Introduction
Fusarium species are ubiquitous fungi
extensively distributed in soil, plants and various organic
substrates. This genus is an important plant pathogen, which causes
different diseases and is responsible for important economic losses
on crops. In humans, the Fusarium species causes a broad
range of diseases, including superficial, locally invasive, or
disseminated infections. Disseminated infections occur almost
exclusively in severely immunocompromised patients and, currently,
disseminated infections are the second most common mold that causes
invasive fungal infections in immunosuppressed hosts, and is
associated with high morbidity and mortality rates (1,2).
Furthermore, the Fusarium species causes allergic diseases,
such as sinusitis in immunocompetent individuals and mycotoxicosis
following ingestion of food that is contaminated with
toxin-producing Fusarium (3,4). This genus
contains >70 species (5); a
literature review of 259 cases of fusariosis between 2001 and 2005
demonstrated that 12 species were associated with infection. The
F. solani species complex was the most common (50% of
cases), followed by the F. oxysporum species complex (20% of
cases) and F. verticillioides and F. moniliforme (10%
of cases for each) (6).
Morphological identification of the Fusarium
species is the primary, but most difficult, step in the detection
procedure. However, for the species that cannot be reliably
recognized by morphological characterization, additional analysis,
such as DNA sequencing and species-specific polymerase chain
reaction (PCR) assays, must be performed.
Translation elongation factor (TEF) 1-α consistently
presents as a single-copy gene in the Fusarium genus. This
gene demonstrates a high level of sequence polymorphism among the
closely associated species of Fusarium, even compared with
the intron-rich portions of protein-coding genes, such as
β-tubulin, calmodulin and histone H3. Therefore, TEF has become the
choice marker as a single-locus detection tool in Fusarium
(7,8).
The strategy that was developed in the present study consisted of
novel PCR-restriction fragment length polymorphism (RFLP) analysis
for detecting DNA polymorphisms in the TEF-1α gene and for
discrimination of the Fusarium genus.
Materials and methods
Microorganisms
Fifty strains of Fusarium spp. (including
environmental, clinical and reference isolates) were used in the
present study. The following reference strains were used: F.
solani complex PTCC 5284, F. solani complex PTCC 5285,
F. oxysporum complex IBRC-M 30067, F. oxysporum
complex PTCC 5115, F. verticillioides PFCC 53–131, F.
verticillioides PFCC 15–89, F. proliferatum PFCC 48–125,
F. proliferatum PFCC 12–86 and F. fujikuroi PTCC
5144. The environmental strains were obtained from soil, and two
strains used in the present study were clinical, which included
F. solani complex PTCC 5284 and B988.
DNA extraction
Thick spore suspension (1 ml) was inoculated in
Ehrlenmeyer flasks containing yeast extract peptone dextrose medium
and incubated on an incubator shaker at 200 rpm under agitation for
72 h at 25°C for mycelia growth. The harvested mycelia were washed
with 0.5 M EDTA and sterile dH2O. The mycelia were
ground into a fine powder using liquid nitrogen and a mortar and
pestle.
Approximately 100 mg powdered mycelium was
transferred into a 1.5-ml tube containing 400 µl lysis buffer (100
mM Tris-HCl, pH 8.0, 30 mM EDTA, pH 8.0, 5% SDS w/v). After
microtubes were boiled at 100°C for 20 min, 3 M acetate potassium
(150 µl) was added to each tube. The suspension was maintained at
−20°C for 10 min and centrifuged at 14,000 × g in 4°C for 10 min.
The supernatant was carefully transferred to a fresh 1.5-ml
Eppendorf tube and 250 µl phenol:chloroform:isoamyl alcohol
(25:24:1, v/v) was added. The microtube was vortexed briefly and
centrifuged at 4°C at 14,000 × g for 10 min. After transferring the
supernatant to a 1.5-ml microtube, 250 µl chloroform:isoamyl
alcohol was added. The tubes were briefly vortexed and centrifuged
at 4°C at 14,000 × g for 10 min. The supernatant was transferred to
a fresh microtube, an equal volume of ice-cold 2-propanol was
added, maintained at −20°C for 10 min and centrifuged at 14,000 × g
for 10 min. The upper aqueous phase was discarded and the pellet
was washed with 70% ethanol (300 µl). The ethanol was discarded and
the DNA pellets were air dried and resuspended in 50 µl
dH2O.
PCR amplification
The primer sets, Fu3f (5′-GGTATCGACAAGCGAACCAT-3′)
and Fu3r (5′-TAGTAGCGGGGAGTCTCGAA-3′) was used to amplify an
~420-bp DNA fragment of the TEF-1α gene (9). PCR reactions were performed with a volume
of 50 µl, comprised of 5 µl 10X reaction buffer, 2.2 mM
MgCl2, 200 µM each dNTP, 2.5 units of Taq DNA
polymerase (CinnaGen, Tehran, Iran), 30 ng template DNA and 50 pmol
of each primer.
An initial denaturation step for 5 min at 94°C was
followed by 30 cycles of denaturation at 94°C for 1 min, annealing
at 58°C for 1 min and extension at 68°C for 2 min. The amplified
PCR product (5 µl) was electrophoresed on 1% agarose gel in TAE
buffer at 100 V for 1 h and stained with ethidium bromide. The PCR
amplification of TEF-1α gene resulted in an ~420-bp fragment.
RFLP analysis
Digestion with one restriction enzyme was not
sufficient to discriminate the 420-bp DNA fragment of the TEF-1α
gene in the Fusarium species. Therefore, double digestion
with two restriction enzymes, XhoI and SduI (Thermo
Fisher Scientific, Inc., Waltham, MA, USA) was used for
discrimination. The restriction digestion reaction was performed in
a total volume of 20 µl containing 5 units of each enzyme, 2 µl
Buffer O (Thermo Fisher Scientific, Inc.), 5 µl PCR product, and
Ultrapure water (CinnaGen, Karaj, Iran) to reach a volume of 20 µl.
Digested PCR products were electrophoresed at 50 V for 3 h on 2%
agarose gel in TAE buffer and stained with ethidium bromide.
Results
PCR amplification of the TEF-1α
gene
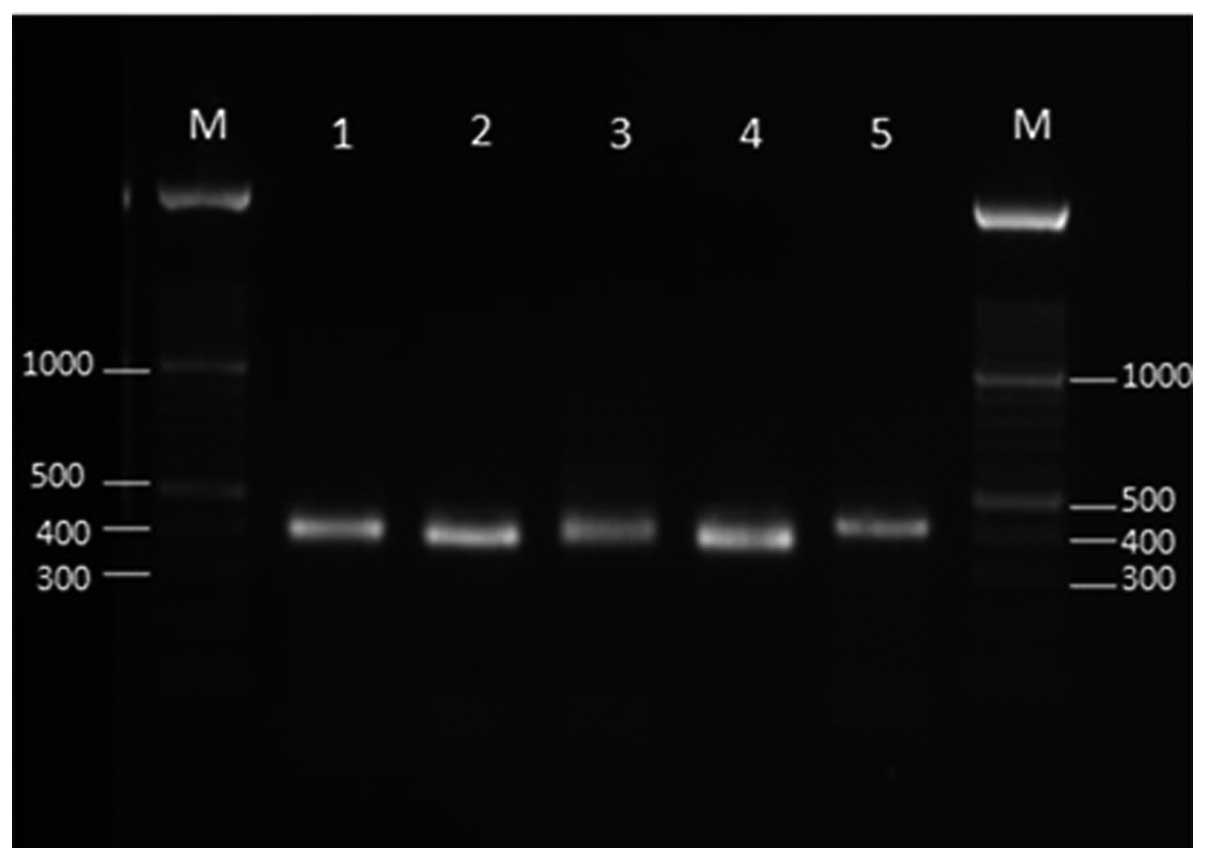
The PCR amplification of TEF-1α gene with Fu3f and
Fu3r primers produced a unique band of ~420 bp for all tested
Fusarium isolates (Fig. 1). The
TEF-1α gene fragment was sequenced for certain isolates, including
the reference strains. The BLAST search in NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi) demonstrated
the TEF-1α gene fragment from five clinically important
Fusarium reference strains, including F. solani
species complex, F. oxysporum species complex, F.
verticillioides, F. proliferatum and F. fujikuroi
exhibited 99% homology with the associated sequences deposited in
the GenBank database.
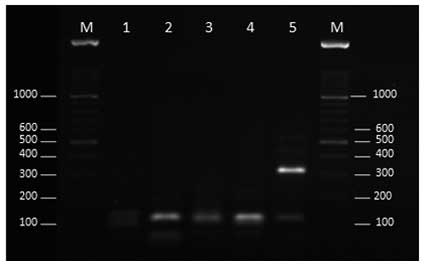
Restriction patterns for the Fusarium
strains
Double digestion of the fragment with restriction
enzymes, XhoI and SduI clearly discriminated the
F. solani species complex, F.oxysporum species
complex, F. verticillioides, F. proliferatum and
F. fujikuroi from each other (Table
I and Fig. 2).
 | Table I.Restriction fragment size (bp) of the
Fusarium species TEF-1α gene, double digested with two
restriction enzymes, XhoI and SduI. |
Table I.
Restriction fragment size (bp) of the
Fusarium species TEF-1α gene, double digested with two
restriction enzymes, XhoI and SduI.
| Fusarium
species | TEF-1α fragment prior
to digestion (bp) | XhoI and
SduI (bp) |
|---|
| F. oxysporum
species complex | 420 | 45,62,103,170 |
| F.
verticillioides | 420 |
6,30,56,47,55,186 |
| F.
proliferatum | 420 | 25,168,187 |
| F.
fujikuroi | 420 | 27,62,99,192 |
| F. solani
species complex | 420 | 308,110 |
The restriction patterns of one clinical and six
environmental Fusarium strains following double digestion
using XhoI and SduI are presented in Fig. 3. The digestion of the 420-bp fragment
from these strains demonstrated different patterns. Strains E4, E16
and E25 were sequenced. A BLAST search showed that strains E4 and
E16 exhibited 100% homology with the F. equiseti and F.
solani species complex, respectively and strain E25 exhibited
99% homology with F. incarnatum. Therefore, the restriction
pattern strain E16 (Fig. 3) was
similar to the F. solani complex PTCC 5284 (Fig. 2).
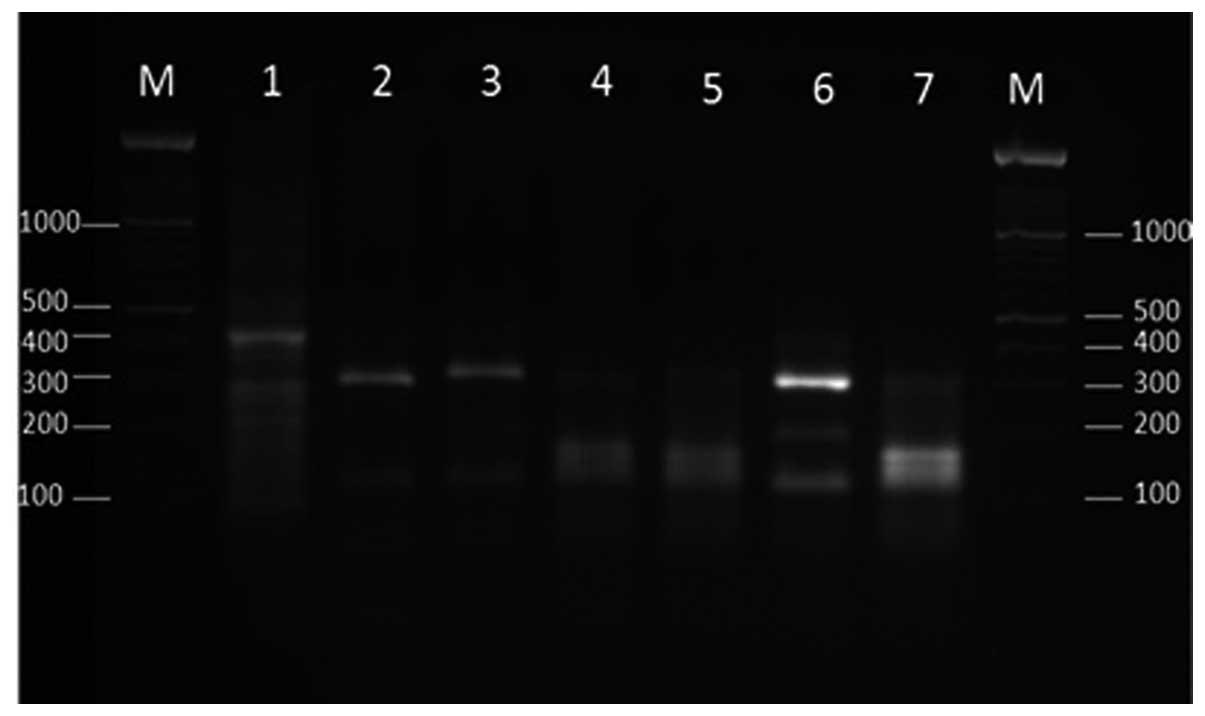 | Figure 3.Agarose gel electrophoresis of
transcription elongation factor-1α gene products (420 bp) of the
Fusarium species (lane 1, clinical isolate; lanes 2–7,
environmental isolates) following double digestion with XhoI
and SduI. Lane M, 100-bp ladder; lane1, B988; lane 2, E4;
lane 3, E16; lane 4, E17; lane 5, E18; lane 6, E20; lane 7,
E25. |
Discussion
Identification of filamentous fungi at the species
level using classical techniques, such as morphological methods, is
difficult and time-consuming. Novel rapid techniques are required
in order to verify the Fusarium genus on time, particularly
for clinical administration of patients. Rapid molecular
approaches, such as PCR, DNA hybridization and DNA microarray have
been developed and they may replace the classical methods. The
major advantages of molecular approaches are their specificity and
that they are completely discriminative even for closely associated
species (8,9).
The majority of molecular techniques are PCR-based,
where the primers are typically directed to conserved regions of
the ribosomal DNA gene, particularly towards the internal
transcribed spacer (ITS) regions. With regard to Fusarium
spp., analysis of ITS sequencing is considered unreliable for
detection of strains, as they contain two paralogous, discrepant
ITS sequence forms, which may cause confusion (10,11). The
TEF-1α gene has shown optimal results for the identification of
Fusarium spp. (12–14).
Guevara-Suarez et al (15) used a TEF-1α gene fragment and performed
a multi-locus sequence analysis of the ITS region with the
RNA-dependent polymerase subunit II (Rpb2) genes, and recognized
the phylogenetic species and circulating haplotypes for
Fusarium isolates from onychomycosis. The pathogenic
isolates to the pecan tree were identified, based on the TEF-1α
gene, as belonging to the F. chlamydosporum species complex,
F. graminearum species complex, F. proliferatum, and
F. oxysporum (16). A
TEF-1α-RFLP technique was described for the identification of the
three clades of F. oxysporum (17). The particularly effective TEF-1α gene
of the Fusarium spp. encouraged the present development of a
PCR-RFLP technique as an advanced, simple and reliable method for
determination and discrimination of the clinically important
Fusarium species.
In the current study, molecular identification was
performed using the TEF-1α gene and RFLP, and it was possible to
discriminate between all five clinically important Fusarium
species. However, further analyses are required for discrimination
between other Fusarium species.
The Primer set, TEF-Fu3 resulted in an ~420-bp
product for five of the Fusarium species, including F.
solani species complex, F. oxysporum species complex,
F. verticillioides, F. proliferatum and F.
fujikuroi. RFLP, using double digestion with two restriction
enzymes, XhoI and SduI differentiated between the
species. This method may facilitate detection, verify the
Fusarium genus, and be applied for disease control. This
PCR-RLFP method is rapid, economical and efficient for detection
and discrimination of the Fusarium genus.
Acknowledgements
The present study was supported by the Health
Research Institute, Infectious and Tropical Diseases Research
Center, Ahvaz Jundishapur University of Medical Sciences (Ahvaz,
Iran) (grant no. 92118).
References
|
1
|
Consigny S, Dhedin N, Datry A, Choquet S,
Leblond V and Chosidow O: Successsful voriconazole treatment of
disseminated fusarium infection in an immunocompromised patient.
Clin Infect Dis. 37:311–313. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nucci M and Anaissie E: Cutaneous
infection by Fusarium species in healthy and immunocompromised
hosts: Implications for diagnosis and management. Clin Infect Dis.
35:909–920. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Twarużek M, Soszczyńska E, Winiarski P,
Zwierz A and Grajewski J: The occurrence of molds in patients with
chronic sinusitis. Eur Arch Otorhinolaryngol. 271:1143–1148. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Beccari G, Caproni L, Tini F, Uhlig S and
Covarelli L: Presence of fusarium species and other toxigenic fungi
in malting barley and multi-mycotoxin analysis by liquid
chromatography-high resolution mass spectrometry. J Agric Food
Chem. 64:4390–4399. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Al-Hatmi AM, Meis JF and de Hoog GS:
Fusarium: Molecular Diversity and Intrinsic Drug Resistance. PLoS
Pathog. 12:e10054642016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nucci M and Anaissie E: Fusarium
infections in immunocompromised patients. Clin Microbiol Rev.
20:695–704. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Geiser DM, Jiménez-Gasco MM, Kang S,
Makalowska I, Veeraraghavan N, Ward TJ, Zhang N, Kuldau GA and
O'Donnell K: FUSARIUM-ID v. 1.0: A DNA sequence database for
identifying Fusarium. Eur J Plant Pathol. 110:473–479. 2004.
View Article : Google Scholar
|
|
8
|
Hsuan HM, Salleh B and Zakaria L:
Molecular identification of Fusarium species in Gibberella
fujikuroi species complex from rice, sugarcane and maize from
Peninsular Malaysia. Int J Mol Sci. 12:6722–6732. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Arif M, Chawla S, Zaidi NW, Rayar JK,
Variar M and Singh US: Development of specific primers for genus
Fusarium and F. solani using rDNA sub-unit and transcription
elongation factor (TEF-1α) gene. Afr J Biotechnol. 11:444–447.
2012.
|
|
10
|
Dornbusch HJ, Buzina W, Summerbell RC,
Lass-Flörl C, Lackner H, Schwinger W, Sovinz P and Urban C:
Fusarium verticillioides abscess of the nasal septum in an
immunosuppressed child: Case report and identification of the
morphologically atypical fungal strain. J Clin Microbiol.
43:1998–2001. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
O'Donnell K and Cigelnik E: Two divergent
intragenomic rDNA ITS2 types within a monophyletic lineage of the
fungus Fusarium are nonorthologous. Mol Phylogenet Evol. 7:103–116.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Palacios SA, Susca A, Haidukowski M, Stea
G, Cendoya E, Ramírez ML, Chulze SN, Farnochi MC, Moretti A and
Torres AM: Genetic variability and fumonisin production by Fusarium
proliferatum isolated from durum wheat grains in Argentina. Int J
Food Microbiol. 201:35–41. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Watanabe M, Yonezawa T, Sugita-Konishi Y
and Kamata Y: Utility of the phylotoxigenic relationships among
trichothecene-producing Fusarium species for predicting their
mycotoxin-producing potential. Food Addit Contam Part A Chem Anal
Control Expo Risk Assess. 30:1370–1381. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Scheel CM, Hurst SF, Barreiros G, Akiti T,
Nucci M and Balajee SA: Molecular analyses of Fusarium isolates
recovered from a cluster of invasive mold infections in a Brazilian
hospital. BMC Infect Dis. 13:492013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Guevara-Suarez M, Cano-Lira JF, de García
MC, Sopo L, De Bedout C, Cano LE, García AM, Motta A, Amézquita A,
Cárdenas M, et al: Genotyping of Fusarium isolates from
onychomycoses in Colombia: Detection of two new species within the
Fusarium solani species complex and in vitro antifungal
susceptibility testing. Mycopathologia. 181:165–174. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lazarotto M, Milanesi PM, Muniz MF,
Reiniger LR, Beltrame R, Harakava R and Blume E: Morphological and
molecular characterization of Fusarium spp pathogenic to pecan tree
in Brazil. Genet Mol Res. 13:9390–9402. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bogale M, Wingfield BD, Wingfield MJ and
Steenkamp ET: Species-specific primers for Fusarium redolens and a
PCR-RFLP technique to distinguish among three clades of Fusarium
oxysporum. FEMS Microbiol Lett. 271:27–32. 2007. View Article : Google Scholar : PubMed/NCBI
|