Introduction
Colorectal cancer (CRC) remains one of the leading
cancers worldwide. According to the newly issued cancer statistics
in China, between 2000 and 2011, CRC ranked as the third and fifth
cause of mortality among females and males, respectively (1). In clinic, the management of CRC at an
early stage continues to be the main approach to reduce mortality
and morbidity (2). Nevertheless, early
and accurate diagnosis have posed a challenge for the diagnosis of
CRC due to a lack of high sensitivity and specific tumor markers or
detection techniques in clinic (3).
Epigenetic modifications such as gene or DNA
methylation in tumors have placed increased attention on the early
identification of CRC. Among the methylated genes in CRC, septin-9
has been highlighted as an ideal candidate biomarker (4–19). Septin-9
was initially identified in myeloid neoplasia (20). It acts as a suppressor gene in numerous
cancer types (21,22). In previous years, an increasing number
of studies have focused on the investigation of plasma methylated
septin-9 for the detection of CRC (4–17).
Consequently, several types of assay kits regarding methylated
septin-9 are being developed and have become commercial products.
Nevertheless, results from single studies are often inconsistent
due to limited sample size and single-center design. For instance,
some studies reported that septin-9 testing presented a limited
sensitivity from 51 to 56% (14,15). By
constrast, some research presented an estimated sensitivity of
septin-9 methylation up to 90% (6).
In the present study, we conducted a comprehensive
meta-analysis and assessed the overall diagnostic efficacy of
plasma methylated septin-9 for CRC detection.
Materials and methods
Search strategy
Two reviewers independently searched the published
articles through the online PubMed/Medline, BioMed Central and CNKI
databases up to January 31st, 2017. The search terms were utilized
in a single or parallel pattern as (‘septin-9’ or ‘septin 9’) and
(‘colon cancer’ or ‘colorectal neoplasm’ or ‘CRC’ or ‘colorectal
carcinoma’ or ‘carcinoma of colon’) and/or (‘sensitivity’ or
‘specificity’ or ‘diagnosis’ or ‘accuracy’ or ‘ROC’ or ‘AUC’. We
also manually searched the references in the studies for
retrieval.
Eligibility criteria
Inclusion criteria for the meta-analysis were: i)
Studies estimated the diagnostic feature of plasma septin-9 for
CRC; ii) studies with sufficient information to generate the
statistical elements as true positive, true negative, false
positive and false negative; and iii) the control sources were from
cancer-free individuals.
Studies were excluded based on the following
criteria: i) studies with unclear definition of the control sources
or the paired controls were other types of tumors; ii) extracted
data were insufficient to establish the 2×2 table; and iii) basic
studies, review articles, letters and conference articles.
Data extraction
Essential data from enrolled articles were extracted
twice by two authors and the information included: The name of the
author, publication data, study population, sample numbers, control
types, measure method, cut-off value, sensitivity, and specificity.
The two-stage study comprised both training and validation cohorts,
and the data from each group were considered to be independent and
were meta-analyzed as individual studies. Any disagreements on data
extraction were solved by group discussion.
Study bias assessment
Study bias of the included articles was judged
according to the Quality Assessment of Studies of Diagnostic
Accuracy included in the Systematic Reviews (QUADAS) II checklist
(23), wherein, the concerns for risk
of bias and applicability were rated as ‘low’, ‘high’ or ‘unclear’,
with an evaluation score of ‘1’, ‘0’ and ‘0’, respectively.
Statistical analyses
The quantitative meta-analyses were carried out
based on Stata 12.0 (StataCorp LP, College Station, TX, USA) and
Meta-disc 1.4 (XI Cochrane Colloquium, Barcelona, Spain) programs
using a bivariate quantitative model. Heterogeneity among the
studies was estimated utilizing Spearman's correlation coefficient,
I-squared (I2) and Chi-squared (χ2) Q tests.
Either P<0.05 or I2>50% were deemed as
statistically different (24). If
significant heterogeneity existed among studies, a random-effect
model was employed for the aggregation of the effect sizes;
otherwise, a fixed-effect model was used (25). Potential sources of heterogeneity were
traced by influence analysis and meta-regression test. Bias of the
publications was determined using Deek's funnel plot asymmetry
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Article filtration and quality
assessment
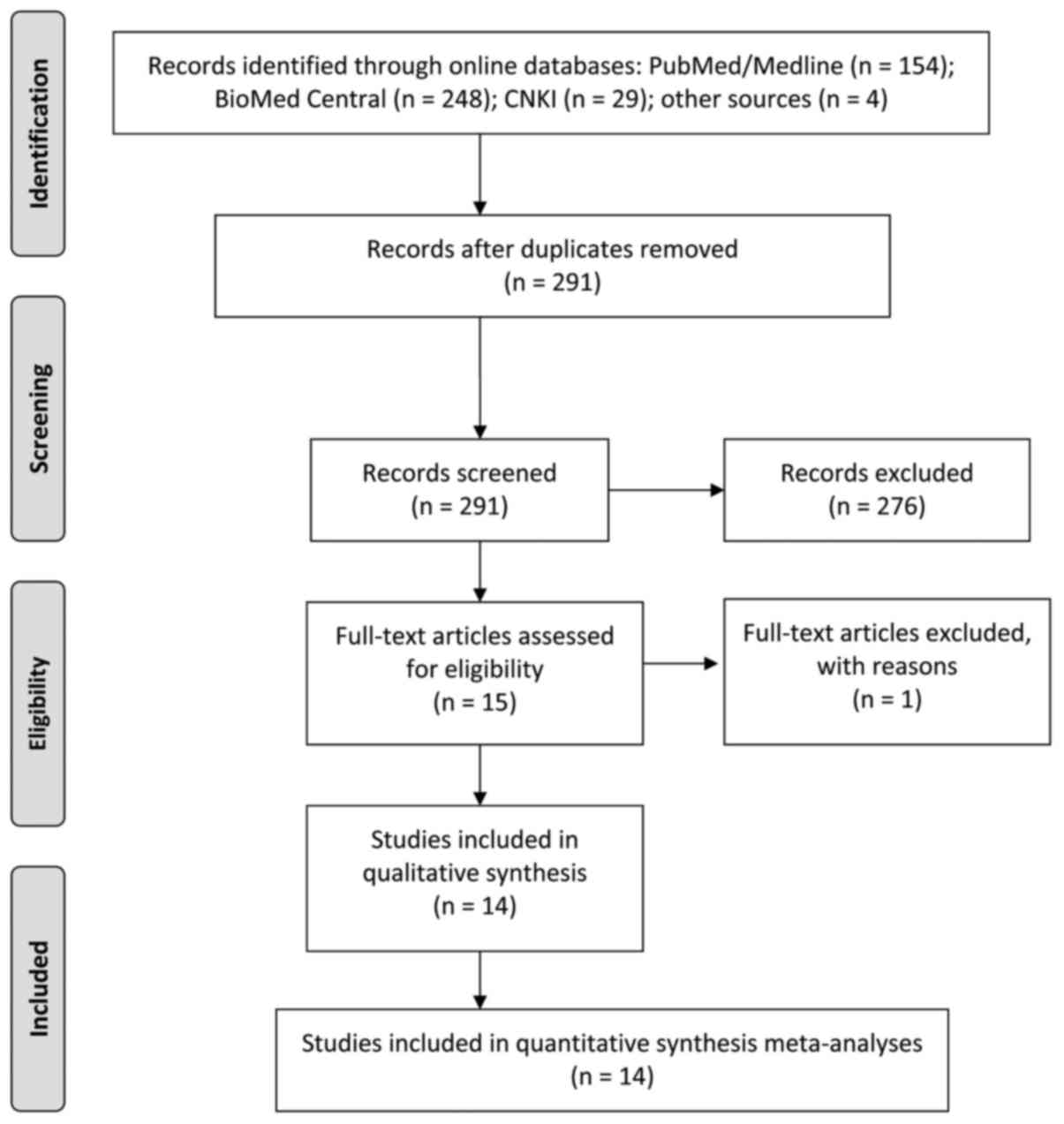
Fig. 1 shows the
procedure of article filtration based on the PRISMA statement: In
total, 291 citations were obtained from the online databases
according to the specified criteria, of which 276 records were
excluded after a careful review of the title and abstracts. The
following 15 eligible studies then received full text evaluation
and 1 of them was eliminated due to lack of relevance (26). Finally, 14 cohorts comprising 23 single
studies were enrolled for the meta-analysis (4–17).
The study quality of the included cohorts was
assessed by the QUADAS II tool (23).
Fig. 2 plots the proportions of
studies with low, high, or unclear concerns regarding risk of bias
and applicability and the included studies showed low risk of
bias.
Study features
In the current study, 1,462 CRC patients and 4,703
CRC-free subjects were enrolled for the final meta-analyses. The
CRC patients enrolled had not previously undergone treatment and
the final diagnoses were confirmed histologically via the surgical
specimen. The control sources comprised healthy participants,
hyperplastic polyps and individuals with normal colonoscopy
examinations. The patient size varied from 30 to 252 and the
control size varied from 9 to 1,500. Blood samples were collected
and tested in plasma and the methylated status of septin-9 was
examined via the qPCR. The reference genes involved,
β-actin, CFF1 and HB14 were employed for
determining the validity of the results. Study ethnicity comprised
Asian, American and European. The main characteristics of included
studies are summarized in Table I.
 | Table I.Main features of the included studies
for plasma septin-9 methylation in the identification of CRC. |
Table I.
Main features of the included studies
for plasma septin-9 methylation in the identification of CRC.
| Author | Year | Area | Patient vs.
controls | Patient/control
size | Sample type | Test method | Reference gene | Cut-off value | (Refs.) |
|---|
| Potter et
al | 2014 | Germany | CRC vs. | 44/1500 | Plasma | RT-qPCR | β-actin | CP<45.0 | (4) |
|
|
|
| Non-CRC |
|
|
|
|
|
|
| Lee et al | 2013 | Korea | CRC vs. Normal | 101/96 | Plasma | RT-qPCR | β-actin | 1/3 algorithms | (5) |
| Tóth et
al | 2012 |
Hungary | CRC vs. Normal | 93/94 | Plasma | RT-qPCR | β-actin | CP<40.5 | (6) |
| Warren et
al | 2011 | America | CRC vs. Non-CRC | 50/94 | Plasma | RT-qPCR | β-actin | CP<39.0 | (7) |
| Ahlquist et
al | 2012 | America | CRC vs. Non-CRC | 52/48 | Plasma | RT-qPCR | β-actin | Unclear | (8) |
| Grützmann et
al | 2008 | America | CRC vs. Non-CRC | 252/183 | Plasma | RT-qPCR | CFF1 and HB14 | 2/3 algorithms | (9) |
| Chen et
al | 2016 | Taiwan | CRC vs. Normal | Sep-51 | Plasma | RT-qPCR | β-actin | Unclear | (10) |
| He et
al | 2010 | China | CRC vs. Normal | 182/170 | Plasma | RT-qPCR | β-actin | Unclear | (11) |
| Jin et
al | 2015 | China | CRC vs.
Non-CRC | 135/341 | Plasma | RT-qPCR | Unclear | Unclear | (12) |
| Johnson et
al | 2014 | America | CRC vs.
Non-CRC | 101/200 | Plasma | RT-qPCR | Unclear | Unclear | (13) |
| Church et
al | 2013 | America | CRC vs.
Non-CRC. | 53/1457 | Plasma | RT-qPCR | β-actin | CP<50.0 1/3 and
2/3 | (14) |
| deVos et
al | 2009 | America | CRC vs Non-CRC | Training: 97/172
Test: 90/155 | Plasma | RT-qPCR | β-actin | algorithms | (15) |
| Tänzer et
al | 2010 | Germany | CRC vs.
Non-CRC | 33/34 | Plasma | RT-qPCR | Unclear | 1/3 and 2/3
algorithms | (16) |
| Ørntoft et
al | 2015 | Denmark | CRC vs.
Non-CRC | 150/150 | Plasma | RT-qPCR | β-actin | 1/3 and 2/3
algorithms | (17) |
Heterogeneity
Heterogeneity underlying eligible studies was
mirrored by the examination of threshold and non-threshold effects
among studies (25,27). As shown in Table II, Spearman's correlation coefficient
of the overall effect size revealed a P>0.05, suggesting that no
significant heterogeneity generated from the threshold effect.
Moreover, I2 and χ2-based Q tests were
conducted to evaluate heterogeneity caused by the non-threshold
effect. As the data indicated, either the overall pooled analysis
(Q=111.58, P<0.01, I2=77.6%) or the stratified
analyses (Table II) presented
significant heterogeneity from the threshold effect. As a result,
we selected a random-effect model for the aggregation of the effect
sizes.
 | Table II.Exploration of study heterogeneity
using Meta-disc 1.4 software. |
Table II.
Exploration of study heterogeneity
using Meta-disc 1.4 software.
|
|
|
|
| Heterogeneity
sources |
|---|
|
|
|
|
|
|
|---|
| Tests | Spearman's
correlation coefficient | Cochran's Q
test | I2 test
(%) | Threshold
effect | Non-threshold
effect |
|---|
| Overall | 0.065a | 111.58b | 77.6 | No | Yes |
|
| P=0.745 | P<0.01 |
|
|
|
| Outliers
elimination | 0.235a | 92.18b | 77.2 | No | Yes |
|
| P=0.291 | P<0.01 |
|
|
|
| Ethnicity |
|
|
|
|
|
|
Asian | −0.200a | 19.70b | 84.8 | No | Yes |
|
| P=0.800 | P=0.0002 |
|
|
|
|
American | 0.574a | 41.40b | 63.8 | No | Yes |
|
| P=0.800 | P=0.0003 |
|
|
|
|
European | −0.886a | 43.15b | 88.9 | Yes | Yes |
|
| P=0.019 | P<0.01 |
|
|
|
Pooled diagnostic performance
For efficacy when differentiating CRC patients from
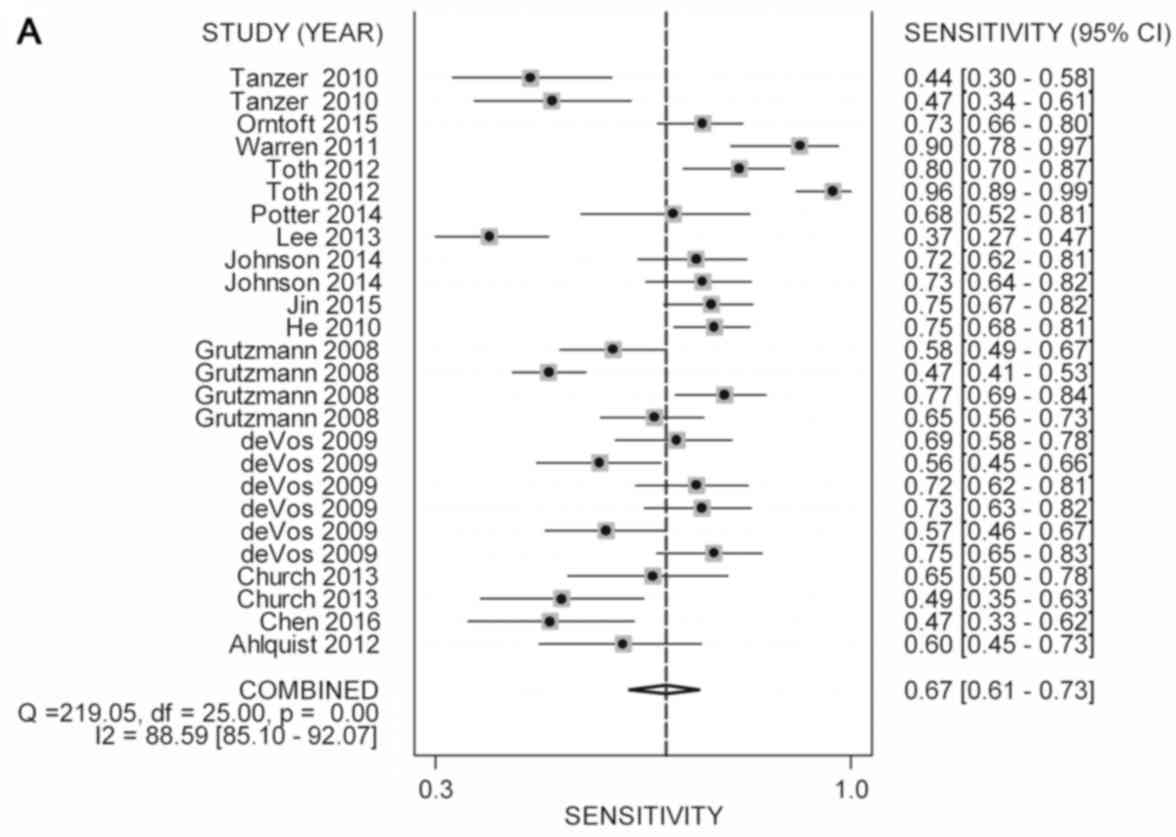
non-CRC ones, methylated septin-9 in plasma retained a combined
sensitivity of 0.67 (95% CI, 0.61–0.73) and specificity of 0.89
(95% CI, 0.86–0.92) (Fig. 3A and B and
Table III). Moreover, the pooled
positive likelihood ratio (PLR), negative likelihood ratio (NLR),
diagnostic odds ratio (DOR) and AUC were 6.26 (95% CI, 4.76–8.22),
0.37 (95% CI, 0.31–0.44), 16.93 (95% CI, 11.56–24.77) and 0.87,
respectively (Fig. 3C and Table III).
 | Table III.The pooled analyses of diagnostic
efficacy of methylated septin-9 in confirming colorectal
cancer. |
Table III.
The pooled analyses of diagnostic
efficacy of methylated septin-9 in confirming colorectal
cancer.
| Analysis | Pooled sensitivity
(95% CI) | Pooled specificity
(95% CI) | Pooled PLR (95%
CI) | Pooled NLR (95%
CI) | Pooled DOR (95%
CI) | AUC |
|---|
|
| (95%
CI) | (95% CI) | (95% CI) | (95% CI) | (95% CI) |
|
| Overall | 0.67 | 0.89 | 6.26 | 0.37 | 16.93 | 0.87 |
|
| (0.61–0.73) | (0.86–0.92) | (4.76–8.22) | (0.31–0.44) | (11.56–24.77) |
|
| Outliers
excluded | 0.70 | 0.90 | 6.68 | 0.33 | 20.1 | 0.88 |
|
| (0.64–0.76) | (0.86–0.92) | (5.08–8.77) | (0.27–0.40) | (13.92–29.04) |
|
| Ethnicity |
|
|
|
|
|
|
|
Asian | 0.64 | 0.90 | 6.89 | 0.42 | 17.23 | 0.94 |
|
| (0.59–0.68) | (0.88–0.92) | (3.42–13.89) | (0.22–0.80) | (5.68–52.27) |
|
|
American | 0.64 | 0.89 | 5.99 | 0.38 | 17.18 | 0.85 |
|
| (0.62–0.67) | (0.88–0.90) | (4.72–7.61) | (0.33–0.45) | (12.78–23.10) |
|
|
European | 0.72 | 0.80 | 3.75 | 0.33 | 14.1 | 0.85 |
|
| (0.68–0.76) | (0.78–0.82) | (2.05–6.85) | (0.18–0.61) | (4.17–47.72) |
|
Subgroup analyses
In the stratified studies analyzed according to
ethnicity, the data demonstrated that European-based septin-9
methylation test had the optimal sensitivity of 0.72 (95% CI,
0.68–0.76), whereas the Asian-based analysis achieved the highest
specificity of 0.90 (95% CI, 0.88–0.92) (Table III). In addition, testing of septin-9
in Americans conferred a sensitivity of 0.64 (95% CI, 0.62–0.67) at
a specificity of 0.89 (95% CI, 0.88–0.90), corresponding to an AUC
of 0.85, PLR of 5.99 (95% CI, 4.72–7.61), NLR of 0.38 (95% CI,
0.33–0.45) and DOR of 17.18 (95% CI, 12.78–23.10) (Table III).
Influence analysis and
meta-regression
Influence analysis and meta-regression test were
applied to deeply analyze the characteristics of included studies
to better explain the sources of heterogeneity. As indicated by
Fig. 4, four individual studies were
evaluated as outliers. We further adjusted the analyses by removing
the outlier studies. Accordingly, the combined effects size of
sensitivity elevated from 0.67 to 0.70, NLR was reduced from 0.37
to 0.33 and I2 increased from 77.6 to 77.2%. On the
other hand, the univariate meta-regression test was applied to
further trace the underlying sources of study heterogeneity. We
conducted the test relying on six predefined covariates: Study
ethnicity (Asian vs. American vs. European), CRC cases (<100 vs.
≥100), control size (control <100 vs. control ≥100), reference
gene (β-actin vs. other), test algorithms (1/3 vs 2/3 algorithm)
and article quality (QUADAS scores) (28). However, all these factors showed a low
likelihood of causes of heterogeneities (Table IV).
 | Table IV.Exploration of the potential sources
of heterogeneity by the meta-regression test. |
Table IV.
Exploration of the potential sources
of heterogeneity by the meta-regression test.
| Study
characteristic | P-value | RDOR (95% CI) |
|---|
| Study ethnicity
(Asian vs. American vs. European) |
0.0972 | 1.70
(0.90–3.22) |
| CRC cases (<100
vs. ≥100) |
0.6843 | 0.82
(0.29–2.29) |
| Control size
(control <100 vs. control ≥100) |
0.2159 | 1.85
(0.68–5.02) |
| Reference gene
(β-actin vs. other) | 0.887 | 1.09
(0.31–3.79) |
| Study quality
(QUADAS scores) |
0.7141 | 0.88
(0.43–1.81) |
| Test algorithms
(1/3 vs. 2/3 algorithm) |
0.1603 | 0.72
(0.45–1.15) |
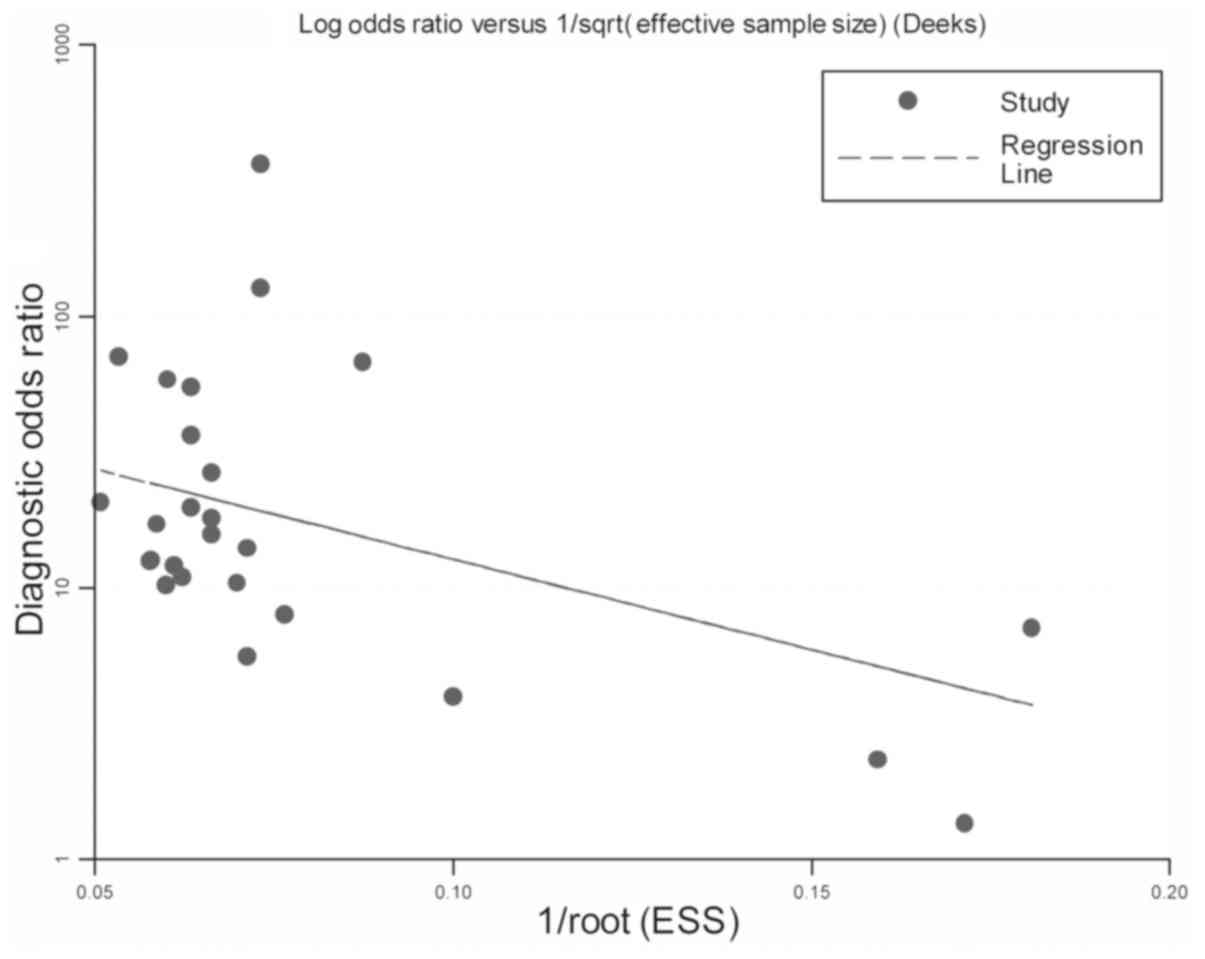
Publication bias
Risk of bias among publications was examined by the
Deeks' funnel plot asymmetry test among the included studies. In
Fig. 5, the slope coefficient of the
regression line exhibited a P-value of 0.193, suggesting that no
clear bias existed among the eligible studies due to
publication.
Discussion
Methylated septin-9 has recently been developed as a
novel, non-invasive biomarker for CRC diagnosis (2,3). The
technique has developed rapidly for the testing of plasmic septin-9
methylation and several kinds of assay kits have been developed and
become commercial products (22). The
current meta-analysis assessed the diagnostic utility of plasma
methylated septin-9 as a serological marker for the identification
of CRC.
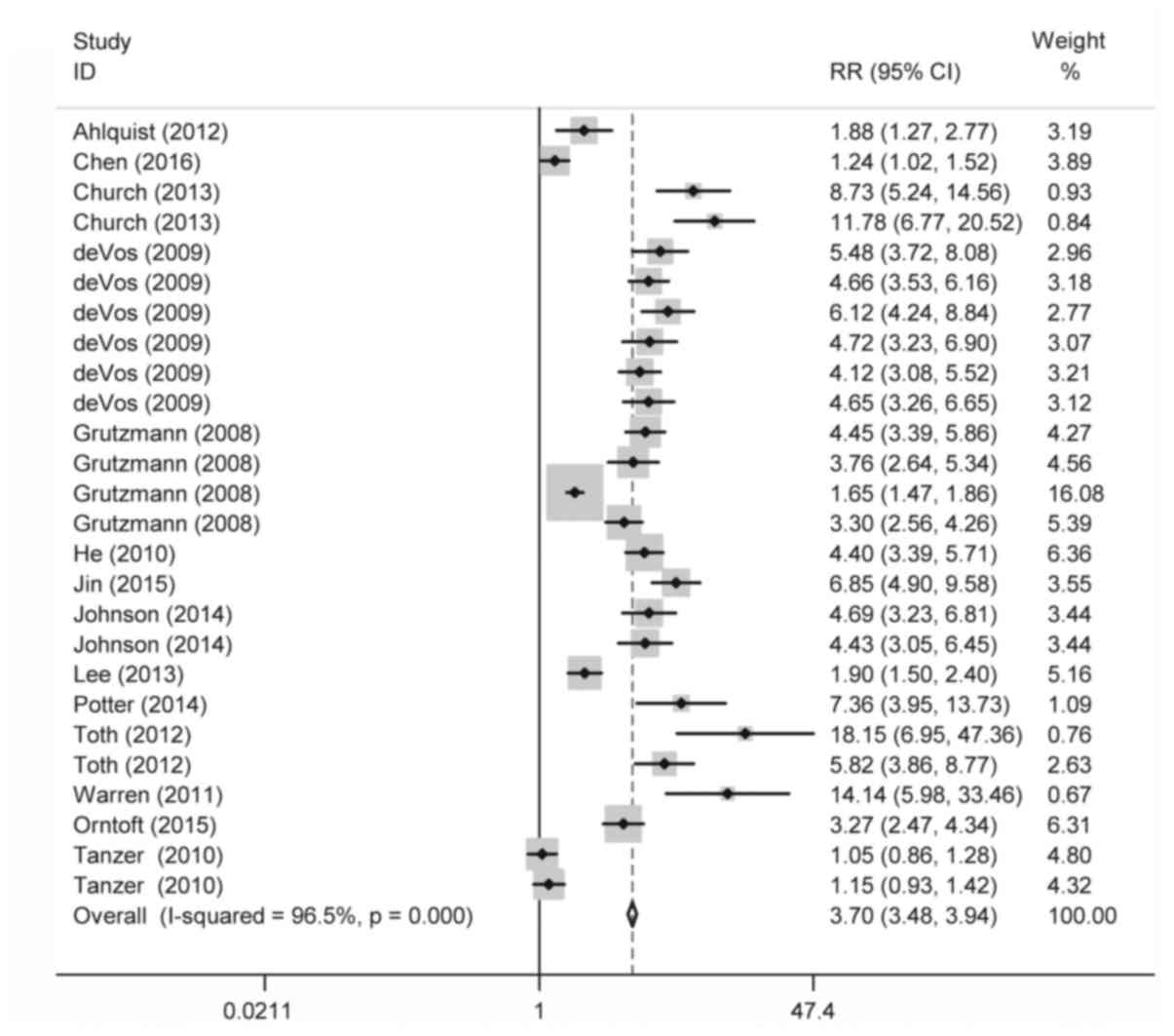
Our analysis revealed that the overall testing of
methylated septin-9 in plasma retained a relatively low sensitivity
of 0.67, but maintained a high specificity of 0.89. In the SROC
curve analysis, the combined AUC was estimated to be 0.87,
revealing a relatively high efficacy for the septin-9 methylation
testing in the diagnosis of CRC. The DOR is also recommended as an
important indicator in mirroring the overall diagnostic performance
(24). In the present study, the DOR
of plasmic septin-9 methylation test was shown to be 16.93,
suggesting a powerful discriminatory performance in confirming CRC.
Moreover, a pooled PLR of 6.26 means that testing of methylated
septin-9 in plasma reached a ratio of 6.26 between the true and
false-positive rate. For the false-negative rate reflected by NLR
(25), the value was estimated to be
0.37, which is not low enough to eliminate CRC. In general, our
data have shown that analysis of methylated septin-9 in plasma
achieved an overall high efficacy and is acceptable as a routine
biomarker for CRC detection.
Recent evidence has substantiated an independent
association between ethnicity and DNA methylation status (29). We therefore further conducted a
stratified analysis according to ethnicity. Our findings revealed
that testing of septin-9 methylation in Asians and Europeans
achieved a better efficacy than in Americans. Nevertheless, our
analysis stratified by ethnicity yielded a small study size and
exhibited high heterogeneity. Thus, more investigations are
required to confirm our findings.
On the other hand, we observed a large degree of
heterogeneity in our meta-analyses. Heterogeneity can be
interpreted by both threshold and non-threshold effects (25,27). The
threshold effect can be caused by different cut-off value settings
as well as objective methods. In the present study, the P-values
from Spearman's correlation coefficient were >0.05, suggesting
that there was no clear heterogeneity from the threshold effect.
Nevertheless, significant heterogeneity from the non-threshold
effect seemed to be present in all the meta-analyses as well as in
the subgroup analyses. The causes of heterogeneity from the
non-threshold effect can be interpreted by the different disease
conditions and other concomitant diseases among the participants.
In addition, different test conditions involved in the detection
technique, operators as well as standard tests also constitute
major causes of non-threshold effect (25). In the present study, we hypothesized
that the outlier studies, ethnicity, case or control size,
reference gene, cut-off value setting and study quality may
contribute to the sources of heterogeneity. As a result, we first
conducted influence analysis and identified four outlier studies.
After removing the outliers, the pooled sensitivity was elevated
from 0.67 to 0.70, NLR was reduced from 0.37 to 0.33, but
I2 was elevated from 77.6 to 77.2%. Accordingly, we
further conducted a meta-regression test and found that variations
such as ethnicity, case or control size, reference gene, test
algorithms and article quality were not likely to be sources of
heterogeneity among eligible studies.
Our analysis has several limitations. Firstly, a
large number of heterogeneities were observed in our analyses,
which compromised the pooled accuracy of the effect sizes.
Secondly, the sample numbers in the subgroup studies were small and
the findings require verification. Finally, the control types were
complicated and thus the meta-analyzed data may not completely
mirror the real diagnostic efficacy of plasma septin-9 methylation
for CRC detection.
Despite these limitations, plasma methylated
septin-9 testing showed a relatively high accuracy and may be
optimal for CRC diagnosis. Further investigations are required to
confirm the preliminary evidence from the present study.
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kolligs FT: Diagnostics and epidemiology
of colorectal cancer. Visc Med. 32:158–164. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nguyen MT and Weinberg DS: Biomarkers in
colorectal cancer screening. J Natl Compr Canc Netw. 14:1033–1040.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Potter NT, Hurban P, White MN, Whitlock
KD, Lofton-Day CE, Tetzner R, Koenig T, Quigley NB and Weiss G:
Validation of a real-time PCR-based qualitative assay for the
detection of methylated SEPT9 DNA in human plasma. Clin Chem.
60:1183–1191. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lee HS, Hwang SM, Kim TS, Kim DW, Park DJ,
Kang SB, Kim HH and Park KU: Circulating methylated septin 9
nucleic acid in the plasma of patients with gastrointestinal cancer
in the stomach and colon. Transl Oncol. 6:290–296. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tóth K, Sipos F, Kalmár A, Patai AV,
Wichmann B, Stoehr R, Golcher H, Schellerer V, Tulassay Z and
Molnár B: Detection of methylated SEPT9 in plasma is a reliable
screening method for both left-and right-sided colon cancers. PLoS
One. 7:e460002012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Warren JD, Xiong W, Bunker AM, Vaughn CP,
Furtado LV, Roberts WL, Fang JC, Samowitz WS and Heichman KA:
Septin 9 methylated DNA is a sensitive and specific blood test for
colorectal cancer. BMC Med. 9:1332011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ahlquist DA, Taylor WR, Mahoney DW, Zou H,
Domanico M, Thibodeau SN, Boardman LA, Berger BM and Lidgard GP:
The stool DNA test is more accurate than the plasma septin-9 test
in detecting colorectal neoplasia. Clin Gastroenterol Hepatol.
10:272–277. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Grützmann R, Molnar B, Pilarsky C,
Habermann JK, Schlag PM, Saeger HD, Miehlke S, Stolz T, Model F,
Roblick UJ, et al: Sensitive detection of colorectal cancer in
peripheral blood by septin 9 DNA methylation assay. PLoS One.
3:e37592008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen CH, Yan SL, Yang TH, Chen SF, Yeh YH,
Ou JJ, Lin CH, Lee YT and Chen CH: The relationship between the
methylated septin-9 DNA blood test and stool occult blood test for
diagnosing colorectal cancer in Taiwanese people. J Clin Lab Anal.
Jul 8–2016.(Epub ahead of print).
|
|
11
|
He Q, Chen HY, Bai EQ, Luo YX, Fu RJ, He
YS, Jiang J and Wang HQ: Development of a multiplex MethyLight
assay for the detection of multigene methylation in human
colorectal cancer. Cancer Genet Cytogenet. 202:1–10. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jin P, Kang Q, Wang X, Yang L, Yu Y, Li N,
He YQ, Han X, Hang J, Zhang J, et al: Performance of a
second-generation methylated SEPT9 test in detecting colorectal
neoplasm. J Gastroenterol Hepatol. 30:830–833. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Johnson DA, Barclay RL, Mergener K, Weiss
G, König T, Beck J and Potter NT: Plasma Septin9 versus fecal
immunochemical testing for colorectal cancer screening: A
prospective multicenter study. PLoS One. 9:e982382014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Church TR, Wandell M, Lofton-Day C, Mongin
SJ, Burger M, Payne SR, Castaños-Vélez E, Blumenstein BA, Rösch T,
Osborn N, et al: PRESEPT Clinical Study Steering Committee,
Investigators and Study Team: Prospective evaluation of methylated
SEPT9 in plasma for detection of asymptomatic colorectal cancer.
Gut. 63:317–325. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
deVos T, Tetzner R, Model F, Weiss G,
Schuster M, Distler J, Steiger KV, Grützmann R, Pilarsky C,
Habermann JK, et al: Circulating methylated SEPT9 DNA in plasma is
a biomarker for colorectal cancer. Clin Chem. 55:1337–1346. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tänzer M, Balluff B, Distler J, Hale K,
Leodolter A, Röcken C, Molnar B, Schmid R, Lofton-Day C, Schuster T
and Ebert MP: Performance of epigenetic markers SEPT9 and ALX4 in
plasma for detection of colorectal precancerous lesions. PLoS One.
5:e90612010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ørntoft MB, Nielsen HJ, Ørntoft TF and
Andersen CL; and Danish Study Group on Early Detection of
Colorectal Cancer, . Performance of the colorectal cancer screening
marker Sept9 is influenced by age, diabetes and arthritis: A nested
case-control study. BMC Cancer. 15:8192015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Powrózek T, Krawczyk P, Kucharczyk T and
Milanowski J: Septin 9 promoter region methylation in free
circulating DNA-potential role in noninvasive diagnosis of lung
cancer: Preliminary report. Med Oncol. 31:9172014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lyu N, Yao H, Xiao T, Gao Y and Wu L:
[Protein levels and its clinical significance of septin-9 and
clusterin in peripheral blood of epithelial ovarian cancer
patients). Zhonghua Fu Chan Ke Za Zhi. 50:679–684. 2015.(In
Chinese). PubMed/NCBI
|
|
20
|
Kreuziger LM, Porcher JC, Ketterling RP
and Steensma DP: An MLL-SEPT9 fusion and t(11;17) (q23;q25)
associated with de novo myelodysplastic syndrome. Leuk Res.
31:1145–1148. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bennett KL, Karpenko M, Lin MT, Claus R,
Arab K, Dyckhoff G, Plinkert P, Herpel E, Smiraglia D and Plass C:
Frequently methylated tumor suppressor genes in head and neck
squamous cell carcinoma. Cancer Res. 68:4494–4499. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Molnár B, Tóth K, Barták BK and Tulassay
Z: Plasma methylated septin 9: A colorectal cancer screening
marker. Expert Rev Mol Diagn. 15:171–184. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Whiting PF, Rutjes AW, Westwood ME,
Mallett S, Deeks JJ, Reitsma JB, Leeflang MM, Sterne JA and Bossuyt
PM: QUADAS-2 Group: QUADAS-2: A revised tool for the quality
assessment of diagnostic accuracy studies. Ann Intern Med.
155:529–536. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang Y, Pei F, Wang X, Sun Z, Hu C and Dou
H: Diagnostic accuracy of fecal lactoferrin for inflammatory bowel
disease: A meta-analysis. Int J Clin Exp Pathol. 8:12319–12332.
2015.PubMed/NCBI
|
|
25
|
Cao FF, Yu S, Jiang ZY and Bao YX:
Diagnostic accuracy of Golgi protein 73 in primary hepatic
carcinoma using ELISA: A systematic review and meta-analysis. Clin
Lab. 60:587–597. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tóth K, Galamb O, Spisák S, Wichmann B,
Sipos F, Valcz G, Leiszter K, Molnár B and Tulassay Z: The
influence of methylated septin 9 gene on RNA and protein level in
colorectal cancer. Pathol Oncol Res. 17:503–509. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang C, Zhao K and Rong Q: Diagnostic
value of fecal MicroRNAs for colorectal cancer: A meta-analysis.
Clin Lab. 61:1845–1853. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cui Z, Chen Y, Xiao Z, Hu M, Lin Y, Chen Y
and Zheng Y: Long noncoding RNAs as auxiliary biomarkers for
gastric cancer screening: A pooled analysis of individual studies.
Oncotarget. 7:25791–25800. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhu H, Bhagatwala J, Huang Y, Pollock NK,
Parikh S, Raed A, Gutin B, Harshfield GA and Dong Y: Correction:
Race/ethnicity-specific association of vitamin D and global DNA
methylation: Cross-sectional and interventional findings. PLoS One.
11:e01625822016. View Article : Google Scholar : PubMed/NCBI
|