Introduction
Uterine leiomyomas (ULs) are benign tumors arising
from smooth muscle cells of the uterine myometrium. ULs are
characterized by the high smooth muscle cell (SMCs) proliferation
and marked quantities of extracellular matrix proteins,
predominantly collagen type I and III (1). UL development is considered a multistep
and multi-factorial process, for which growth factors, cytokines,
and ovarian steroid hormones are key in its formation and
development (2). Different studies
have shown that UL originate from somatic mutation in myometrial
cells, which results in progressive loss of growth regulation
(3). These tumors proliferate from a
single progenitor cell and grow as abnormal clones of cells. ULs
are monoclonal, and different leiomyomas arising within one uterus
are not clonally linked (4). In
addition, different rates of growth are due to chromosomal
abnormalities within individual tumors (5). The occurrence of multiple ULs indicates a
genetic predisposition for UL formation. There is a ~2.5-fold
increased risk of developing UL in first-degree relatives of women
with these types of tumor (6).
Abnormal cell proliferation is an important step in
tumor development. There are various proteins in the cell cycle
pathway that affect cell proliferation and tumorigenesis. Among
these proteins, cyclins are particularly important in this process
(7). Cyclins are a family of proteins
that affect cell progression through the cell cycle by triggering
cyclin-dependent kinase (CDK) enzymes. Cyclin D1 (CCND1) is a key
regulatory protein that binds to CDK4 and CDK6 enzymes, and
functions during the transition from the G1 to the S phase
(8). Increased expression of the
CCND1 gene may disrupt normal cell cycle control, and prompt
cells to transition from the G1/S checkpoint of the cell
cycle resulting in tumor formation (8). Various studies have demonstrated that
overexpression of the CCND1 protein is associated with cell
proliferation and different types of cancer, including endometrial
(9), cervical (10), lung (11)
and breast (12) cancer.
Betticher et al (13) described a G to A single nucleotide
polymorphism (G870A) at the exon4/intron4 splicing region of the
CCND1 gene. The A allele has a longer half-life than the G
allele, which is hypothesized to increase CCND1 levels resulting in
the proliferation of abnormal cells and the escape of these cells
from apoptosis (13). To the best of
our knowledge, there is only one published report that has
indicated the effect of the CCND1 G870A polymorphism on UL
susceptibility in obese Korean women (14), although there are different reports
regarding the association between CCND1 gene polymorphisms, and
tumors and cancers.
Thus, the present study aimed to assess the possible
association between the CCND1 G870A polymorphism and UL in a
southeastern Iranian population.
Materials and methods
Study population
A total of 154 Iranian women (aged 37.9±8.9 years)
with UL in their pre-menopause stage who had undergone myomectomy
or hysterectomy and 197 Iranian healthy women (aged 37.5±5.5 years)
in their pre-menopause stage were enrolled in the study. All
individuals were recruited from the Department of Obstetrics and
Gynecology of Ali-ebn-Abitaleb Educational Hospital of Zahedan
University of Medical Sciences (Zahedan, Iran) from July 2012 to
October 2013. UL was confirmed pathologically in all of the women.
The control group was selected from women that were referred for
routine yearly check-ups and undergone the Pap smear test. The UL
and control groups were matched with respect to age, body mass
index (BMI), and ethnicity (Fars or Balouch). Upon sonography or
examination, the participants in the control group exhibited no
evidence of UL and their Pap smear test was negative. Women with
systemic disease and history of malignancy were excluded from the
present study. The participants provided their informed consent
prior to participating in the study and the study protocol was
approved by the Ethics Committee of Zahedan University of Medical
Sciences (Zahedan, Iran) and conducted in accordance with the
Declaration of Helsinki.
Genomic DNA extraction and
genotyping
Venous blood (2 ml) was drawn from each participant.
Genomic DNA was isolated from 200 µl EDTA-treated whole blood using
the salting out phenol chloroform method and stored at 20°C until
analysis. Polymerase chain reaction-restriction fragment length
polymorphism (PCR-RFLP) was performed for genotyping the
CCND1 G870A polymorphism.
The 167-bp fragment containing the CCND1
G870A polymorphism (rs9344) was amplified using primers as
previously described (13). The PCR
conditions were as follows: 6 min at 95°C followed by 30 cycles of
denaturation at 96°C for 30 sec, annealing at 58°C for 30 sec,
extension at 72°C for 30 sec, and a final extension at 72°C for 6
min. NciI restriction enzyme (Fermentas; Thermo Fisher
Scientific, Inc., Pittsburgh, PA, USA) was used to digest the PCR
products at 37°C overnight. The digested products were
electrophoresed on 3% agarose gel at 90 V for 80 min and visualized
by ethidium bromide staining.
In silico analysis
DNA sequences of CCND1 gene were obtained
from The National Center for Biotechnology Information (accession
no. CR536538.1). The coding sequence domain of CCND1 was translated
using the ExPASy server (http://web.expasy.org/translate/). Mode 2 of the
RNAsnp (http://rth.dk/resources/rnasnp/) was used to evaluate
the effects of G870A transition on the CCND1-mRNA structure. RNAsnp
is a server that predicts the effect of single nucleotide
polymorphisms (SNPs) on the local RNA secondary structure based on
the RNA folding algorithms (15). In
the graphic summary of the RNAsnp report, the local region, which
detects maximum structural change, is colored according to the
P-value. If P>0.2 the region is colored black, indicating that
the structural change that occurred is not significant.
Statistical analysis
All statistical analyses were conducted using SPSS
version 20 (IBM SPSS, Armonk, NY, USA). Demographic characteristics
of the groups were analyzed through independent Student's t-test or
Fisher's exact test. Logistic regression analysis assessed the
independent effect of each polymorphism risk on UL. Odds ratios
(ORs) and 95% confidence intervals (CIs) were considered for
significant allelic and genotypic associations. Using the
χ2 test, genotype frequencies of the polymorphism were
evaluated for deviation from Hardy-Weinberg equilibrium. P<0.05
was considered to indicate statistically significant
differences.
Results
Study characteristics
The demographic characteristics of the women with UL
and the control group subjects are provided in Table I. The age, BMI and parity (total number
of births after 20 weeks) were not significantly different between
the UL women and control subjects. No significant differences in
menstrual histories, including age of menarche, menstrual cycle and
duration of menses were identified among the women with UL and the
control subjects. The frequency of bleeding and pain was observed
to be more frequent in women with UL (P<0.0001).
 | Table I.Clinical and demographic
characteristics of the UL women and control group subjects. |
Table I.
Clinical and demographic
characteristics of the UL women and control group subjects.
| Parameter | UL women (n=154) | Control subjects
(n=197) | P-value |
|---|
| Age (years) | 37.9±8.9 | 37.5±5.5 | 0.625 |
| Body mass index
(kg/m2) | 25.6±5.3 | 25.3±4.6 | 0.588 |
| Parity (n) | 3.4±2.1 | 3.5±2.6 | 0.708 |
| Age of menarche
(years) | 13.4±1.3 | 13.2±1.6 | 0.123 |
| Duration of menses
(days) | 6.0±1.6 | 5.9±1.6 | 0.817 |
| Menstrual cycle
(days) | 28.6± 2.2 | 28.5±2.9 | 0.686 |
| Bleeding, n (%) | 93 (60.0) | 9 (4.5) | <0.001 |
| Pain, n (%) | 43 (28.0) | 13 (7.0) | <0.001 |
Genotypes and UL risk
The allelic and genotypic frequencies of the
CCND1 G870A polymorphism in women with UL and the control
subjects are presented in Table II.
The CCND1 G870A polymorphism conformed with Hardy-Weinberg
equilibrium (P>0.05). The frequency of GG, GA, and AA genotypes
were 20, 45.5 and 34.5%, respectively in the women with UL, and
25.4, 53.3 and 21.3%, respectively in the control group. The
frequency of the AA genotype was significantly higher in women with
UL compared with the control subjects, although the frequency of
the GA genotype was not statistically different between the two
groups before and after adjusting for age, BMI, and ethnicity
(dominant model); thus, the risk of UL was 1.4-fold greater in
women with the AA genotype when compared to the GG genotype before
and after adjusting for age, BMI, and ethnicity (OR, 1.4; 95% CI,
1.1–2; P=0.02). Furthermore, the frequency of the CCND1 870A
allele was significantly higher in the women with UL when compared
with the control subjects (57 vs. 48%; P=0.02).
 | Table II.Comparison of the genotypic and
allelic frequency of the cyclin D1 (rs9344) polymorphism in UL
women and the control group. |
Table II.
Comparison of the genotypic and
allelic frequency of the cyclin D1 (rs9344) polymorphism in UL
women and the control group.
| Polymorphism | Control subjects
(n=197) | UL women (n=154) | P-value | OR (95% CI) | P-valuea | OR (95%
CI)a |
|---|
| Cyclin D1
(rs9344) |
|
|
|
|
|
|
| GG, n (%) | 50 (25.4) | 31 (20) | − | 1 | − | 1 |
| GA, n (%) | 105 (53.3) | 70 (45.5) | 0.79 | 0.9 (0.5−1.6) | 0.89 | 1 (0.6–1.7) |
| AA, n (%) | 42 (21.3) | 53 (34.5) | 0.02 | 1.4 (1.1–1.9) | 0.02 | 1.4 (1.1–2) |
| Allele |
|
|
|
|
|
|
| G, n (%) | 205 (52) | 132 (43) | − | 1 | − | − |
| A, n (%) | 189 (48) | 176 (57) | 0.02 | 1.5 (1.1–2) | − | − |
Structural analysis
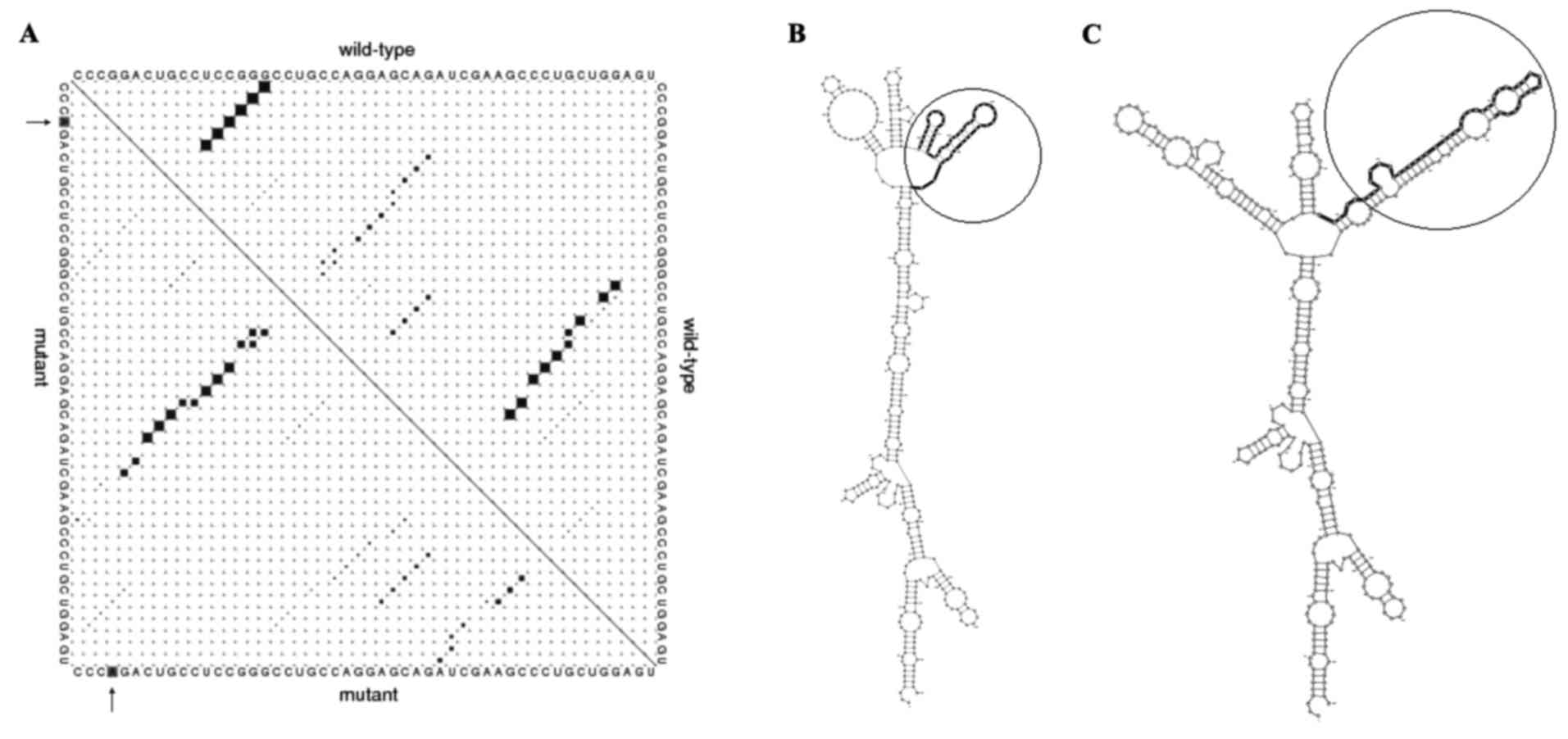
In silico analysis indicated that rs9344
(G>A) polymorphism is located at positions 950, 723 and 241 of
the transcript, CDS and CCND1 protein, respectively, which alters
the CCG codon to CCA. Since these two codons translated to proline,
this mutation does not change the protein features of CCND1,
including molecular weight, isoelectric point and physicochemical
properties (data not shown). Predicting 870AA effects on the local
RNA secondary structure of CCND1 revealed that the SNPs cause
fundamental changes to the secondary structure of mRNA (distance,
0.1922; P=0.1488; P<0.2 indicates significant structural change)
(Fig. 1). The base pair probabilities
of the local region detected with maximum differences locate
between base pairs 523 and 888 (Fig.
1A). Furthermore, the optimal secondary structure of global
wild-type and mutant sequences are presented in Fig. 1B and C.
Discussion
In the current study, a higher frequency of the
CCND1 870AA genotype was identified in women with UL when
compared to the GG genotype, thus, CCND1 870AA homozygous
polymorphism may increase the risk of UL.
CCND1 is an important protein in regulation of the
cell cycle, causing the transition from the G1 to the S
phase of the cell cycle. CCND1 regulates cell cycle progression by
activating CDK4 and CDK6 (7,8) and the activated enzymes phosphorylate the
retinoblastoma (Rb) protein. The phosphorylation of Rb releases the
transcriptional factor, E2F, which subsequently triggers numerous
downstream genes that are necessary for the cell cycle (16).
Altered expression levels of cell cycle regulatory
proteins, such as CCND1, are associated with cell proliferation and
are present in different types of human tumor and cancer, including
endometrial (9,17) and cervical (18) cancer, and endometriosis and adenomyosis
(19). An elevated expression level of
the CCND1 protein was observed in leiomyomas compared with the
adjacent myometrium during all menstrual cycle phases in a study by
Kovács et al (20). There are
several genetic variants in cell cycle regulatory proteins, which
affect protein expression and prevent or stimulate cell
proliferation, increasing the susceptibility to tumor development.
Hence, molecular epidemiologic studies are considered to be useful
strategies for investigating the association between genetic
variations and tumorigenesis (21).
In 1995, Betticher et al (13) described a G870 to A SNP at codon 242 on
exon4 in the CCND1 gene, which affected splicing of the
CCND1 transcript. CCND1 mRNA exhibits different splicing, in which
translation of the transcripts produces various proteins with
dissimilar c-terminal domains. The CCND1 G870A polymorphism
may control splicing of the transcripts, and the 870A allele
generates the truncated transcript, encoding a protein with a
longer half-life; however, the G870 allele produces the full
transcript (13). Therefore, different
studies have considered the CCND1 870A allele as a
susceptible variant in tumorigenesis. Although, there are numerous
reports on the correlation between the CCND1 G870A
polymorphism and different types of cancer (22,23), to the
best of our knowledge, there is only one published study regarding
the association between the CCND1 G870A polymorphism and UL
susceptibility in obese Korean women (14). In 2008, Han et al (14) demonstrated that the AA genotype and A
allele of the CCND1 G870A polymorphism may increase UL risk
in Korean women with a BMI >25 kg/m (2), confirming the results of the current
study.
Although G870A transition does not change amino acid
substitution in the CCND1 protein, this SNP may alter the
CCND1-mRNA structure and function. The mRNAs containing different
nucleotides at an SNP position may differ in their interactions
with cellular molecules involved in mRNA transport, maturation,
translation or degradation (24).
Analysis of the effects of G870A transition on the secondary
structure of mRNA indicated that this SNP may alter the structure
and minimum free energy of mRNA. Therefore, G870A may alter
CCND1 gene expression levels. Although the current results
of in silico analysis improve our understanding of the
effects of G870A on the CCND1 mRNA, further studies are required to
evaluate the influence of this SNP on UL susceptibility.
There were certain limitations of the current study.
As the conclusion is based on a small number of patients and
control subjects, the statistical power is not considered to be
particularly high. In addition, the study would be more valuable if
myomatous and intact adjacent tissue samples had been investigated.
Finally, as there are different ethnic groups in Iran, further
studies are required with greater sample sizes from each ethnic
group to confirm or refute our findings.
In conclusion, the findings of the present study
revealed that the CCND1 870AA genotype and CCND1 870A
allele were associated with a predisposition for UL, and that this
polymorphism may be a risk factor for UL. Further studies with
greater sample sizes from different populations are required to
validate the current findings.
Acknowledgements
The present study was extracted from an MS thesis
(registered no. 6169) at Zahedan University of Medical Sciences
(Zahedan, Iran). The authors would like to thank the Deputy of
Research Affairs at Zahedan University of Medical Sciences for
funding this project.
References
|
1
|
Stewart EA, Friedman AJ, Peck K and Nowak
RA: Relative overexpression of collagen type I and collagen type
III messenger ribonucleic acids by uterine leiomyomas during the
proliferative phase of the menstrual cycle. J Clin Endocrinol
Metab. 79:900–906. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Barbarisi A, Petillo O, Di Lieto A, Melone
MA, Margarucci S, Cannas M and Peluso G: 17-beta estradiol elicits
an autocrine leiomyoma cell proliferation: Evidence for a
stimulation of protein kinase-dependent pathway. J Cell Physiol.
186:414–424. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jeon YT, Kim JW, Park NH, Song YS, Kang SB
and Lee HP: DNA repair gene XRCC1 Arg399Gln polymorphism is
associated with increased risk of uterine leiomyoma. Hum Reprod.
20:1586–1589. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gittenberger-de Groot AC, DeRuiter MC,
Bergwerff M and Poelmann RE: Smooth muscle cell origin and its
relation to heterogeneity in development and disease. Arterioscler
Thromb Vasc Biol. 19:1589–1594. 1999. View Article : Google Scholar
|
|
5
|
Medikare V, Kandukuri LR, Ananthapur V,
Deenadayal M and Nallari P: The genetic bases of uterine fibroids;
a review. J Reprod Infertil. 12:181–191. 2011.PubMed/NCBI
|
|
6
|
Vikhlyaeva EM, Khodzhaeva ZS and
Fantschenko ND: Familial predisposition to uterine leiomyomas. Int
J Gynaecol Obstet. 51:127–131. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Galderisi U, Jori FP and Giordano A: Cell
cycle regulation and neural differentiation. Oncogene.
22:5208–5219. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sherr CJ: Cancer cell cycles. Science.
274:1672–1677. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shevra CR, Ghosh A and Kumar M: Cyclin D1
and Ki-67 expression in normal, hyperplastic and neoplastic
endometrium. J Postgrad Med. 61:15–20. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bae DS, Cho SB, Kim YJ, Whang JD, Song SY,
Park CS, Kim DS and Lee JH: Aberrant expression of cyclin D1 is
associated with poor prognosis in early stage cervical cancer of
the uterus. Gynecol Oncol. 81:341–347. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ayed A and Adesina A: Prognostic
significance of cyclin D1 expression in resected stage I, II
non-small cell lung cancer in Arabs. Interact Cardiovasc Thorac
Surg. 5:47–51. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Abramson VG, Troxel AB, Feldman M, Mies C,
Wang Y, Sherman L, McNally S, Diehl A and Demichele A: Cyclin D1b
in human breast carcinoma and coexpression with cyclin D1a is
associated with poor outcome. Anticancer Res. 30:1279–1285.
2010.PubMed/NCBI
|
|
13
|
Betticher DC, Thatcher N, Altermatt HJ,
Hoban P, Ryder WD and Heighway J: Alternate splicing produces a
novel cyclin D1 transcript. Oncogene. 11:1005–1011. 1995.PubMed/NCBI
|
|
14
|
Han SS, No JH, Jeon YT, Kim JW, Park NH,
Song YS, Kang SB and Lee HP: Association of cyclin D1 G870A
polymorphism with uterine leiomyoma in women whose body mass index
values are above 25 kg/m2. Hum Reprod. 23:525–529. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sabarinathan R, Tafer H, Seemann SE,
Hofacker IL, Stadler PF and Gorodkin J: The RNAsnp web server:
Predicting SNP effects on local RNA secondary structure. Nucleic
Acids Res. 41:W475–W479. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mallya SM and Arnold A: Cyclin D1 in
parathyroid disease. Front Biosci. 5:D367–D371. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Catalano S, Giordano C, Rizza P, Gu G,
Barone I, Bonofiglio D, Giordano F, Malivindi R, Gaccione D,
Lanzino M, et al: Evidence that leptin through STAT and CREB
signaling enhances cyclin D1 expression and promotes human
endometrial cancer proliferation. J Cell Physiol. 218:490–500.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li HY, Xu Q, Zhu T, Zhou JH, Deng DR, Wang
SX, Lu YP and Ma D: Expression and clinical significance of Pin1
and Cyclin D1 in cervical cancer cell lines and cervical epithelial
tissues. Ai Zheng. 25:367–372. 2006.(In Chinese). PubMed/NCBI
|
|
19
|
Goumenou AG, Matalliotakis IM, Tzardi M,
Fragouli IG, Mahutte NG and Arici A: p16, retinoblastoma (pRb), and
cyclin D1 protein expression in human endometriotic and adenomyotic
lesions. Fertil Steril 85 (Suppl 1). 1204–1207. 2006. View Article : Google Scholar
|
|
20
|
Kovács KA, Oszter A, Göcze PM, Környei JL
and Szabó I: Comparative analysis of cyclin D1 and oestrogen
receptor (alpha and beta) levels in human leiomyoma and adjacent
myometrium. Mol Hum Reprod. 7:1085–1091. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Velez AM Abreu and Howard MS:
Tumor-suppressor Genes, Cell Cycle Regulatory Checkpoints, and the
Skin. N Am J Med Sci. 7:176–188. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dai X, Zhang X, Wang B, Wang C, Jiang J
and Wu C: Association Between Polymorphism rs678653 in Human Cyclin
D1 Gene (CCND1) and Susceptibility to Cancer: A Meta-Analysis. Med
Sci Monit. 22:863–874. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang M, Zhu H, Hu T, Liu S and Wang H:
Association of CCND1 gene polymorphism with cervical cancer
susceptibility in Caucasian population: A meta-analysis. Int J Clin
Exp Med. 8:12983–12988. 2015.PubMed/NCBI
|
|
24
|
Shen LX, Basilion JP and Stanton VP Jr:
Single-nucleotide polymorphisms can cause different structural
folds of mRNA. Proc Natl Acad Sci USA. 96:7871–7876. 1999.
View Article : Google Scholar : PubMed/NCBI
|