder cancer (MIBC) at the time of diagnosis (of stage T2 and
above) and they have a less favorable prognosis with 5-year
survival <50% (6,7). Despite improvements in surgical
techniques and postoperative recovery pathways, this complex
procedure remains highly challenging, and the treatment has not
advanced for several decades (8,9).
Furthermore, the high cost of surgery and management of subsequent
complications, chemotherapy, surveillance imaging and high
end-of-life costs contribute to the substantial financial burden of
advanced disease (10). Therefore,
development of early biological markers for diagnosis of MIBC is of
importance.
MicroRNA (miRNA) is endogenous RNA of ~22
nucleotides that targets mRNA for cleavage or translational
repression. miRNA families are responsible for a number of
physiological processes, including cell growth/differentiation,
maintenance of internal environmental stabilization, immune
response and embryonic development of different cancers (11–14).
Increasing evidence has suggested that miRNAs can be transmitted
through extracellular vesicles, such as exosomes, that are devoted
to cell-cell contact and influence, and additionally they are
convenient for detection (15,16). Thus far, various miRNAs have been
identified to have prognostic values with bladder cancer, in NIBC
and MIBC (17–19).
Currently, there are few meta-analyses published on
the diagnostic performance of miRNA assays for MIBC. Therefore, the
present study performed a meta-analysis to review and assess the
overall diagnostic values of miRNA assays for MIBC.
Materials and methods
Publication search
To identify all the potentially eligible studies on
miRNA polymorphisms and cancer risk, we carried out a systematic
search on PubMed, Web of Science and Chinese National Knowledge
Infrastructure, covering all studies published between January 1,
2000 and January 25, 2016, using the search terms: (‘miR’ or
‘miRNAs’ or ‘microRNAs’) and (‘diagnostic value’ or ‘diagnoses’ or
‘receiver operating characteristics’ or ‘ROC curve’ or ‘sensitivity
and specificity’) and (‘muscle-invasive bladder cancer,
muscle-invasive urinary bladder neoplasms, muscle-invasive
urothelial cancer, MIBC, MIUCC’). References of the retrieved
studies and review studies were also screened. Qualified studies
had to meet all the following standards: i) Diagnosis of MIBC in
histology, ii) utility of miRNA expression profiles (from tissue or
blood or urine) for urological cancers diagnosis. The exclusion
criteria included: i) Reviews, case reports, and meta analyses, ii)
studies not related to MIBC and the diagnostic value of miRNA for
urological cancers, iii) studies without valid data.
Data extraction and quality
assessment
Quality assessment was performed for each included
study by independent reviewers using the Quality Assessment of
Diagnostic Accuracy Studies (QUADAS-2) tool (20). The QUADAS-2 tool contains seven
questions, and each one should be answered with ‘yes’ (1 score),
‘unclear’ or ‘no’ (0 score). All questions were given equal weight,
resulting in a maximum possible score of 7. Conflicting evaluation
was resolved following a full discussion.
The assessment consisted of four domains: Patient
selection, index test, reference standard, and flow and timing. The
first three domains were assessed in terms of applicability. Each
of the four domains was assessed via the risk of bias. Assessments
were labeled as ‘high’, ‘low’ or ‘unclear’, corresponding to high
risk, low risk and unclear, respectively.
Statistical analysis
All the statistical analyses were performed using
Rev Man 5.3 software (Copenhagen: The Nordic Cochrane Centre, The
Cochrane Collaboration, 2014). The sensitivity and specificity data
of miRNAs associated with the predicted and/or diagnostic value of
MIBC were extracted from each study. First, the results of
sensitivity, specificity, positive and negative likelihood ratio
(PLR and NLR, respectively), diagnostic odds ratio (DOR) and 95%
confidence intervals (CIs) were calculated using the random-effect
model. Subsequently, the summary receiver operator characteristic
(SROC) curve was created and the area under the SROC curve (AUC)
was calculated. PLR was on behalf of the odds of positive test
results of MIBC patients, while NLR reflected the odds of positive
results in those without MIBC. DOR was the outcome of the
combination of PLR and NLR (DOR = PLR/NLR). In addition, the
heterogeneity between studies was evaluated through χ2
test and I2 test. If the tests show a P<0.1 or
I2>50%, the existence of significant heterogeneity
would be verified (21). Subsequently,
meta-regression and subgroup analyses were undertaken to explore
the sources of between-study heterogeneity. Furthermore, Deeks'
funnel plots were adopted to evaluate the publication bias.
The percentages of patients in each subgroup were
calculated for the categorical variables using unpaired Student's
t-test, χ2 test or Fisher's exact test,
appropriately.
Results
Patient characteristics
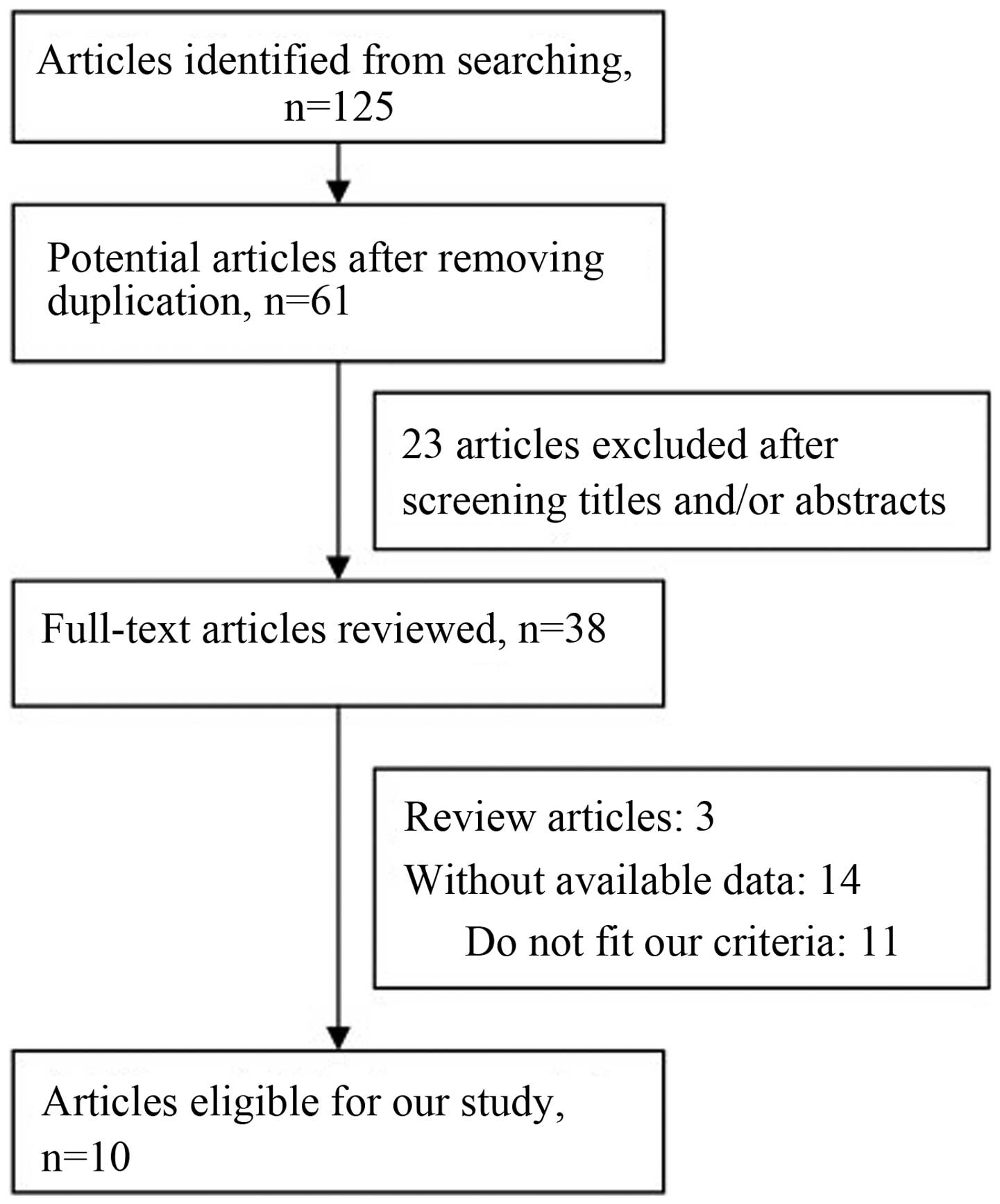
The flow graph of study selections is shown in
Fig. 1. A total of 125 potentially
relevant studies were selected with an established search strategy.
Following a detailed evaluation, 10 studies (22–31) were
used for the meta-analysis. The main characteristics of the
included studies are summarized in Table
I. Among the 10 studies, the total number of patients and
controls were 577 and 412, respectively. Four studies were
conducted in Asian populations, while the remaining studies were
conducted in Caucasian populations. The diagnostic performances of
single and multiple miRNAs have been investigated among those
included studies.
 | Table I.Main characteristics of the 10 studies
included in the meta-analysis. |
Table I.
Main characteristics of the 10 studies
included in the meta-analysis.
|
|
|
| Sample size, n | Mean age, year | Mean ratio, % |
|
|
|
|
|
|
|---|
|
|
|
|
|
|
|
|
|
|
|
|
|
|---|
| First author,
year | Country | Ethnicity | Case | Control | Case | Control | Case | Control | Control | Specimen | miRNA profiling | Regulated
features | Score | Refs. |
|---|
| Veerla, 2009 | Sweden | Caucasian | 17 | 17 | NA | NA | NA | NA | Normal | Tissue | miR-100, miR-125b,
miR-199b, miR-222 | Up | 5 | (22) |
| Pignot, 2012 | France | Caucasian | 80 | 11 | 70 | 67 | 78.8 | 79.5 | Normal | Tissue | miR-9, miR-182,
miR-200b | Up | 7 | (23) |
|
|
|
|
|
|
|
|
|
|
|
| miR-1, miR-133a,
miR-133b | Down |
|
|
|
|
|
|
|
|
|
|
|
|
|
| miR-143, miR-145,
miR-204 | Down |
|
|
|
|
|
|
|
|
|
|
|
|
|
| miR-199a, miR-199b,
miR-1281 | Down |
|
|
| Pignot, 2012 |
France | Caucasian | 21 | 5 | 68 | 66 | 90.5 | 100.0 | Normal | Tissue | miR-19A, miR-20A,
miR-92A | Up | 6 | (24) |
| Adam, 2013 | Germany | Caucasian | 10 | 18 | 62 | 55 | 70.0 |
46.0 | Normal | Blood | miR-200b, miR-541,
miR-566 | Up | 7 | (25) |
|
|
|
|
|
|
|
|
|
|
|
| miR-543,
miR-544, | Up |
|
|
|
|
|
|
|
|
|
|
|
|
|
| miR-604,
miR-940-p |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| miR-33b,
miR-92b,6 | Down |
|
|
|
|
|
|
|
|
|
|
|
|
|
| miR-1246,
miR-182 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| miR-25, miR-148b,
miR-487 | Down |
|
|
| Ratert, 2013 | Germany | Caucasian | 15 | 42 | 74 | 68 | 80.0 |
83.0 | NMIBC, normal |
| miR-141,
miR-205 | Up | 6 | (26) |
| Li, 2014 | China | Asian | 15 | 15 | NA | NA | NA | NA | Normal | Tissue | miR-34a | Up | 5 | (27) |
| Avgeris, 2015 | Greece | Caucasian | 45 | 39 | NA | NA | 84.3 | NA | Normal | Tissue | miR-143, miR-145,
miR-224 | Up | 5 | (28) |
| Wang, 2015 | China | Asian | 144 | 169 | NA | NA | 68.2 | NA | Normal | Urine | miR-124 | Up | 6 | (29) |
| Xu, 2015 | China | Asian | 202 | 40 | 68 | 64 | 74.3 |
65.0 | Normal | Tissue | let-7c,
mir-125b-1 | Up | 7 | (30) |
|
|
|
|
|
|
|
|
|
|
|
| mir-193a,
mir-99a | Up |
|
|
| Fang, 2015 | China | Asian | 28 | 56 | 75 | 74 | 71.4 |
64.2 | Normal | Tissue | mir-205 | Up | 6 | (31) |
Quality assessment of studies
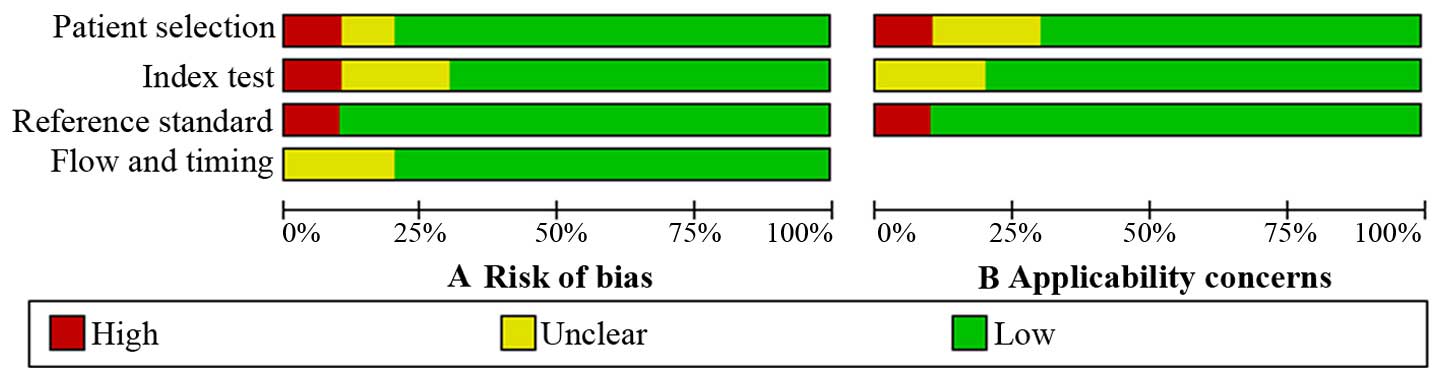
The results of the QUADAS-2 assessment are shown as
a bar graph in Fig. 2. The majority of
all included studies fulfilled the majority of the items in
QUADAS-2, which indicated that the general quality of the included
studies is good.
Diagnostic accuracy
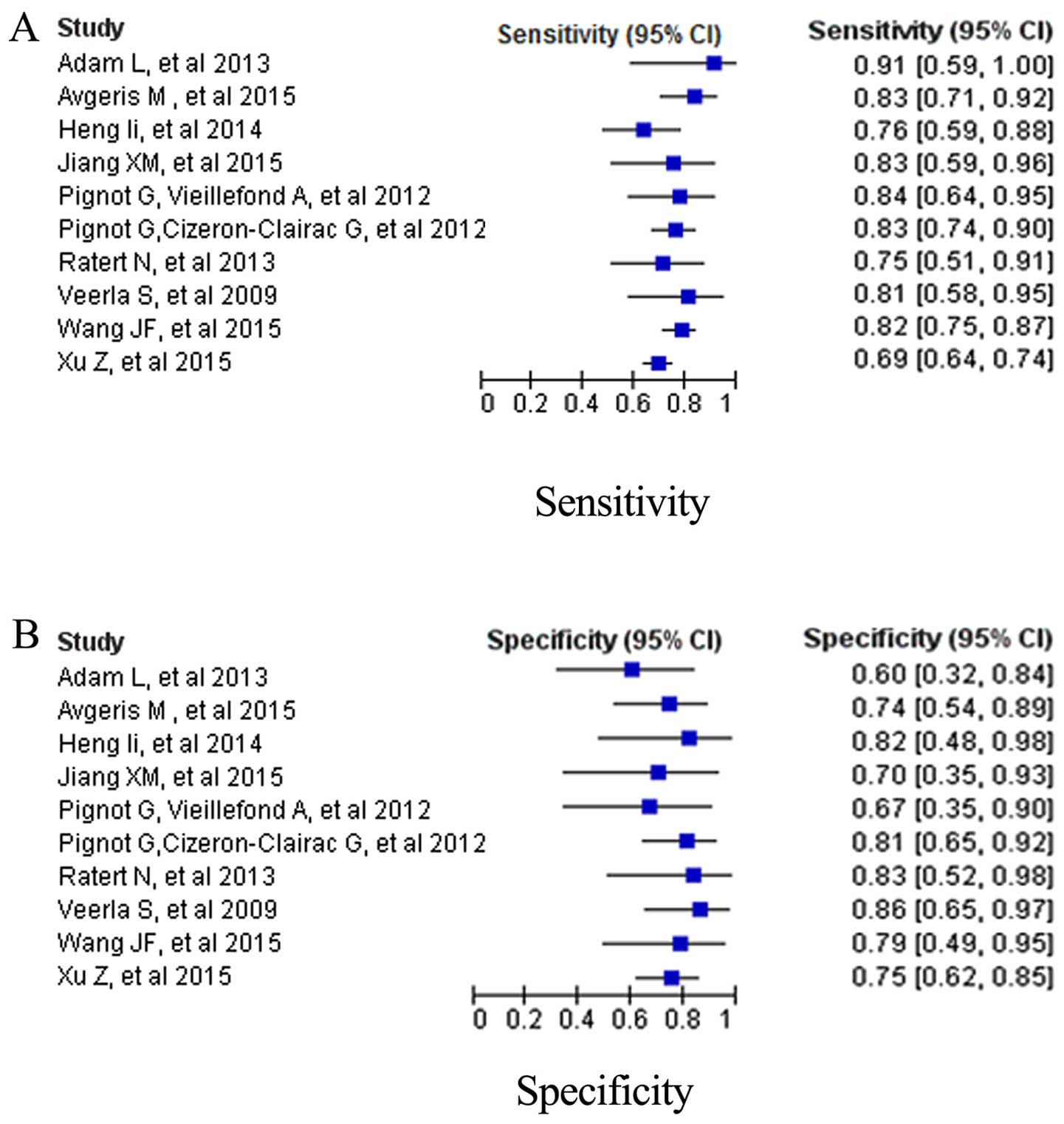
Due to the existence of significant heterogeneity
between studies in sensitivity and specificity (I2=81.3%
and I2=77.8%), the random effects model was adopted.
Fig. 3 depicts the forest plots of
data from the miRNA panels subgroup and mean sensitivity and
specificity. The pooled results of diagnostic criteria and their
95% CIs are listed in Table II. The
overall sensitivity, specificity, PLR, NLR and DOR were 0.78
(0.69–0.86), 0.77 (0.72–0.81), 2.9 (95% CI, 2.1–3.8), 0.31 (95% CI,
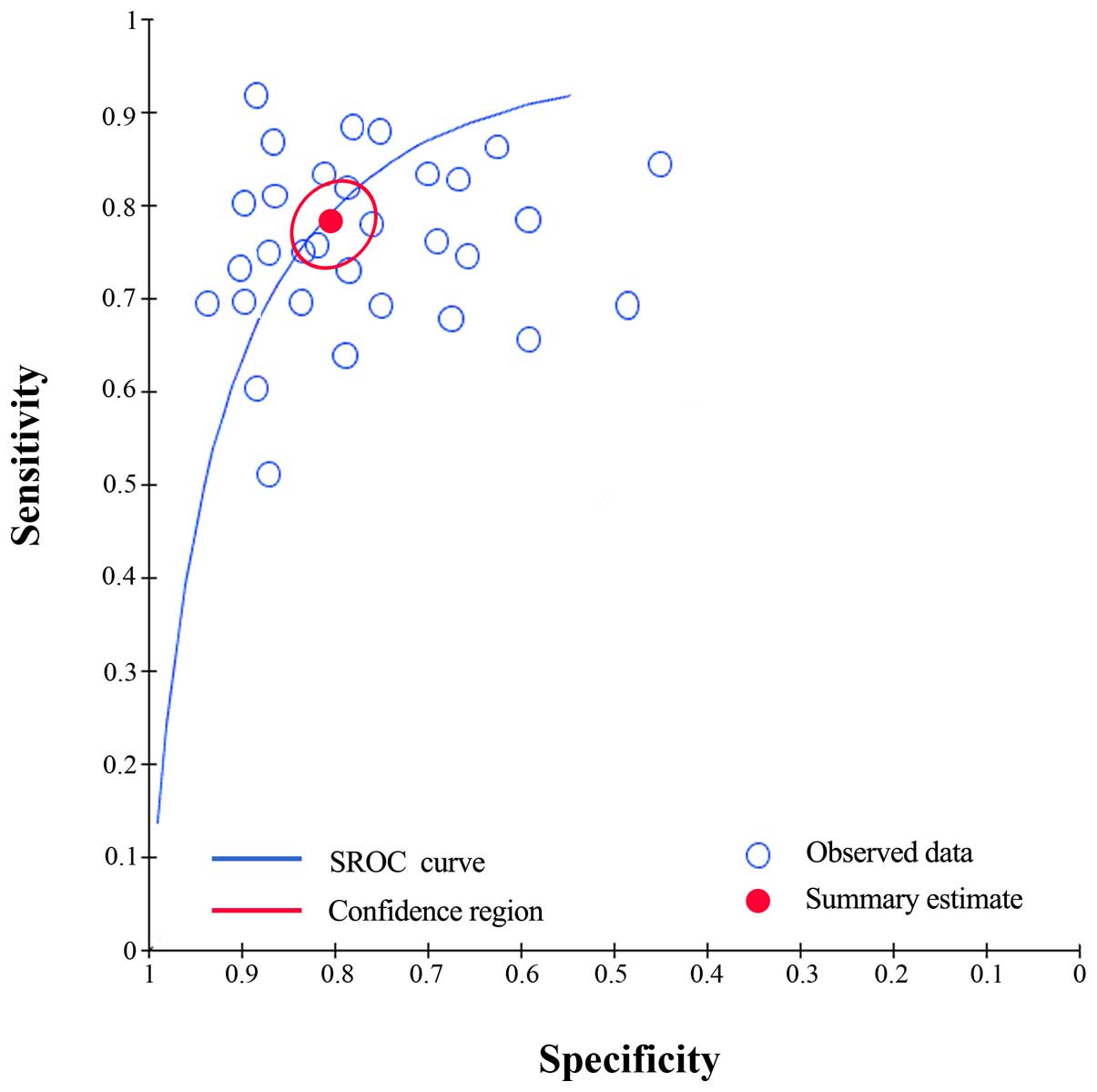
0.27–0.35), 7 (95% CI, 4–13), respectively. Moreover, the SROC
curve was generated and the AUC was calculated as 0.80 (95% CI,
0.69–0.87) (Fig. 4), which implied a
relatively high diagnostic accuracy.
 | Table II.Summary estimates of diagnostic
criteria and the 95% confidence intervals (CIs). |
Table II.
Summary estimates of diagnostic
criteria and the 95% confidence intervals (CIs).
| Analysis | Sensitivity (95%
CI) | Specificity (95%
CI) | PLR (95% CI) | NLR (95% CI) | DOR (95% CI) | AUC (95% CI) |
|---|
| Ethnicity |
|
|
|
|
|
|
|
Caucasian | 0.71
(0.66–0.75) | 0.76
(0.67–0.83) | 2.4 (2.0–2.8) | 0.34
(0.22–0.46) | 6
(4–11) | 0.74
(0.69–0.78) |
|
Asian | 0.67
(0.63–0.72) | 0.75
(0.73–0.82) | 2.5 (2.1–3.0) | 0.32
(0.21–0.42) | 7
(5–12) | 0.82
(0.77–0.86) |
| miRNA
profiling |
|
|
|
|
|
|
| Single
miRNA | 0.70
(0.65–0.74) | 0.73
(0.67–0.79) | 2.6 (2.3–2.9) | 0.38
(0.29–0.47) | 5
(3–10) | 0.79
(0.76–0.82) |
|
Multiple miRNA | 0.81
(0.72–0.91) | 0.84
(0.75–0.93) | 4.2 (2.7–5.7) | 0.26
(0.15–0.37) | 16 (9–24) | 0.86
(0.83–0.89) |
| Overall | 0.78
(0.69–0.86) | 0.77
(0.72–0.81) | 2.9 (2.1–3.8) | 0.31
(0.27–0.35) | 7
(4–13) | 0.80
(0.69–0.87) |
Subgroup analyses
A subgroup analyses was also performed to identify
potential sources of heterogeneity (Table
II). For single miRNA profiling assays, sensitivity,
specificity and AUC values were 0.70 (0.65–0.74), 0.73 (95% CI,
0.67–0.79), and 0.79 (95% CI, 0.76–0.82), respectively. For
multiple miRNAs, the sensitivity (SEN), specificity (SPE) and AUC
values were 0.81 (95% CI, 0.72–0.91), 0.84 (95% CI, 0.75–0.93) and
0.86 (95% CI, 0.83–0.89), respectively. These data indicated that
multiple miRNAs profiling were more accurate than single miRNA
profiling. The studies based on Caucasian populations had a pooled
sensitivity of 0.71 and a specificity of 0.76, while studies based
on Asian populations had a relatively lower sensitivity (0.67) and
specificity (0.75). The subgroup analyses suggested that the
ethnicity and miRNA profiling had an evident influence on the
diagnostic accuracy. Therefore, these factors may be the potential
sources of heterogeneity.
Publication bias
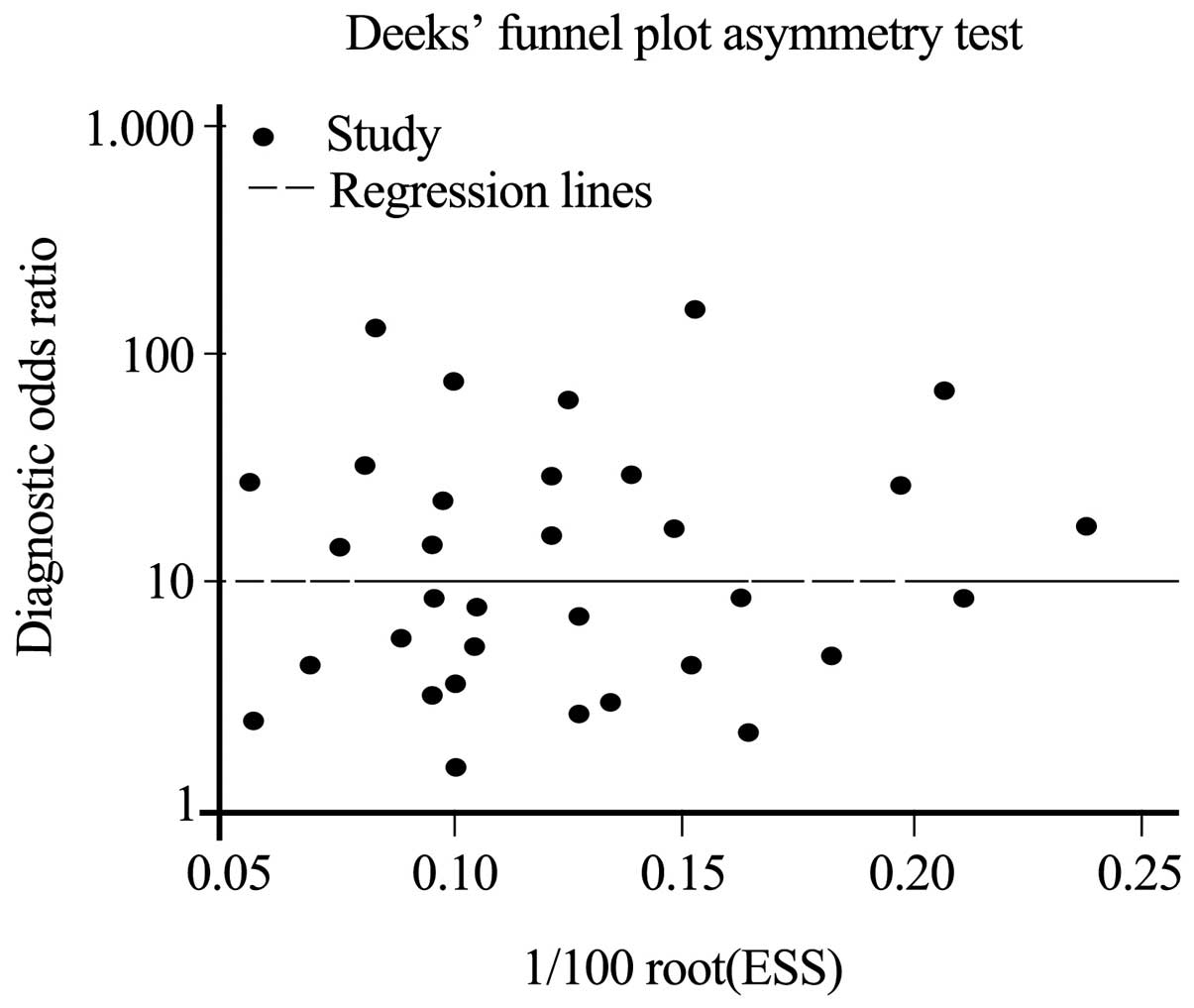
Finally, the Deeks' funnel plot asymmetry test was
conducted to evaluate the publication bias. The P-value of 0.7
suggested that no significant publication bias existed among the
studies (Fig. 5).
Discussion
Although clear progression in surgery and chemical
treatment has been achieved in MIBC, the prognosis of the patients
is poor (6,7). As aforementioned, approximately a quarter
of bladder cancer patients have MIBC at diagnosis, and the critical
factor is how to detect and diagnose MIBC as early as possible.
Thus, novel and reliable biomarkers for MIBC detection are urgently
required.
As the existing biomarkers do not exhibit high
sensitivity and specificity in MIBC detection, miRNAs have been
reported as markers of MIBC in numerous studies. Pignot et
al (23) reported that miR-9,
miR-182 and miR-200b were associated with MIBC aggressiveness, and
with recurrence-free and overall survival in univariate analysis
and multivariate analysis. Avgeris et al (28) reported that high miR-143/145 levels
could predict the inferior overall survival for MIBC effectively
and the progression of superficial tumors independently. Xu et
al (30) identified four specific
miRNAs (let-7c, mir-125b-1, mir-193a, and mir-99a) in association
with the progression and aggressiveness of MIBC via microarray
analysis. Several genome-wide profiling studies have been reported
and identified specific miRNA alterations in bladder cancer
(32–34). Those results suggested a promising
prognostic value of these miRNAs markers.
However, as the association between miRNAs and MIBC
are inconsistent and the studies are designed differently,
comparing the wide ranges of diagnostic performance is difficult.
Therefore, the present study aimed to summarize the result of
individual studies and investigate the diagnostic value of miRNAs
for MIBC.
Following analysis, the overall sensitivity and
specificity of miRNAs was 0.78 and 0.77, which indicated accuracy
of miRNAs to detect MIBC. The pooled PLR was 2.9 and the pooled NLR
was 0.31, respectively. The AUC was 0.80 and DOR was 7. These data
suggested that miRNAs had a relatively high diagnostic accuracy. As
heterogeneity between studies could affect the results of the
meta-analysis, subgroup analyses will aid to understand these
influences. Therefore, subgroup analyses were performed based on
ethnicity and miRNA profiling. Subgroups of miRNA profiling
indicated that multiple miRNA assays (SEN, SPE and AUC of 0.81,
0.84 and 0.86, respectively) had a higher diagnostic performance
than those of single miRNA assays (SEN, SPE and AUC of 0.70, 0.73
and 0.79, respectively). No significant different was observed for
the miRNA expression profile test between the Asian and Caucasian
groups.
There were several limitations in the meta-analysis.
First, the included studies were based on limited sample size, and
if the specimen of miRNA could be divided into blood, urine, tissue
groups, do analysis and comparison respectively, the results may be
more significant and accurate. Second, the majority of the included
studies did not differentiate the grade of MIBC, and therefore,
subgroup analyses based on these variables were restricted due to
limited reported data. Third, as the majority of the studies did
not provide further miRNA research, the changes of miRNA in
different stages of MIBC, following treatment in different methods
or in patients who received treatment experiencing MIBC again could
not be verified.
In conclusion, the present study analyzed the pooled
data of SEN, SPE, PLR, NLR, DOR and AUC from 10 studies. miRNA
assays could serve as markers for MIBC diagnosis, particularly the
combined usage of miRNA, and have a good potential as an accurate
biomarker to diagnose of MIBC. However, the clinical application of
miRNA profiling for MIBC diagnosis remains validating in future
studies.
Glossary
Abbreviations
Abbreviations:
|
MIBC
|
muscle-invasive bladder cancer
|
|
NIBC
|
non-invasive bladder cancer
|
|
PLR
|
positive likelihood ratio
|
|
NLR
|
negative likelihood ratio
|
|
DOR
|
diagnostic odds ratio
|
|
SROC
|
summary receiver operator
characteristic
|
|
SEN
|
sensitivity
|
|
SPE
|
specificity
|
|
AUC
|
area under SROC curve
|
References
|
1
|
Knowles MA and Hurst CD: Molecular biology
of bladder cancer: New insights into pathogenesis and clinical
diversity. Nat Rev Cancer. 15:25–41. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xu S, Zhang GM, Guan FJ, Dong DH, Luo L,
Li B, Ma XC, Zhao J and Sun LJ: The association between metabolic
syndrome and the risk of urothelial carcinoma of the bladder: A
case-control study in China. World J Surg Oncol. 13:2362015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rocken C and Behrens HM: Validating the
prognostic and discriminating value of the TNM-classification for
gastric cancer - a critical appraisal. Eur J Cancer. 51:577–586.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Eble JN, Sauter G, Epstein JI and
Sesterhen IA: World Health Organization Classification of Tumours.
Pathology and genetics of tumours of the urinary system and male
genital organs. IARC Press. (Lyon). 2004.
|
|
6
|
Soloway MS: Bladder cancer: Lack of
progress in bladder cancer - what are the obstacles? Nat Rev Urol.
10:5–6. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chamie K and Litwin MS: Quality of bladder
cancer care in the USA. Expert Rev Pharmacoecon Outcomes Res.
11:619–621. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Corcoran AT, Handorf E, Canter D,
Tomaszewski JJ, Bekelman JE, Kim SP, Uzzo RG, Kutikov A and
Smaldone MC: Variation in performance of candidate surgical quality
measures for muscle-invasive bladder cancer by hospital type. BJU
Int. 115:230–237. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jayaratna IS, Navai N and Dinney CP: Risk
based neoadjuvant chemotherapy in muscle invasive bladder cancer.
Transl Androl Urol. 4:273–282. 2015.PubMed/NCBI
|
|
10
|
Johnson DC, Greene PS and Nielsen ME:
Surgical advances in bladder cancer: At what cost? Urol Clin North
Am. 42:235–252, ix. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Beitzinger M and Meister G: Preview.
MicroRNAs: From decay to decoy. Cell. 140:612–614. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kent OA and Mendell JT: A small piece in
the cancer puzzle: MicroRNAs as tumor suppressors and oncogenes.
Oncogene. 25:6188–6196. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Maida Y, Takakura M, Nishiuchi T,
Yoshimoto T and Kyo S: Exosomal transfer of functional small RNAs
mediates cancer-stroma communication in human endometrium. Cancer
Med. 5:304–314. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kim MS, Haney MJ, Zhao Y, Mahajan V,
Deygen I, Klyachko NL, Inskoe E, Piroyan A, Sokolsky M, Okolie O,
et al: Development of exosome-encapsulated paclitaxel to overcome
MDR in cancer cells. Nanomedicine. 12:655–664. 2016.PubMed/NCBI
|
|
17
|
Catto JW, Miah S, Owen HC, Bryant H, Myers
K, Dudziec E, Larré S, Milo M, Rehman I, Rosario DJ, et al:
Distinct microRNA alterations characterize high- and low-grade
bladder cancer. Cancer Res. 69:8472–8481. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pignot G, Cizeron-Clairac G, Vacher S,
Susini A, Tozlu S, Vieillefond A, Zerbib M, Lidereau R, Debre B,
Amsellem-Ouazana D, et al: MicroRNA expression profile in a large
series of bladder tumors: Identification of a 3-miRNA signature
associated with aggressiveness of muscle-invasive bladder cancer.
Int J Cancer. 132:2479–2491. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Segersten U, Spector Y, Goren Y, Tabak S
and Malmström PU: The role of microRNA profiling in prognosticating
progression in Ta and T1 urinary bladder cancer. Urol Oncol.
32:613–618. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Whiting PF, Rutjes AW, Westwood ME,
Mallett S, Deeks JJ, Reitsma JB, Leeflang MM, Sterne JA and Bossuyt
PM: QUADAS-2 Group: QUADAS-2: A revised tool for the quality
assessment of diagnostic accuracy studies. Ann Intern Med.
155:529–536. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dinnes J, Deeks J, Kirby J and Roderick P:
A methodological review of how heterogeneity has been examined in
systematic reviews of diagnostic test accuracy. Health Technol
Assess. 9:1–113, iii. 2005. View
Article : Google Scholar
|
|
22
|
Veerla S, Lindgren D, Kvist A, Frigyesi A,
Staaf J, Persson H, Liedberg F, Chebil G, Gudjonsson S, Borg A, et
al: miRNA expression in urothelial carcinomas: Important roles of
miR-10a, miR-222, miR-125b, miR-7 and miR-452 for tumor stage and
metastasis, and frequent homozygous losses of miR-31. Int J Cancer.
124:2236–2242. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pignot G, Cizeron-Clairac G, Vacher S,
Susini A, Tozlu S, Zerbib M, Lidereau R, Debre B, Amsellem-Ouazana
D and Bieche I: MicroRNA expression profile in a large series of
bladder tumors: Identification of a 3-miRNA signature predictive of
aggressiveness and prognosis of muscle-invasive bladder cancer. Eur
Urol Suppl. 11:e1692012. View Article : Google Scholar
|
|
24
|
Pignot G, Vieillefond A, Vacher S, Zerbib
M, Debre B, Lidereau R, Amsellem-Ouazana D and Bieche I: Hedgehog
pathway activation in human transitional cell carcinoma of the
bladder. Br J Cancer. 106:1177–1186. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Adam L, Wszolek MF, Liu CG, Jing W, Diao
L, Zien A, Zhang JD, Jackson D and Dinney CP: Plasma microRNA
profiles for bladder cancer detection. Urol Oncol. 31:1701–1708.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ratert N, Meyer HA, Jung M, Lioudmer P,
Mollenkopf HJ, Wagner I, Miller K, Kilic E, Erbersdobler A, Weikert
S, et al: miRNA profiling identifies candidate mirnas for bladder
cancer diagnosis and clinical outcome. J Mol Diagn. 15:695–705.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li H, Yu G, Shi R, Lang B, Chen X, Xia D,
Xiao H, Guo X, Guan W, Ye Z, et al: Cisplatin-induced epigenetic
activation of miR-34a sensitizes bladder cancer cells to
chemotherapy. Mol Cancer. 13:82014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Avgeris M, Mavridis K, Tokas T,
Stravodimos K, Fragoulis EG and Scorilas A: Uncovering the clinical
utility of miR-143, miR-145 and miR-224 for predicting the survival
of bladder cancer patients following treatment. Carcinogenesis.
36:528–537. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang J, Zhang X, Wang L, Dong Z, Du L,
Yang Y, Guo Y and Wang C: Downregulation of urinary cell-free
microRNA-214 as a diagnostic and prognostic biomarker in bladder
cancer. J Surg Oncol. 111:992–999. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xu Z, Yu YQ, Ge YZ, Zhu JG, Zhu M, Zhao
YC, Xu LW, Yang XB, Geng LG, Dou QL, et al: MicroRNA expression
profiles in muscle-invasive bladder cancer: Identification of a
four-microRNA signature associated with patient survival. Tumour
Biol. 36:8159–8166. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fang Z, Dai W, Wang X, Chen W, Shen C, Ye
G and Li L: Circulating miR-205: A promising biomarker for the
detection and prognosis evaluation of bladder cancer. Tumour Biol.
37:8075–8082. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Catto JW, Abbod MF, Wild PJ, Linkens DA,
Pilarsky C, Rehman I, Rosario DJ, Denzinger S, Burger M, Stoehr R,
et al: The application of artificial intelligence to microarray
data: Identification of a novel gene signature to identify bladder
cancer progression. Eur Urol. 57:398–406. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Silva-Santos RM, Costa-Pinheiro P, Luis A,
Antunes L, Lobo F, Oliveira J, Henrique R and Jerónimo C: MicroRNA
profile: A promising ancillary tool for accurate renal cell tumour
diagnosis. Br J Cancer. 109:2646–2653. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jiang X, Du L, Wang L, Li J, Liu Y, Zheng
G, Qu A, Zhang X, Pan H, Yang Y, et al: Serum microRNA expression
signatures identified from genome-wide microRNA profiling serve as
novel noninvasive biomarkers for diagnosis and recurrence of
bladder cancer. Int J Cancer. 136:854–862. 2015. View Article : Google Scholar : PubMed/NCBI
|