Introduction
Bipolar disorder is a polygenic, common and chronic
psychiatric disorder that has a lifetime risk of 1% worldwide
(1). It is also termed manic
depression due to the dramatic mood changes, such as extreme mania
or severe depression, that patients experience. It is usually
accompanied by thinking and behavioral disturbances, and often by
psychotic features, such as delusions or hallucinations. These
episodic mood changes occur at an extreme level that significantly
affects the individual's social and business life (2).
Studies on families and with twins have demonstrated
that bipolar disorder is highly heritable. First-degree relatives
of affected individiuals are associated with a 5–10-fold increased
risk of bipolar disorder compared to that of the general
population. The risk may increase up to the 40–70-fold in
monozygous twins (2). Despite this
strong familiality, identification of bipolar disorder
susceptibility genes has been challenging due to the multifactorial
genetic architecture of the disease. Multiple candidate genes have
been proposed by linkage analysis and genome-wide association
studies (GWAs) although these results have not been consistently
replicated (3,4).
GWAs are systematic and objective studies based on
the ‘common disease, common variant’ hypothesis, which allow us to
identify population specific and disease-associated variants
(5). Attempts to identify risk genes
for common disorders, such as bipolar disorder and schizophrenia in
humans have began to provide important findings. It is known that
schizophrenia and bipolar disorder overlap epidemiologically,
symptomatically and genetically (6). A
large scale GWAs demonstrated that schizophrenia and bipolar
disorder patients carry the same disease-associated variants, such
as in the zinc finger protein 804A and calcium voltage-gated
channel subunit α1 C (CACNA1C) genes (2).
Stefansson et al (7) combined single nucleotide polymorphism
(SNP) data from various GWAs for schizophrenia and conducted a
meta-analysis of the most significant associatied signals. The
genome-wide scan results of 12,945 schizophrenia cases and 34,591
controls were analyzed. Significant associations were identified on
chromosomes 6p21.3–22.1 (major histocompatibility complex, five
SNPs; rs6913660, rs13219354, rs6932590, rs13211507 and rs3131296),
11q24.2 (neurogranin, rs12807809) and 18q21.2 (transcription factor
4; TCF4, rs9960767) (7). One of these
risk genes, TCF4, is also involved in normal brain development and
TCF4 mutations have been associated with Pitt-Hopkins syndrome, a
rare developmental disorder characterized by severe motor and
mental retardation (8), additionally
TCF4 deletions have been identified as risk factors for
autistic-like behaviours (9).
The TCF4 gene is a basic helix-loop-helix (bHLH)
transcription factor, which is also known to regulate the
expression of many other genes that are involved in cell
differentiation, cell survival and neurodevelopment (10). These associations between TCF4 and
neurodevelopmental diseases have resulted in the present evaluation
of whether the rs9960767 variant of the TCF4 gene is also
associated with bipolar disorder as a neuropsychiatric disorder. In
the present study, the allele and genotype frequencies of the TCF4
gene rs9960767 variant were analyzed in 95 bipolar disorder
patients and 108 healthy control subjects from a Turkish population
using polymerase chain reaction-restriction fragment length
polymorphism (PCR-RFLP) analysis. For this method, the PCR primers
were designed by the present authors and a suitable restriction
enzyme was selected for analysis of the polymorphic gene site. The
primers and enzyme may be used for further investigations of this
particular SNP (rs9960767).
Materials and methods
Patients
The present study involved 95 patients with bipolar
disorder [55 females and 40 males; aged 21–75 years (mean age, 38
years)] and 108 voluntary healthy controls [62 females and 46
males; aged 19–72 years (mean age, 36 years)] were included. All
subjects were recruited from the Department of Psychiatry, Faculty
of Medicine, University of Kocaeli (Kocaeli, Turkey) and diagnosis
of the bipolar patients was based on the criteria of the Diagnostic
and Statistical Manual of Mental Disorders. All subjects provided
written informed consent and the Institutional Review Board
approved the study.
Genotyping
Genomic DNA was isolated from all subjects according
to the conventional salting-out method (11). Genotype and allele frequencies for the
TCF4 rs9960767 variant were analyzed using a PCR-RFLP method
designed in our laboratory. Forward and reverse primers, which
cover the polymorphic site were designed in the present study and
purchased from Integrated DNA Technologies (Coralville, IA, USA).
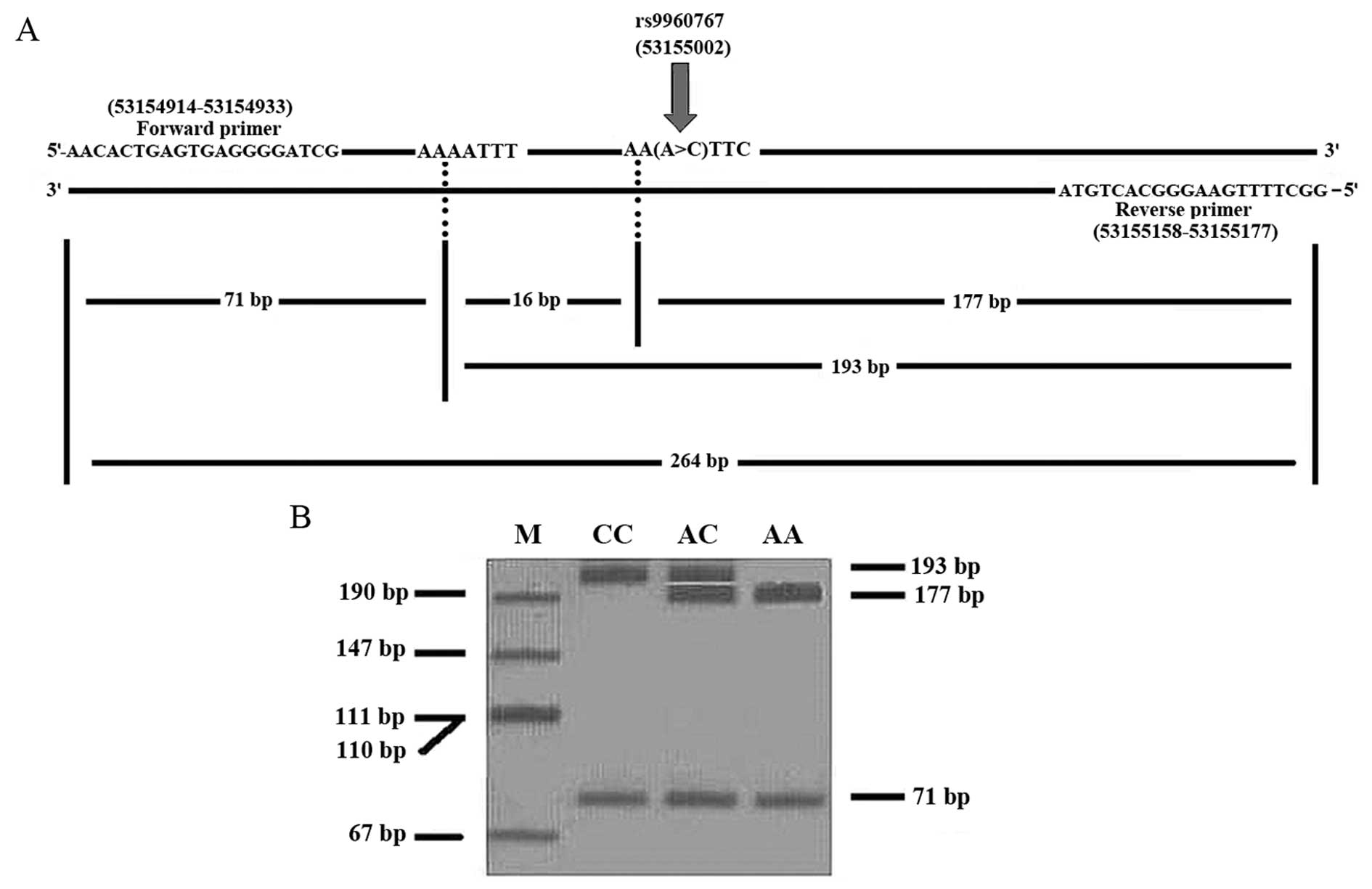
The 264-bp fragment was amplified with 10 pmol each of the forward
primer 5′-AACACTGAGTGAGGGGATCG-3′ and the reverse primer
5′-GGCTTTTGAAGGGCACTGTA-3′. The PCR reaction was performed using a
thermal cycler (T100; Bio-Rad Laboratories, Inc., Hercules, CA,
USA). The PCR thermal conditions were as follows: Denaturation at
95°C for 5 min, followed by 35 cycles of 95°C for 1 min, 57°C for
30 sec, 72°C for 1 min and a final extension step at 72°C for 10
min. The digestion of the amplified 264-bp fragment with the
ApoI restriction endonuclease was conducted at 37°C
overnight. Digestion fragments were electrophoresed at 20 W for 40
min on an 8% polyacrylamide gel, subsequently Silver Staining was
conducted and scanning was performed using a CanoScan N670U (Canon,
Inc., Tokyo, Japan). The ApoI digestion of the PCR product
produced 193- and 71-bp fragments for the CC genotype, 177-, 71-
and 16-bp fragments for the AA genotype and 193-, 177-, 71- and
16-bp fragments for the AC genotype (Fig.
1).
Statistical analysis
The Hardy-Weinberg equilibrium (HWE) was verified
for the two groups. Statistical analyses were performed using the
SPSS software package, version 21.0 (IBM SPSS, Armonk, NY, USA).
Allelic distributions and genotype frequencies were compared using
the χ2 test and Student's t-test. The relative risk
(odds ratio; OR) analysis was performed with 2×2 crosstabulation
and binary logistic regression model for gender. P<0.05 was
considered to indicate a statistically significant difference.
Results
TCF4 gene rs9960767 allele and genotype frequencies
were analyzed for 95 bipolar disorder patients and 108 healthy
control subjects. A allele frequencies were 95.8% in the patients
and 94.4% in the control subjects. The genotype frequencies of the
TCF4 gene rs9960767 variant were as follows: AA, 91.6% and AC, 8.4%
in the patients (the CC genotype was not observed in the patients);
AA, 89.8%; AC, 9.3% and CC, 0.9% in the control subjects. No
statistically significant association was identified between the
patients and the control subjects (χ2=0.937; P=0.626).
Gender specific statistical analysis was also performed, however,
no significant association was identified according to the gender
distrubition. All cases and controls were in HWE (P>0.05;
Table I).
 | Table I.Allele and genotype frequencies of the
TCF4 gene rs9960767 polymorphism in patients with bipolar disorder
and control subjects. |
Table I.
Allele and genotype frequencies of the
TCF4 gene rs9960767 polymorphism in patients with bipolar disorder
and control subjects.
| Gene | Cases, n (%) | Controls, n (%) | χ2 | P-value | OR; 95% CI |
|---|
| TCF4 (rs9960767) |
95 (100.0) | 108
(100.0) | 0.937 | 0.626 |
|
| AA | 87
(91.6) | 97
(89.8) | 0.185 | 0.667 | 1,233
(0,474–3,207) |
| CC | 0 (−) | 1
(0.9) | 0.884 | 0.347 | – |
| AC | 8
(8.4) | 10 (9.3) | 0.044 | 0.834 | 0,901
(0,340–2,385) |
| Allele frequency |
|
| A
allele |
(95.8) |
(94.4) | 0.884 | 0.347 | – |
| C
allele |
(4.2) |
(5.6) | 0.185 | 0.667 | 0.811
(0.312–2.108) |
| HWE exact
(P-value) |
1.000 | 0.275 |
|
|
|
Discussion
It is known that bipolar disorder (also termed manic
depression) is strongly influenced by genetic factors (12). It is a common, complex and polygenic
disease that shows multifactorial inheritance. There are numerous
methods that have been developed to investigate complex disorders.
One of the most promising methods is with GWA, due to the power of
identifying the common genetic variants that are involved in major
complex disorders, such as schizophrenia and bipolar disorder
(13). The genetic and symptomatic
overlaps between these disorders resulted in the examination of
mutual variants in the present study. Stefansson et al
(7) found a significant association
with various polymorphisms in a large GWA study. The samples were
as follows: European samples, 2,663 schizophrenia patients and
13,498 control subjects (SGENE-plus samples); European follow-up
samples, 4,999 schizophrenia patients and 15,555 control subjects;
and 5,283 schizophrenia patients and 5,088 control subjects from
the International Schizophrenia Consortium and the Molecular
Genetics of Schizophrenia study. TCF4 gene rs9960767 is one of the
risk variants and was found to be more frequent in patients with
schizophrenia [P=4.1×10-9; OR=1.23, 95% confidence interval
(CI)=1.15–1.32]. This association was further confirmed in a
large-scale meta-analysis of schizophrenia GWAs
(P=4.2×10−9; OR=1.20, 95% CI=1.13–1.27) (14).
TCF4 gene (also termed immunoglobulin transcription
factor 2) is located on chromosome 18q21.2. This gene encodes a
bHLH transcription factor, which recognizes a 5′-CANNTG-3′ motif in
Ephrussi-box (E-box) found in immunoglobluin enhancers. TCF4
binding activates transcription in E-box. TCF4 is expressed in the
nervous system and initiates neuronal differantiation. Multiple
alternatively spliced transcipt variants have been described that
encode different proteins. rs9960767 is an intronic variant found
in the TCF4 gene that results in A>C transversion (dbSNP). Li
et al (15) attempted to
investigate nine polymorphisms in a large Han Chinese population;
four of the SNPs were not identified to be polymorphic in the Han
Chinese population, which is consistent with the HapMap data. The
authors genotyped 2,496 schizophrenia patients and 5,184 control
subjects, however, did not find any A>C transversion even in a
single sample for rs9960767 (15).
There are numerous cognitive-based studies
evaluating the association between cognitive performance and the
TCF4 genotype in psychiatric disorders. Lennertz et al
(16) investigated the correlation
between neurocognitive functions and schizophrenia. In total, 401
schizophrenia patients were genotyped for TCF4 rs9960767 and the
Rey Auditory Verbal Learning Test was performed with all subjects.
No significant difference for verbal memory was identified between
the AA genotype and C risk allele carrier subjects although the C
risk allele carriers performed better than the non-carriers in
recognition memory (P=0.049) (16).
Quednow et al (17) performed a
sensorimotor gating analysis in cases of schizophrenia and healthy
control subjects. The TCF4 genotype was found to modulate
sensorimotor gating, which was reduced in C risk allele carriers
(17). Albanna et al (18) genotyped TCF4 rs9960767 in 173 patients
with first-episode psychosis. Cognitive tests were also performed
on all subjects. AA carriers showed improved performance (P=0.038)
in problem solving and reasoning compared with the C risk allele
carriers (CC/CA) (18). These studies
demonstrated that TCF4 may influence cognitive function and
contribute to psychiatric disorders.
TCF4 variations are known to be involved in a rare
neurodevelopmental disease termed Pitt-Hopkins syndrome, which
shows an autosomal dominant transmission and is charactarized by
developmental delay, typical facial features, and severe mental and
motor retardation (19). A
translocation that contains exon four of the TCF4 gene was also
found to be associated with milder mental retardation when compared
with patients with Pitt-Hopkins syndrome (20).
The TCF4 gene is associated with schizophrenia, a
common disease and Pitt-Hopkins syndrome, which is rare. It
interacts with other genes and forms a transcriptional network to
regulate differentiation in various cell types. The TCF4 gene has
been identified as a target for the microRNA 137 (miR137) gene
product, miRNA137, which is asociated with schizophrenia and severe
cognitive abnormalities (21). miR137
is involved in neurogenesis and neuronal differentiation. Specific
software programmes, such as TargetScan (22) and PicTar (23), were used to analyze the targets of
miR137, and four genes (TCF4, CACNA1C, CUB and Sushi multiple
domains 1 and WW domain binding protein 1-like) were detected in
the study by the Schizophrenia Psychiatric GWAS Consortium
(24). Overexpression or knockdown of
the miR137 gene also affect expression of the TCF4 gene that
confirmed the association between TCF4 and miR137 (25).
Although the TCF4 rs9960767 variant is in an
intronic region, it may affect transcriptional regulation. It
remains unclear how the TCF4 gene is involved in psychiatric
disorders, although its role in neuronal differantiation may be an
indication of the pathogenesis of neuropsychiatric diseases.
Population size is important, particularly in case-control studies,
for increasing statistical power. In the present study, a CC
genotype was not available, which was due to the relatively small
population size.
In conclusion, this is the first study, to the best
of our knowledge, to investigate the association between the TCF4
gene rs9960767 variant and bipolar disorder. It was found that the
TCF4 gene rs9960767 was not a genetic risk factor for bipolar
disorder in the Turkish population that was evaluated. Neither
allelic, nor genotypic associations have been found in overall
population or in terms of gender. Larger populations may be
required for corroboration of these study findings. Further
genetic, biological and cognitive studies are required to better
understand the complex mechanisms underlying the effect of TCF4 on
neural functions with regard to psychiatric diseases.
Acknowledgements
The present study was supported by Kocaeli
University (Kocaeli, Turkey; grant no. 2011/68).
References
|
1
|
Craddock N, O'Donovan MC and Owen MJ: The
genetics of schizophrenia and bipolar disorder: dissecting
psychosis. J Med Genet. 42:193–204. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Craddock N and Sklar P: Genetics of
bipolar disorder. Lancet. 381:1654–1662. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hou L, Bergen SE, Akula N, Song J, Hultman
CM, Landén M, Adli M, Alda M, Ardau R, Arias B, et al: Genome-wide
association study of 40,000 individuals identifies two novel loci
associated with bipolar disorder. Hum Mol Genet. Jun 21–2016.(Epub
ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kao CF, Chen HW, Chen HC, Yang JH, Huang
MC, Chiu YH, Lin SK, Lee YC, Liu CM, Chuang LC, et al:
Identification of susceptible loci and enriched pathways for
bipolar II disorder using genome-wide association studies. Int J
Neuropsychopharmacol. Aug 2–2016.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Owen MJ, Williams HJ and O'Donovan MC:
Schizophrenia genetics: Advancing on two fronts. Curr Opin Genet
Dev. 19:266–270. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Craddock N and Owen MJ: The Kraepelinian
dichotomy - going, going but still not gone. Br J Psychiatry.
196:92–95. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Stefansson H, Ophoff RA, Steinberg S,
Andreassen OA, Cichon S, Rujescu D, Werge T, Pietiläinen OP, Mors
O, Mortensen PB, et al: Genetic Risk and Outcome in Psychosis
(GROUP): Common variants conferring risk of schizophrenia. Nature.
460:744–747. 2009.PubMed/NCBI
|
|
8
|
Amiel J, Rio M, de Pontual L, Redon R,
Malan V, Boddaert N, Plouin P, Carter NP, Lyonnet S, Munnich A, et
al: Mutations in TCF4, encoding a class I basic helix-loop-helix
transcription factor, are responsible for Pitt-Hopkins syndrome, a
severe epileptic encephalopathy associated with autonomic
dysfunction. Am J Hum Genet. 80:988–993. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
O'Donnell L, Soileau B, Heard P, Carter E,
Sebold C, Gelfond J, Hale DE and Cody JD: Genetic determinants of
autism in individuals with deletions of 18q. Hum Genet.
128:155–164. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Forrest MP, Waite AJ, Martin-Rendon E and
Blake DJ: Knockdown of human TCF4 affects multiple signaling
pathways involved in cell survival, epithelial to mesenchymal
transition and neuronal differentiation. PLoS One. 8:e731692013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Miller SA, Dykes DD and Polesky HF: A
simple salting out procedure for extracting DNA from human
nucleated cells. Nucleic Acids Res. 16:12151988. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Goes FS: Genetics of bipolar disorder:
recent update and future directions. Psychiatr Clin North Am.
39:139–155. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
McCarthy MI, Abecasis GR, Cardon LR,
Goldstein DB, Little J, Ioannidis JP and Hirschhorn JN: Genome-wide
association studies for complex traits: consensus, uncertainty and
challenges. Nat Rev Genet. 9:356–369. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Steinberg S, de Jong S, Andreassen OA,
Werge T, Børglum AD, Mors O, Mortensen PB, Gustafsson O, Costas J,
Pietiläinen OP, et al: Irish Schizophrenia Genomics Consortium;
GROUP; Wellcome Trust Case Control Consortium 2: Common variants at
VRK2 and TCF4 conferring risk of schizophrenia. Hum Mol Genet.
20:4076–4081. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li T, Li Z, Chen P, Zhao Q, Wang T, Huang
K, Li J, Li Y, Liu J, Zeng Z, et al: Common variants in major
histocompatibility complex region and TCF4 gene are significantly
associated with schizophrenia in Han Chinese. Biol Psychiatry.
68:671–673. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lennertz L, Rujescu D, Wagner M, Frommann
I, Schulze-Rauschenbach S, Schuhmacher A, Landsberg MW, Franke P,
Möller HJ, Wölwer W, et al: Novel schizophrenia risk gene TCF4
influences verbal learning and memory functioning in schizophrenia
patients. Neuropsychobiology. 63:131–136. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Quednow BB, Ettinger U, Mössner R, Rujescu
D, Giegling I, Collier DA, Schmechtig A, Kühn KU, Möller HJ, Maier
W, et al: The schizophrenia risk allele C of the TCF4 rs9960767
polymorphism disrupts sensorimotor gating in schizophrenia spectrum
and healthy volunteers. J Neurosci. 31:6684–6691. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Albanna A, Choudhry Z, Harvey PO, Fathalli
F, Cassidy C, Sengupta SM, Iyer SN, Rho A, Lepage M, Malla A, et
al: TCF4 gene polymorphism and cognitive performance in patients
with first episode psychosis. Schizophr Res. 152:124–129. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sepp M, Pruunsild P and Timmusk T:
Pitt-Hopkins syndrome-associated mutations in TCF4 lead to variable
impairment of the transcription factor function ranging from
hypomorphic to dominant-negative effects. Hum Mol Genet.
21:2873–2888. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kalscheuer VM, Feenstra I, Van
Ravenswaaij-Arts CM, Smeets DF, Menzel C, Ullmann R, Musante L and
Ropers HH: Disruption of the TCF4 gene in a girl with mental
retardation but without the classical Pitt-Hopkins syndrome. Am J
Med Genet A. 146A:2053–2059. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Siegert S, Seo J, Kwon EJ, Rudenko A, Cho
S, Wang W, Flood Z, Martorell AJ, Ericsson M, Mungenast AE, et al:
The schizophrenia risk gene product miR-137 alters presynaptic
plasticity. Nat Neurosci. 18:1008–1016. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Krek A, Grün D, Poy MN, Wolf R, Rosenberg
L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M,
et al: Combinatorial microRNA target predictions. Nat Genet.
37:495–500. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Schizophrenia Psychiatric Genome-Wide
Association Study (GWAS) Consortium, . Genome-wide association
study identifies five new schizophrenia loci. Nat Genet.
43:969–976. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ripke S, O'Dushlaine C, Chambert K, Moran
JL, Kähler AK, Akterin S, Bergen SE, Collins AL, Crowley JJ, Fromer
M, et al: Multicenter Genetic Studies of Schizophrenia Consortium;
Psychosis Endophenotypes International Consortium; Wellcome Trust
Case Control Consortium 2: Genome-wide association analysis
identifies 13 new risk loci for schizophrenia. Nat Genet.
45:1150–1159. 2013. View
Article : Google Scholar : PubMed/NCBI
|