Introduction
Kinesin-mediated cargo transport is essential for
many cellular functions and the morphogenesis of cells. Kinesin
superfamily proteins (KIFs) are microtubule-dependent motor
proteins involved in the transport of various cargoes, including
membrane vesicles, organelles, proteins complexes, and mRNAs
(1,2).
Kinesin 1 is the first identified member of KIFs responsible for
anterograde axonal transport (3).
Kinesin 1 is a tetrameric protein composed of two KIF5 motors, also
known as kinesin heavy chains (KHCs) and two kinesin light chains
(KLCs) (2–4). The KIF5s contain the motor domain at the
amino N-terminal region, a coiled-coil dimerization domain, and a
carboxyl C-terminal tail region that regulates the motor activity.
KLCs contain the N-terminal α-helix domain that binds to KIF5s, the
central tetratricopeptide repeat (TPR) domains, and the C-terminal
region that negatively regulates the motor domain ATPase activity
of KIF5s (3–5). Three isoforms of KIF5s exist in mammals;
KIF5A and KIF5C are expressed in neurons, whereas KIF5B is
expressed ubiquitously (6). Three
isoforms of KLC have been identified in mammals as follows: The
neuronal tissue-specific KLC1, the ubiquitous KLC2, and the
testis-specific KLC3 (4,7). Certain kinesin 1 cargoes, such as
glutamate receptor-interacting protein 1 and γ-aminobutyric acid
receptor-associated protein interact directly with the tail cargo
binding-domain of KIF5s, while others, including S100 proteins and
Alcadein α bind to the TPR domains of KLCs (8–11).
The TPR domain consisting of multiple tandem repeats
is a well-known module facilitating protein-protein interaction
that organizes complexes involved in a number of biological
processes, including the adaptor function that the binding kinesin
1 to other cargos (12,13). Each TPR motif consists of 34 amino
acids, which form a helix-turn-helix structure (12). The TPR domains of KLCs are highly
conserved across species and are known to be involved in cargo
interaction (5,12–14). The
first protein identified to bind to the TPR domain of KLC1 was the
c-Jun NH2-terminal kinase (JNK)-interacting protein (JIP, also
termed JSAP) group (15,16). The three JIP isoforms in mammals are
scaffolds for the mitogen-activated protein kinase cascade that
activates JNK (17,18).
Although many cargoes, including organelles moved by
kinesin 1 have been identified, it remains unclear as to how
kinesin 1 binds to different cargoes and regulates organelle
transport. In certain cases, cargoes bind to adaptor/scaffolding
proteins that mediate the attachment of kinesin 1 to the cargo
(2). To improve the understanding of
kinesin 1-dependent transport events, it was necessary to further
identify the interacting partners of kinesin 1. In the present
study, proteins that interact specifically with the TPR domain of
KLC1 were screened for and FUN14 domain-containing protein 1
(FUNDC1), which is a mitochondrial outer membrane protein with
three transmembrane domains (19) was
identified. FUNDC1 functions as a receptor of selective mitophagy
in response to hypoxia via interaction with LC3 (19). The interaction between KLC1 and FUNDC1
indicates that KLC1 may serve as a potential competitor to LC3 for
interaction with FUNDC1.
Materials and methods
Plasmid constructs
The mouse KLC1 cDNA fragment corresponding to the
TPR domain (amino acids 80–541) was amplified by polymerase chain
reaction (PCR) from the Marathon-Ready™ cDNA library (Clontech
Laboratories, Inc., Palo Alto, CA, USA) using the appropriate
primers (forward primer: 5′-AGCGAGGCGCAGGTGATGATGGCG-3′, reverse
primer: 5′-AGTGCCATCCCCATTCCACTCTAC-3′). The PCR reaction was
performed in a total volume of 25 µl containing ~50 ng cDNA
library, 200 mM deoxynucleotide triphosphates (dNTPs), 1X reaction
buffer (MBI Fermentas, St. Leon-Rot, Germany), 1.5 mM
MgCl2, and 2-unit Taq polymerase (MBI Fermentas). The
temperature profile for the 30 cycles amplification reaction using
a PerkinElmer's PCR machine (PerkinElmer Applied Biosystems,
Warrington, UK) was as follows: Initial denaturation at 94°C for 4
min, denaturation at 94°C for 30 sec, annealing at 52°C for 50 sec
and extension at 72°C for 2 min, with a final extension at 72°C for
5 min. The amplified fragment was cloned into pLexA (Clontech
Laboratories, Inc.). The resulting recombinant plasmid,
pLexA-6xTPR-KLC1, served as bait plasmid. The full-length cDNA of
mouse LC3B (GeneBank accession number: NM_026160) was amplified by
PCR from the Marathon-Ready™ cDNA library (Clontech Laboratories,
Inc.) using the appropriate primers (forward primer:
5′-ATGCCGTCCGAGAAGACCTTCAAGCAG-3′, reverse primer:
5′-TTACACAGCCATTGCTGTCCCGAATGT-3′) and cloned into pLexA and pB42AD
(Clontech Laboratories, Inc.). The PCR reaction was performed
according to the above-mentioned method. The C-terminal region of
KIF3A (provided by Professor Kozo Kaibuchi; Nagoya University
Graduate School of Medicine, Nagoya, Japan) was amplified by PCR
using the appropriate primers (forward primer:
5′-CGCCAGTTTCAGAAAGAAATCGAA-3′, reverse primer:
5′-TTACTGAAGTAAAGAATCAATTAC-3′) and cloned into pLexA and pB42AD.
The PCR reaction was performed according to the above-mentioned
method.
Screening of KLC1-binding proteins by
yeast two-hybrid assay
The Matchmaker LexA Two-Hybrid system (Clontech
Laboratories, Inc.) was used for screening according to the
manufacturer's instructions. Briefly, pLexA-6xTPR-KLC1 was
transformed into the yeast strain, EGY48 carrying the p8op-lacZ
gene. The transformed EGY48 yeast cells containing pLexA-6xTPR-KLC1
were transformed with the mouse brain cDNA library (9) and grown on synthetic dextrose (SD) plates
supplemented with glucose, but with no histidine, tryptophan, or
uracil (SD/-His/-Trp/-Ura) (Clontech Laboratories, Inc.). The
selection of positive clones was performed on an
SD/-His/-Trp/-Ura/-Leu plate containing galactose, raffinose, BU
salts (Clontech Laboratories, Inc.) and 0.5 ml of X-Gal solution
(20 mg/ml). Plasmids from the positive clones were analyzed by
restriction digestion with EcoRI and XhoI. Unique
inserts were sequenced and DNA sequence analysis was performed
using the BLAST algorithm (National Center for Biotechnology
Information; https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Sequence-verified clones were analyzed again for interaction with
bait in yeast by retransformation.
β-Galactosidase activity in liquid
cultures of yeast
The β-galactosidase activity of yeast was assayed as
described previously (10). Briefly,
mid-log phase yeast cells were collected and permeabilized with
0.1% sodium dodecyl sulfate (SDS) and chloroform. An excess
quantity of o-nitrophenyl-β-D-galactoside was added to the yeast
lysate, the mixture was incubated at 30°C and the reaction was
terminated by increasing the pH to 11 by the addition of 1 M
Na2CO3. Formation of the reaction product,
o-nitrophenol, was determined by measuring absorbance at a
wavelength of 420 nm on a spectrophotometer, and normalizing for
the reaction time and cell density. The units of enzyme activity
were calculated with the following equation: Units=1,000 ×
[(optical density; OD420-1.75 ×
OD550)]/[reaction time (min) × culture volume (ml) ×
OD600]. All experiments were independently performed a
minimum of three times.
Glutathione S-transferase (GST)
pull-down assays
cDNA encoding the full-length FUNDC1 was cloned into
pET41a. The recombinant GST-FUNDC1 fusion protein was expressed in
E. coli strain BL21 GOLD (Stratagene, La Jolla, CA, USA)
following induction with 0.5 mM isopropyl
thio-β-D-galactopyranoside (IPTG) for 3 h. The fusion proteins were
purified by attachment to glutathione-agarose beads (Sigma-Aldrich,
St. Louis, MO, USA) according to the manufacturer's protocol.
Purified His-tagged KLC1 protein was incubated overnight at 4°C
with the glutathione beads coupled with GST alone or GST-FUNDC1
protein. The beads were pelleted by centrifugation, washed three
times with the extraction buffer [1% Triton X-100 in
phosphate-buffered saline (PBS) containing 10 µg/ml each of
aprotinin, leupeptin, and pepstatin and 1 µM phenylmethanesulfonyl
fluoride], and once with PBS. The bound proteins were eluted from
the glutathione beads with 100 µl 1X Laemmli loading buffer. The
pulled-down proteins were analyzed by immunoblotting with anti-KLC1
antibody (1:800, cat. no. ab187179; Abcam, Cambridge, MA, USA).
Cell culture and transfection
Human embryonic kidney (HEK)-293T [American Type
Culture Collection (ATCC) CRL-3216] cells were cultured in
Dulbecco's modified Eagle's medium supplemented with 10% fetal
bovine serum, L-glutamine, 100 U/ml penicillin, and 0.1 mg/ml
streptomycin at 37°C in a humidified 5% CO2 incubator.
Transient transfections were performed using the CaPO4
precipitation method (20).
Immunocytochemistry
HEK-293T cells grown on poly-D-lysine-coated
coverslips were transfected with enhanced green fluorescent protein
(EGFP)-FUNDC1 and KLC1 constructs. Twenty-four hours after
transfection, cells were washed with PBS, fixed with 4%
paraformaldehyde in PBS for 5 min, and permeabilized with 0.2%
Triton X-100 (Sigma-Aldrich) in PBS for 10 min. After blocking with
5% normal goat serum in PBS for 30 min, cells were incubated
overnight at 4°C with anti-KLC1 antibody (1:500, cat. no. ab187179;
Abcam) in PBS containing 1% bovine serum albumin (BSA) and 0.05%
Tween-20 (Sigma-Aldrich). After washing three times with PBS, cells
were incubated for 40 min with Dylight 594-conjugated goat
anti-rabbit IgG antibody (1:800, cat. no. 111-516-046; Jackson
ImmunoResearch Laboratories, Inc., West Grove, PA, USA). After
washing three times with PBS, the cells were mounted with
Fluoromount (DAKO, Santa Clara, CA, USA). Fluorescence images were
acquired on a Zeiss LSM510 META confocal laser-scanning microscope
(Carl Zeiss Inc., Zena, Germany).
Co-immunoprecipitation and immunoblot
analysis
Twenty-four hours after transfection with the
myc-KLC1 and FLAG-FUNDC1 constructs, the HEK-293T cells were rinsed
with ice-cold PBS twice and lysed with ice-cold lysis buffer [PBS
containing 0.5% NP-40 and 1X protease inhibitor cocktail set V (EMD
Millipore, Billerica, MA, USA)] by gentle rotation for 30 min.
Lysates were centrifuged at 16,000 × g for 10 min at 4°C. The
supernatant was incubated with anti-FLAG M2 agarose beads
(Sigma-Aldrich) for 2 h at 4°C with constant shaking. The beads
were collected by centrifugation at 2,000 × g for 30 sec and washed
five times with ice-cold PBS containing 0.5% NP-40. The washed
beads were resuspended with 2X Laemmli loading buffer and the
proteins were eluted and denatured by boiling for 2 min. The
proteins were processed for 10% SDS-PAGE and immunoblot analysis
with antibodies against KLC1 (Abcam), KIF5B (6), LC3B (1:1,000, cat. no. ab48394; Abcam)
and FLAG (1:2,000, cat. no. F7425; Sigma-Aldrich).
Results
Kinesin 1 is significant in fast axonal transport
and is involved in the intracellular trafficking of various cargoes
(1,2).
Certain cargoes, such as JIPs interact with KLC subunits, but not
KHC motor subunits of kinesin 1 (15,21). To
identify proteins that interact with the TPR domain of KLC1, a bait
construct encoding a fusion protein containing the TPR domain of
mouse KLC1 was used for yeast two-hybrid screening. In a screen of
9×106 independent transformants, two positive clones
were obtained. The two clones overlapped at the open reading frame
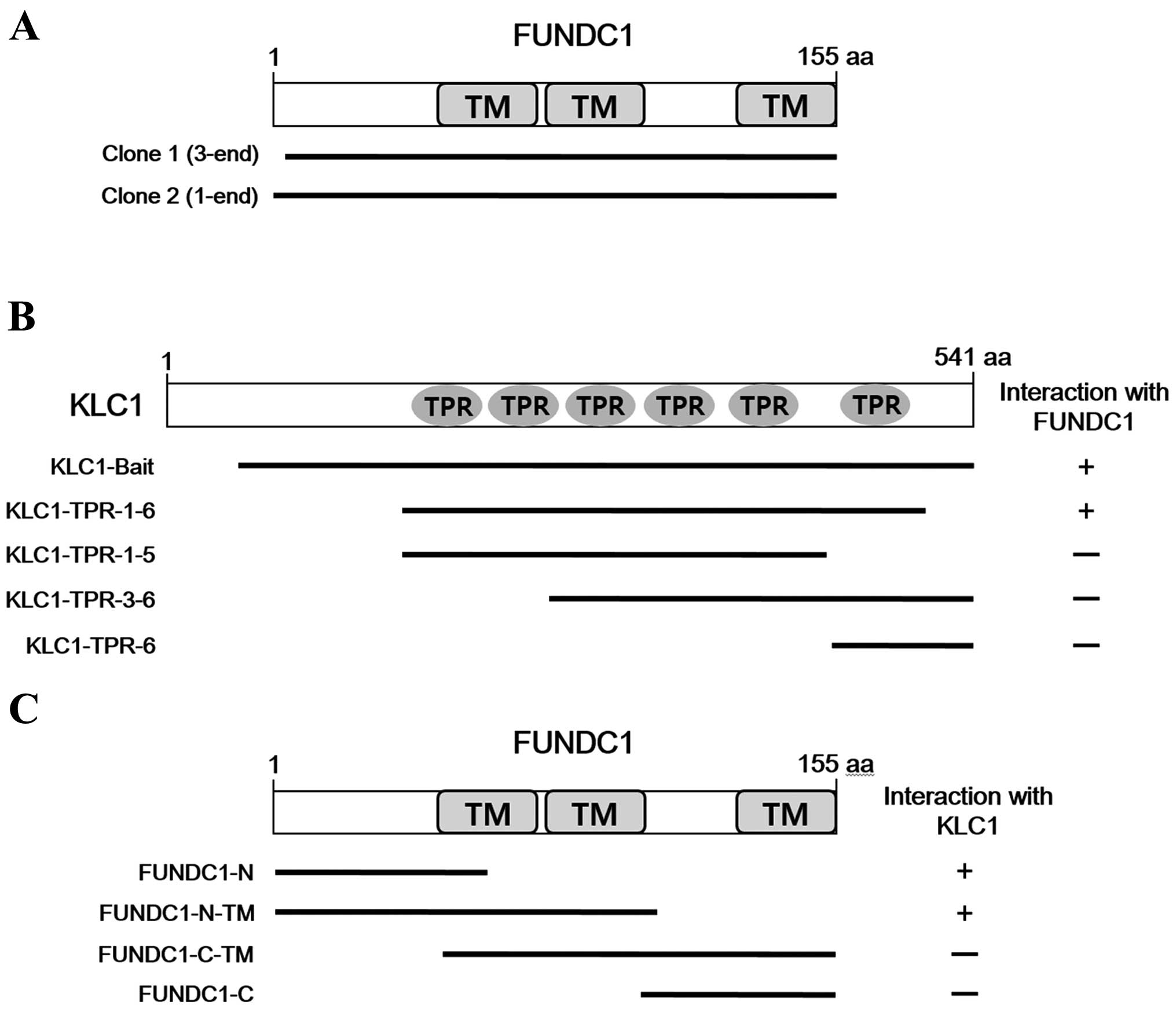
(ORF) of FUNDC1 (Fig. 1A). To identify
the region of KLC1 required for interaction with FUNDC1, various
fragments of KLC1 were constructed and tested for interaction with
FUNDC1 using a yeast two-hybrid system (Fig. 1B). The result indicates that the region
containing all six TPR repeats of KLC1 is required for binding.
FUNDC1 is a mitochondrial outer membrane protein with three
transmembrane domains (19). To
determine the binding domain of FUNDC1 that is required for the
interaction with KLC1, various fragments of FUNDC1 were
constructed. As shown in Fig. 1C, the
N-terminal cytoplasmic region of FUNDC1 interacted with KLC1.
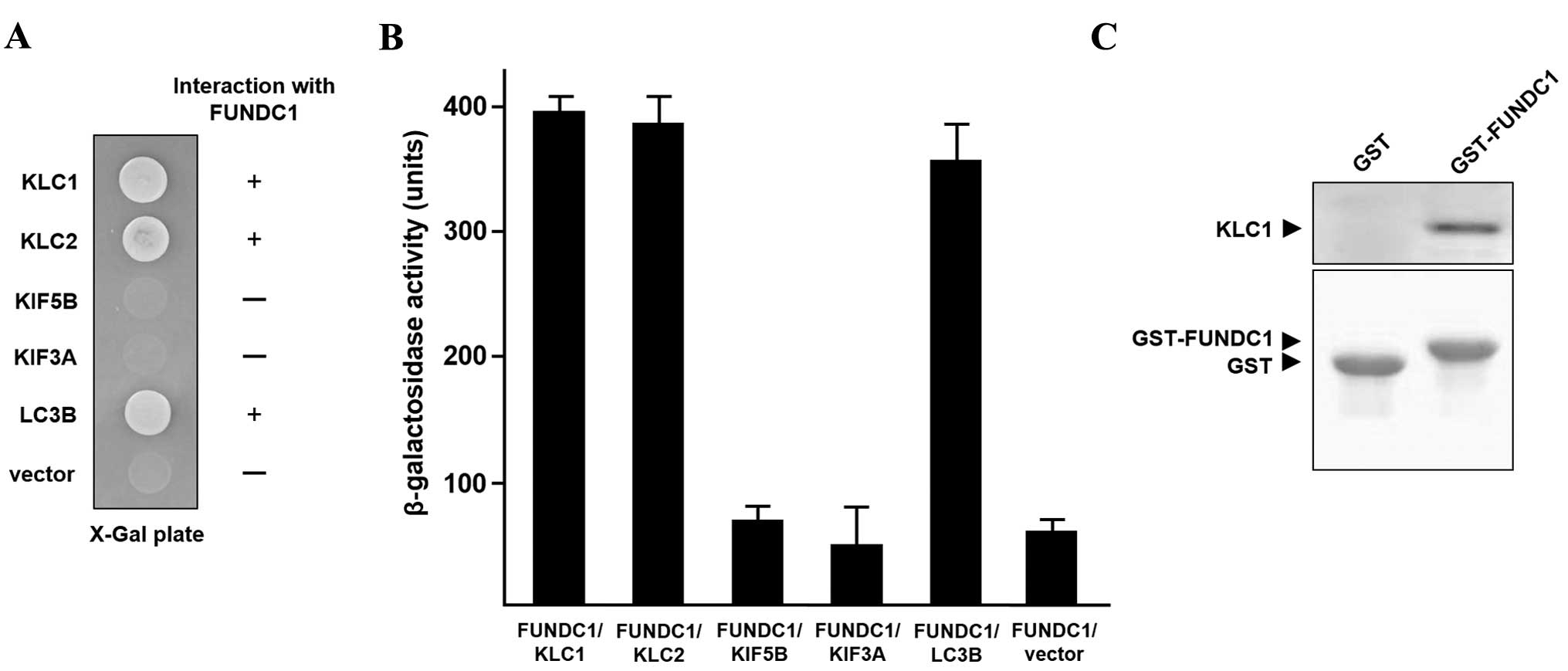
Subsequently, whether KLC2, KIF5B, and KIF3A, a motor subunit of
kinesin 2, interact with FUNDC1 was investigated. As shown in
Fig. 2A, KIF5B and KIF3A did not
interact with FUNDC1, but KLC2 bound to FUNDC1. LC3B, known to
interact with FUNDC1 (19), served as
a positive control. In addition, a quantitative β-galactosidase
assay demonstrated that FUNDC1 bound to KLC1 and KLC2 (Fig. 2B). To demonstrate the direct
interaction between KLC1 and FUNDC1, GST-FUNDC1 and His-KLC1
proteins were prepared and assessed for binding in a GST pull-down
assay. KLC1 interacted with GST-FUNDC1, but not with GST alone
(Fig. 2C). This result indicates that
KLC1 directly interacts with FUNDC1.
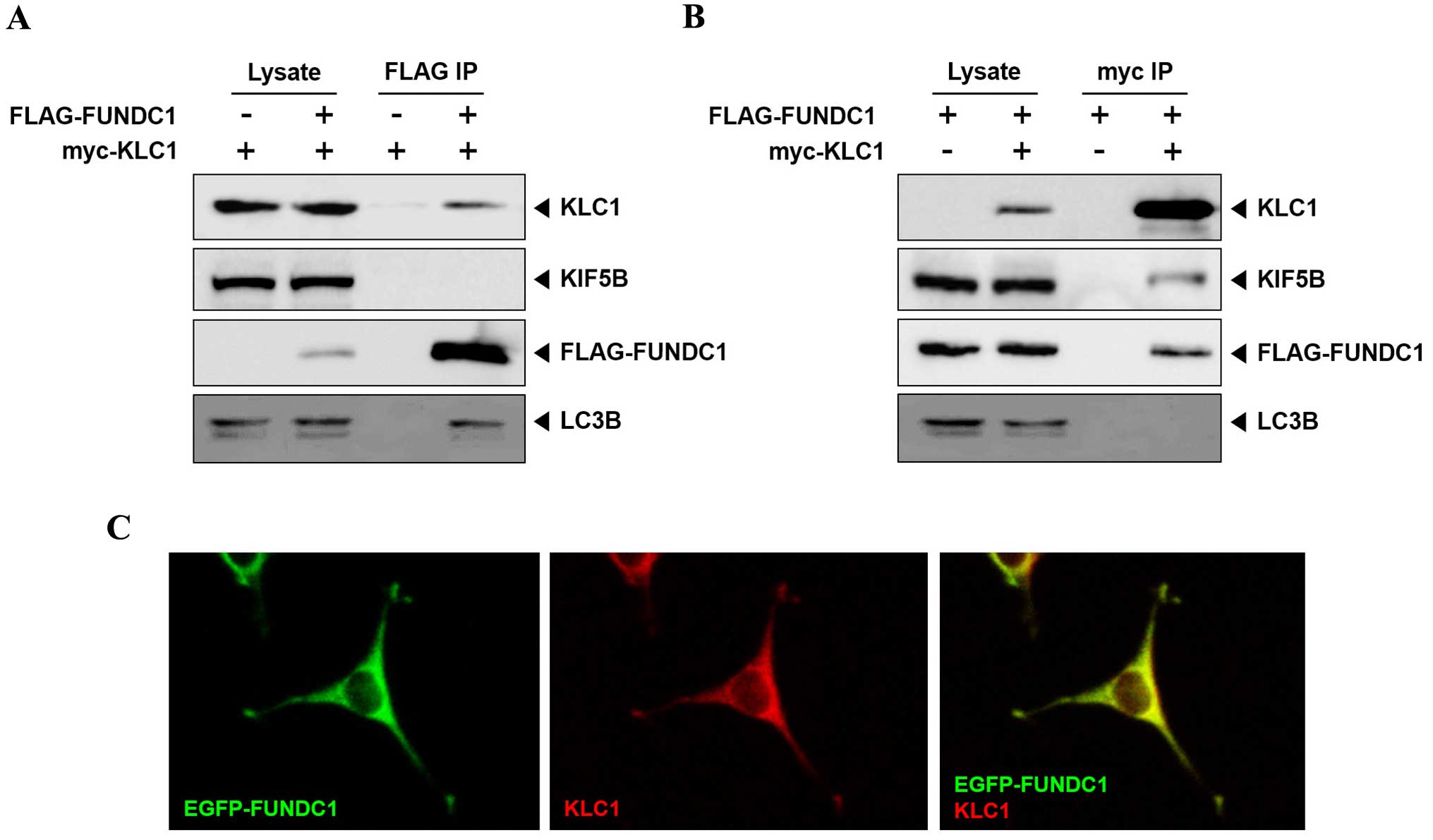
To further confirm the FUNDC1 and KLC1 interaction
in mammalian cells, co-immunoprecipitation from HEK-293T cells that
were transfected with FLAG-FUNDC1 and myc-KLC1 was performed.
Anti-FLAG antibody precipitated KLC1 and endogenous LC3; however,
KIF5B did not (Fig. 3A). Conversely,
anti-myc antibody precipitated KIF5B and FUNDC1, but not LC3
(Fig. 3B). These results indicate that
FUNDC1 interacts with free KLC1, but not with KLC1 bound to KIF5.
In order to address whether KLC1 and FUNDC1 co-localize in cells,
KLC1 was co-expressed with EGFP-FUNDC1 in HEK-293T cells. KLC1 and
FUNDC1 were identified to co-localize at the same region in cells
(Fig. 3C). Taken together, these
results indicate that FUNDC1 is a novel binding partner of
KLC1.
Discussion
Mitochondrial autophagy, mitophagy, is involved in
the removal of dysfunctional mitochondria, and controls
mitochondrial quality and quantity (22,23). FUNDC1
is a receptor of mitophagy in response to hypoxia (19). LC3 functions as a binding partner for
autophagy receptors (22,23). The N-terminal region of FUNDC1 is
exposed to the cytosol and contains a typical LC3-interacting
region (LIR) motif (19). The
conserved LIR motif is essential for the interaction between FUNDC1
and LC3. Deletion or mutations of LIR motif abolish the FUNDC1 and
LC3 interaction, and block the induction of mitophagy (19). In the current study, KLC1 interacted
with the LIR motif-containing region of FUNDC1. Although the
possibility that KLC1 interacts with another N-terminal region than
the LIR motif of FUNDC1 cannot be excluded, this result indicates
that the KLC1 may interfere with LC3 binding to FUNDC1. As shown in
Fig. 3B, LC3B did not co-precipitate
with KLC1-bound FUNDC1, indicating a possible competition between
KLC1 and LC3 for binding to FUNDC1.
The role of adaptor or scaffolding proteins
effectively controls the cargo recognition of motors (2). Cargo-associated KIF5s may not always be
associated with KLCs. The axonal transport of mitochondria is
directly mediated by KLC-independent interaction between the
mitochondrial rho (Miro)-Milton complex and KIF5 (24,25). Milton
acts as an adaptor protein that links KIF5 motor to Miro, a
mitochondrial outer membrane protein (24,25). KLC is
not required for the transport of mitochondria and is absent from
Milton-KIF5 complex (24). Instead,
KLC inhibits the KIF5 and Milton interaction (24). In the present study, FUNDC1 was found
to associate with free KLC1, but not KIF5-bound KLC1 (i.e., the
KLC1 as a subunit of kinesin 1). Thus, it is hypothesized that,
under normoxia, the KLC1 and FUNDC1 interaction may prevent
mitophagy of healthy mitochondria, by inhibiting the FUNDC1
interaction with LC3, and lead to dissociation of KIF5 from KLC1,
allowing free KIF5 to associate with the Miro-Milton complex and
mediate transport of mitochondria to their proper location. In
response to hypoxia, FUNDC1 may be dissociated from KLC1, and
subsequently interact with LC3 and trigger mitophagy of damaged
mitochondria.
Acknowledgements
The present study was supported by the Basic Science
Research Program through the National Research Foundation of Korea
by the Ministry of Education, Science and Technology (grant no.
2015R1D1A1A01056820).
References
|
1
|
Hirokawa N, Niwa S and Tanaka Y: Molecular
motors in neurons: Transport mechanisms and roles in brain
function, development, and disease. Neuron. 68:610–638. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fu MM and Holzbaur EL: Integrated
regulation of motor-driven organelle transport by scaffolding
proteins. Trends Cell Biol. 24:564–574. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hirokawa N: Kinesin and dynein superfamily
proteins and the mechanism of organelle transport. Science.
279:519–526. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vale RD and Fletterick RJ: The design plan
of kinesin motors. Annu Rev Cell Dev Biol. 13:745–777. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yip YY, Pernigo S, Sanger A, Xu M, Parsons
M, Steiner RA and Dodding MP: The light chains of kinesin-1 are
autoinhibited. Proc Natl Acad Sci USA. 113:2418–2423. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kanai Y, Okada Y, Tanaka Y, Harada A,
Terada S and Hirokawa N: KIF5C, a novel neuronal kinesin enriched
in motor neurons. J Neurosci. 20:6374–6384. 2000.PubMed/NCBI
|
|
7
|
Rahman A, Friedman DS and Goldstein LS:
Two kinesin light chain genes in mice. Identification and
characterization of the encoded proteins. J Biol Chem.
273:15395–15403. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Setou M, Seog DH, Tanaka Y, Kanai Y, Takei
Y, Kawagishi M and Hirokawa N: Glutamate-receptor-interacting
protein GRIP1 directly steers kinesin to dendrites. Nature.
417:83–87. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nakajima K, Yin X, Takei Y, Seog DH, Homma
N and Hirokawa N: Molecular motor KIF5A is essential for GABA(A)
receptor transport, and KIF5A deletion causes epilepsy. Neuron.
76:945–961. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shimamoto S, Takata M, Tokuda M, Oohira F,
Tokumitsu H and Kobayashi R: Interactions of S100A2 and S100A6 with
the tetratricopeptide repeat proteins, Hsp90/Hsp70-organizing
protein and kinesin light chain. J Biol Chem. 283:28246–28258.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Araki Y, Kawano T, Taru H, Saito Y, Wada
S, Miyamoto K, Kobayashi H, Ishikawa HO, Ohsugi Y, Yamamoto T, et
al: The novel cargo Alcadein induces vesicle association of
kinesin-1 motor components and activates axonal transport. EMBO J.
26:1475–1486. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhu H, Lee HY, Tong Y, Hong BS, Kim KP,
Shen Y, Lim KJ, Mackenzie F, Tempel W and Park HW: Crystal
structures of the tetratricopeptide repeat domains of kinesin light
chains: Insight into cargo recognition mechanisms. PLoS One.
7:e339432012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kawano T, Araseki M, Araki Y, Kinjo M,
Yamamoto T and Suzuki T: A small peptide sequence is sufficient for
initiating kinesin-1 activation through part of TPR region of KLC1.
Traffic. 13:834–848. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Blatch GL and Lässle M: The
tetratricopeptide repeat: A structural motif mediating
protein-protein interactions. BioEssays. 21:932–939. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Verhey KJ, Meyer D, Deehan R, Blenis J,
Schnapp BJ, Rapoport TA and Margolis B: Cargo of kinesin identified
as JIP scaffolding proteins and associated signaling molecules. J
Cell Biol. 152:959–970. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sato T, Ishikawa M, Mochizuki M, Ohta M,
Ohkura M, Nakai J, Takamatsu N and Yoshioka K: JSAP1/JIP3 and JLP
regulate kinesin-1-dependent axonal transport to prevent neuronal
degeneration. Cell Death Differ. 22:1260–1274. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jaeschke A, Czech MP and Davis RJ: An
essential role of the JIP1 scaffold protein for JNK activation in
adipose tissue. Genes Dev. 18:1976–1980. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ito M, Yoshioka K, Akechi M, Yamashita S,
Takamatsu N, Sugiyama K, Hibi M, Nakabeppu Y, Shiba T and Yamamoto
KI: JSAP1, a novel jun N-terminal protein kinase (JNK)-binding
protein that functions as a Scaffold factor in the JNK signaling
pathway. Mol Cell Biol. 19:7539–7548. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu L, Feng D, Chen G, Chen M, Zheng Q,
Song P, Ma Q, Zhu C, Wang R, Qi W, et al: Mitochondrial
outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in
mammalian cells. Nat Cell Biol. 14:177–185. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wigler M, Silverstein S, Lee LS, Pellicer
A, Cheng Y and Axel R: Transfer of purified herpes virus thymidine
kinase gene to cultured mouse cells. Cell. 11:223–232. 1977.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cyrus BF and Muller WA: A unique role for
endothelial cell kinesin light chain 1, variant 1 in leukocyte
transendothelial migration. Am J Pathol. 186:1375–1386. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wei H, Liu L and Chen Q: Selective removal
of mitochondria via mitophagy: distinct pathways for different
mitochondrial stresses. Biochim Biophys Acta. 1853:2784–2790. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Murrow L and Debnath J: Autophagy as a
stress-response and quality-control mechanism: Implications for
cell injury and human disease. Annu Rev Pathol. 8:105–137. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Glater EE, Megeath LJ, Stowers RS and
Schwarz TL: Axonal transport of mitochondria requires milton to
recruit kinesin heavy chain and is light chain independent. J Cell
Biol. 173:545–557. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lin MY and Sheng ZH: Regulation of
mitochondrial transport in neurons. Exp Cell Res. 334:35–44. 2015.
View Article : Google Scholar : PubMed/NCBI
|