Introduction
Cholangiocarcinoma (CCA) is the second most common
hepatobiliary cancer after hepatocellular carcinoma and accounts
for 10–15% of all liver carcinomas (1,2). It arises
in the biliary tract epithelium (cholangiocyte) and is categorized
into three groups: Intrahepatic, perihilar and distal hepatic
carcinoma. Previous studies in the US and worldwide have
demonstrated that the associated mortality and incidence rates have
increased since 2002 (3,4). Clinically, CCA is a challenge to treat
as patients often do not exhibit clear symptoms, making early
diagnose difficult (5). At present,
there is no specific molecular targeting treatment for CCA.
Conventional chemotherapy is still used but is generally
ineffective in cases of CCA associated with different genetic
variants (6). A greater understanding
of the biology of CCA may improve treatment and diagnosis.
Recent research has focused on target identification
and selective inhibitors of several molecular signaling pathways
(7). In earlier reports, genetic
alterations were identified to be involved in CCA carcinogenesis,
including point mutations in p53, B-Raf proto-oncogene
serine/threonine kinase and V-Ki-ras2 Kirsten rat sarcoma viral
oncogene homolog (8,9). Liver-specific downregulation of Smad
family member 4 and phosphatase and tensin homolog, as established
tumor suppressor genes, may also induce CCA (10,11). In
addition, increased expression of cyclooxygenase-2, receptor
tyrosine-protein kinase c-erbB-2 and interleukin 6 are considered
to be involved in early carcinogenesis in the biliary tract
epithelium (12,13).
Although a variety of anticancer protein inhibitors,
including Notch, Wnt, growth factor receptor and histone
deacetylase inhibitors, are being developed to suppress
oncoproteins, an average time of 7 years is typically required for
drug approval following clinical trial evaluation (14). However, it has been demonstrated that
certain drugs already approved by the US Food and Drug
Administration (FDA) may be used to suppress cancer. For example,
inhibitors of 3-hydroxy-3-methylglutaryl (HMG)-CoA reductase,
including lovastatin, atorvastatin and simvastatin, and of
cyclooxygenase-2 (COX-2), including celecoxib, have
potential preventative effects on CCA carcinogenesis (15,16).
Identifying novel uses for the approved drugs also minimizes the
need for toxicity studies, particularly with regard to
non-antineoplastic drugs.
The study of cancer genomes, and CCA mechanisms in
particular, is still in its infancy. Single gene studies are no
longer feasible for this complex disease. Improved and more
powerful tools for analysis by high-throughput array are required
to provide greater accuracy, precision and cost-effectiveness. Gene
Expression Omnibus (GEO) is a public functional genomics data
repository created to aid the identification and analysis of gene
regulatory and cancer signaling pathways. By also serving as a
gene-drug interaction database, this may further be used to
determine novel therapeutic drugs and targets that may be crucial
to the identification of novel protein inhibitors.
In the present study, gene expression profiles from
the GSE26566 dataset were collected to analyze differential gene
expression, and currently approved drugs associated with the
upregulated genes were searched in drug interaction databases. The
greater availability of gene expression analytical tools and
improved access to information on currently approved treatments may
be useful in developing novel indications for CCA drug
treatment.
Materials and methods
Data collection
The gene expression dataset of normal subjects and
intrahepatic CCA (iHCCA) patients [Gene Expression Omnibus (GEO)
accession no. GSE26566 (17)] were
obtained from the National Center for Biotechnology Information GEO
database (https://www.ncbi.nlm.nih.gov/geo/). A total of 104
cholangiocarcinoma and 6 normal intrahepatic bile duct gene
expression data profiles were collected. This microarray platform
(GLP6104) was collected from one type of platform: Illumina
humanRef-8 v2.0 expression beadchip.
Gene expression analysis
The microarray database GLP6104 platform was
analyzed using GEO2R (version 2.14.1) for the collected datasets.
The data was normalized by log 2 transformation in GEO2R along with
the Benjamini-Hochberg procedure for false discovery rate
correction. Samples were assigned to normal and cholangiocarcinoma
subject groups. Following analysis by GEO2R, the dataset was
displayed in terms of adjusted P-values, P-values, log fold
changes, gene symbols and gene names. The top 15 most up- and
downregulated genes according to most adjusted P-value (from
Student's t-test, P<0.05) were selected as differentially
expressed genes (DEGs).
Gene ontology (GO) analysis
To investigate the DEGs at a functional level, the
Database for Annotation, Visualization and Integrated Discovery
(DAVID; https://david.ncifcrf.gov/) was used
to establish gene clustering following GO analysis, as described
previously (18). The GO was
categorized into cellular component, biological process and
molecular function following standard GO analysis as detailed
previously (19).
Pathway analysis
The Kyoto Encyclopedia of Genes and Genomes (KEGG;
http://www.genome.jp/kegg/) was accessed
for assistance in investigating the dysregulated signaling pathways
in cholangiocarcinoma as detailed previously (20). The DEGs were searched in the database
for investigation of biochemistry pathways that may be involved in
the tumorigenesis and development of cholangiocarcinoma. The up-
and downregulated genes were analyzed by DAVID. The significant
categories were predicted by an Expression Analysis Systematic
Explorer score <0.01 and the minimum number of genes for the
corresponding term >2 were considered.
Protein-protein interaction (PPI)
analysis
PPI network analysis was conducted by using the
search tool for the retrieval of interacting genes/proteins
(STRING; http://string-db.org/) as in previous
study (21). The threshold protein
interaction reliability score was 0.400, which represents a medium
level of confidence (22).
Validation by The Cancer Genome Atlas
(TCGA) Kaplan-Meier-plotter analysis
To confirm the analysis, an overall survival (OS)
Kaplan-Meier computation was performed by using the top 30 DEGs (15
most upregulated and 15 most downregulated). Expression and OS data
of the iHCCA cohort in the TCGA database was assessed and P-values
obtained by log-rank test using the cBioPortal for Cancer Genomics
(http://www.cbioportal.org/) (23–25).
Gene-drug interaction analysis
The drug signatures of upregulated genes and their
interacting genes were identified by searching the drug signatures
database for gene set analysis (DSigDB) (26). The signatures of the database are
grouped into four categories: i) 1,202 FDA-approved drugs retrieved
for bioactivity assay results from PubChem and ChEMBL; ii) 1,220
kinase inhibitors from human kinome profiling databases (Medical
Research Council Kinase Inhibitor database and Harvard Medical
School Library of Integrated Network-based Cellular Signatures
database); iii) 1,309 perturbagens identified from DEGs in the
three cell lines PC3 (prostate cancer), HL60 (leukemia) and SKMEL5
(melanoma) (with >2-fold change compared with controls); and iv)
drug candidates identified by manual curation and text mining from
the Therapeutics Targets Database and the Comparative
Toxicogenomics Database (27–29). The drugs were further categorized
according to the US-FDA database (https://www.accessdata.fda.gov/scripts/cder/daf/)
as FDA-approved non-antineoplastic and neoplastic drugs.
Results
Gene expression analysis
The dataset retrieved from GEO26566 included 104
iHCCA and 6 normal profiles. The top 15 most significantly up- and
downregulated genes, respectively, determined by integrative
analysis in GEOR2, are listed in Table
I. FYVE, RhoGEF and PH domain-containing 6 (FGD6) and
chymotrypsinogen B2 (CTRB2) were the most significantly up-
and downregulated genes, respectively (Table I).
 | Table I.Top 30 most significantly up- and
downregulated differentially expressed genes. |
Table I.
Top 30 most significantly up- and
downregulated differentially expressed genes.
| A, Upregulated
genes |
|---|
|
|---|
| Gene probe ID | Gene symbol | Description | P-value | Log fold
change |
|---|
| 154240685 | FGD6 | FYVE, RhoGEF and PH
domain-containing 6 | 9.58E-09 | 3.86 |
| 349501059 | CHEK1 | Checkpoint kinase
1 | 3.22E-07 | 4.34 |
| 61743966 | CTBP1 | C-terminal-binding
protein 1 | 3.29E-07 | 3.63 |
| 168229167 | BUB1B | BUB1 mitotic
checkpoint serine/threonine kinase B | 6.76E-07 | 5.38 |
| 154800452 | TROAP |
Trophinin-associated protein | 7.45E-07 | 5.41 |
| 157419137 | LAMC2 | Laminin subunit
gamma 2 | 1.57E-06 | 4.8 |
| 19882252 | CST2 | Cystatin-SA | 1.77E-06 | 4.71 |
| 748821156 | CKAP2L |
Cytoskeleton-associated protein
2-like | 2.44E-06 | 3.93 |
| 300794717 | WDHD1 | WD repeat and
HMG-box DNA-binding protein 1 | 4.42E-06 | 4.05 |
| 938148819 | MMP11 | Matrix
metallopeptidase 11 | 1.23E-05 | 6.10 |
| 444189305 | FAM83B | Family with
sequence similarity 83 member B | 3.77E-05 | 5.82 |
| 54607054 | GJB3 | Gap junction
protein beta 3 | 1.36E-04 | 4.90 |
| 148806907 | FNDC1 | Fibronectin type
III-domain containing 1 | 1.45E-04 | 4.41 |
| 379056370 | CDCA8 | Cell division
cycle-associated 8 | 1.66E-04 | 4.01 |
| 748983174 | CNBD2 | Cyclic
nucleotide-binding domain-containing 2 | 2.68E-04 | 6.61 |
|
| B, Downregulated
genes |
|
| Gene probe
ID | Gene
symbol |
Description | P-value | Log fold
change |
|
| 118498349 | CTRB2 | Chymotrypsinogen
B2 | 8.80E-34 | −10.15 |
| 236459772 | CELA2A | Chymotrypsin-like
elastase family member 2A | 3.21E-29 | −9.71 |
| 54607079 | CPB1 | Carboxypeptidase
B1 | 9.23E-29 | −10.98 |
| 38016927 | PLA2G1B | Phospholipase A2
group IB | 8.53E-25 | −10.78 |
| 747165370 | CPA1 | Carboxypeptidase
A1 | 1.09E-21 | −11.95 |
| 58331210 | CELA2B | Chymotrypsin-like
elastase family member 2B | 3.17E-20 | −8.90 |
| 440309868 | CEL | Carboxyl ester
lipase | 5.85E-18 | −9.68 |
| 734703929 |
PNLIPRP1 | Pancreatic
lipase-related protein 1 | 1.14E-14 | −8.37 |
| 236460049 | CELA3A | Chymotrypsin-like
elastase family member 3A | 2.42E-14 | −10.80 |
| 357588512 | CLPS | Colipase | 1.88E-12 | −12.09 |
| 310923201 | PRSS1 | Protease, serine
1 | 2.48E-10 | −11.52 |
| 217416389 | CPA2 | Carboxypeptidase
A2 | 3.24E-10 | −11.27 |
| 62526042 | CTRC | Chymotrypsin 2 | 7.93E-09 | −10.82 |
| 1010226556 | CELA3B | Chymotrypsin-like
elastase family member 3B | 8.45E-07 | −11.10 |
| 1042779786 | PNLIP | Pancreatic
lipase | 1.22E-06 | −12.04 |
Functional enrichment analysis
GO enrichment analysis of DEGs was performed for the
top 30 most significant DEG signatures (15 most upregulated and 15
most downregulated). The DEGs were classified into the three GO
categories of biological process, cellular component and molecular
function. It was identified that the significantly enriched GO
terms for biological process included proteolysis and lipid
catabolic processes, while those for cellular component included
extracellular space and extracellular region; the significantly
enriched GO terms for molecular function included serine-type
endopeptidase activity and triglyceride lipase activity (Table II).
 | Table II.Enriched GO categories of
differentially expressed genes. |
Table II.
Enriched GO categories of
differentially expressed genes.
| A, Biological
process clustering |
|---|
|
|---|
| GO term | Biological
process | Gene count | Genes | P-value |
|---|
| GO:0006508 | Proteolysis | 11 | CPA1, CPA2, CPB1,
CTRC, CELA2A, CELA2B, CELA3A, CELA3B, CTRB2, MMP11, PRSS1 | 1.7E-09 |
| GO:0007586 | Digestion | 4 | CELA3A, CTRB2,
CLPS, PRSS1 | 1.2E-04 |
| GO:0044241 | Lipid
digestion | 3 | CEL, CLPS,
PNLIP | 1.8E-04 |
| GO:0022617 | Extracellular
matrix | 4 | CTRB2, LAMC2,
MMP11, PRSS1 | 2.1E-04 |
| GO:0016042 | Lipid catabolic
process | 4 | CLPS, PNLIPRP1,
PNLIP, PLA2G1B | 3.0E-04 |
| GO:0009235 | Cobalamin metabolic
process | 3 | CTRC, CTRB2,
PRSS1 | 4.8E-04 |
| GO:0030299 | Intestinal
cholesterol absorption | 2 | CEL, PNLIP | 1.4E-02 |
| GO:0006629 | Lipid metabolic
process | 3 | CEL, CLPS,
PNLIPRP1 | 2.4E-02 |
| GO:0007093 | Mitotic cell cycle
checkpoint | 2 | BUB1B, CHEK1 | 4.8E-02 |
| GO:0001523 | Retinoid metabolic
process | 2 | CLPS, PNLIP | 9.0E-02 |
|
| B, Cellular
component |
|
| GO term | Biological
process | Gene
count | Genes | P-value |
|
| GO:0005615 | Extracellular
space | 14 | CEL, CPA1, CPA2,
CPB1, CHEK1, CELA2A, CELA2B, CELA3A, CELA3B, CTRB2, CST2, LAMC2,
PNLIPRP1, PLA2G1B | 4.1E-08 |
| GO:0005576 | Extracellular
region | 13 | CEL, CPA2, CTRC,
CELA2B, CTRB2, CLPS, FNDC1, LAMC2, MMP11, PNLIPRP1, PNLIP, PLA2G1B,
PRSS1 | 2.7E-06 |
| GO:0051233 | Spindle
midzone | 2 | BUB1B, CDCA8 | 3.0E-02 |
|
| C, Molecular
function |
|
| GO term | Biological
process | Gene
count | Genes | P-value |
|
| GO:0004252 | Serine-type
endopeptidase activity | 8 | CTRC, CELA2A,
CELA2B, CELA3A, CELA3B, CTRB2, MMP11, PRSS1 | 1.1E-07 |
| GO:0004806 | Triglyceride lipase
activity | 3 | CEL, PNLIPRP1,
PNLIP | 3.3E-04 |
| GO:0004181 | Metal
carboxypeptidase activity | 3 | CPA1, CPA2,
CPB1 | 8.4E-04 |
| GO:0016298 | Lipase
activity | 2 | CEL, PNLIPRP1 | 1.4E-02 |
| GO:0004180 | Carboxypeptidase
activity | 2 | CPA2, CPB1 | 2.7E-02 |
| GO:0008236 | Serine-type
peptidase activity | 2 | CTRB2, PRSS1 | 9.6E-02 |
Pathway enrichment analysis based on the KEGG
database demonstrated that the DEGs were significantly enriched in
4 terms (Table III). The most
significant pathway was pancreatic secretion (P=1.6E-17).
 | Table III.Enriched Kyoto Encyclopedia of Genes
and Genomes pathways of differentially expressed genes. |
Table III.
Enriched Kyoto Encyclopedia of Genes
and Genomes pathways of differentially expressed genes.
| Term | Pathway | P-value | Gene count | Genes |
|---|
| Hsa:04972 | Pancreatic
secretion | 1.6E-17 | 12 | CEL, CPA1, CPA2,
CPB1, CELA2A, CELA2B, CELA3A, CELA3B, PNLIPRP1, PNLIP, PLA2G1B,
PRSS1 |
| Hsa:04972 | Protein digestion
and absorption | 7.5E-10 | 8 | CPA1, CPA2,
CPB1, CELA2A, CELA2B, CELA3A, CELA3B, PRSS1 |
| Hsa:04975 | Fat digestion and
absorption | 2.0E-06 | 5 | CEL, CLPS,
PNLIPRP1, PNLIP, PLA2G1B |
| Hsa:00561 | Glycerolipid
metabolism | 8.7E-03 | 3 | CEL, PNLIPRP1,
PNLIP |
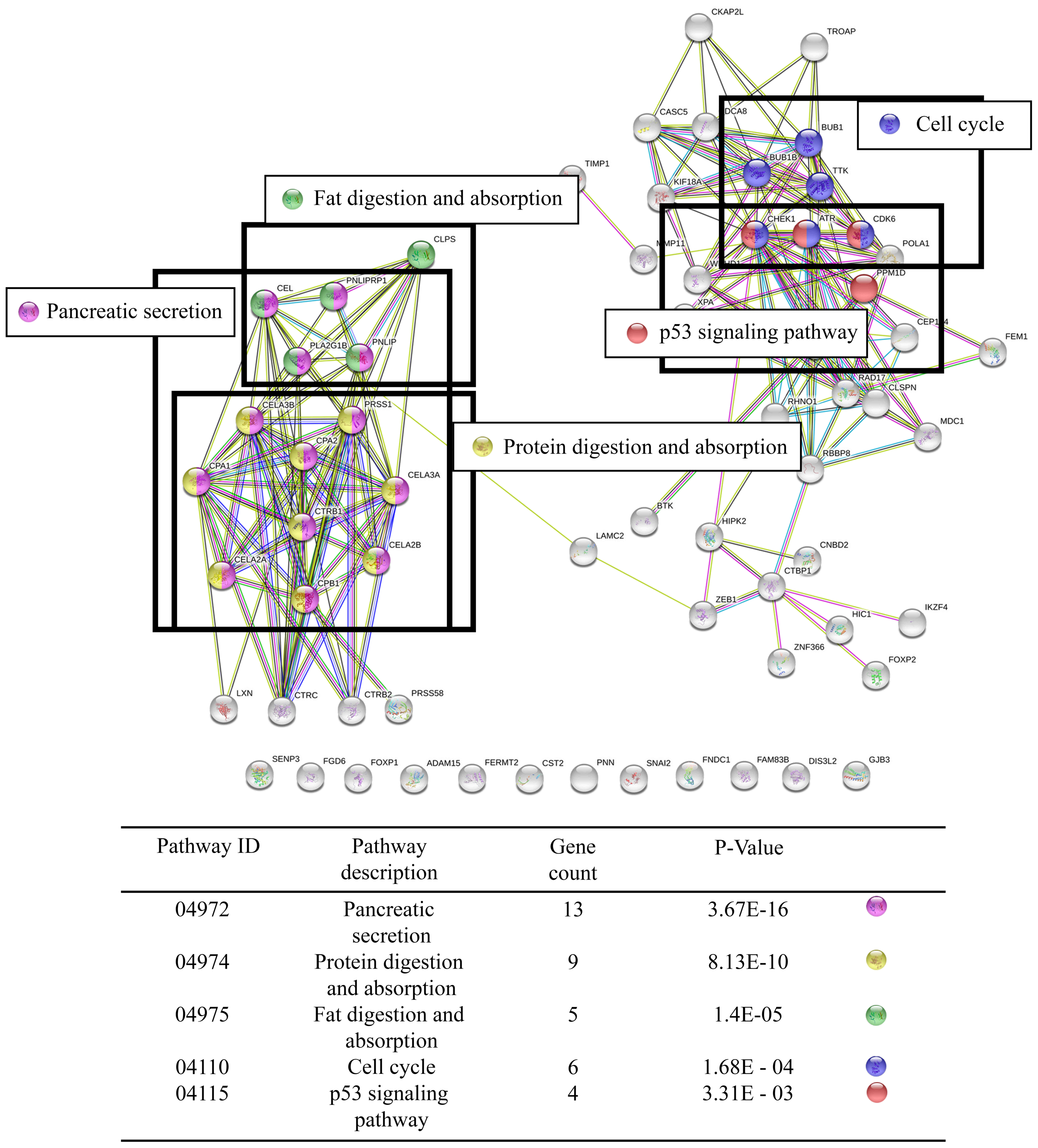
PPI network analysis
To investigate the associations between the DEGs and
signaling pathways, a comprehensive analysis of protein
interactions of the top 15 up- and downregulated genes,
respectively, was conducted using the STRING database. As
illustrated in Fig. 1, direct or
indirect interactions were identified with the exception of 12
genes [Forkhead box p1, ADAM metallopeptidase domain 15, FGD6,
sentrin-specific-peptidase 3, fermitin family member 2,
cystatin-SA, pinin, Snail family transcriptional repressor 2,
fibronectin type III domain containing 1 (FNDC1), family
with sequence similarity 83 member B (FAM83B), DIS3-like
3′-5′ exoribonuclease 2 and gap junction protein beta 3
(GJB3)]. The associations between pathways and gene
expression were categorized into 5 pathways, namely pancreatic
secretion, protein digestion and absorption, lipid digestion and
absorption, cell cycle and p53 signaling pathway.
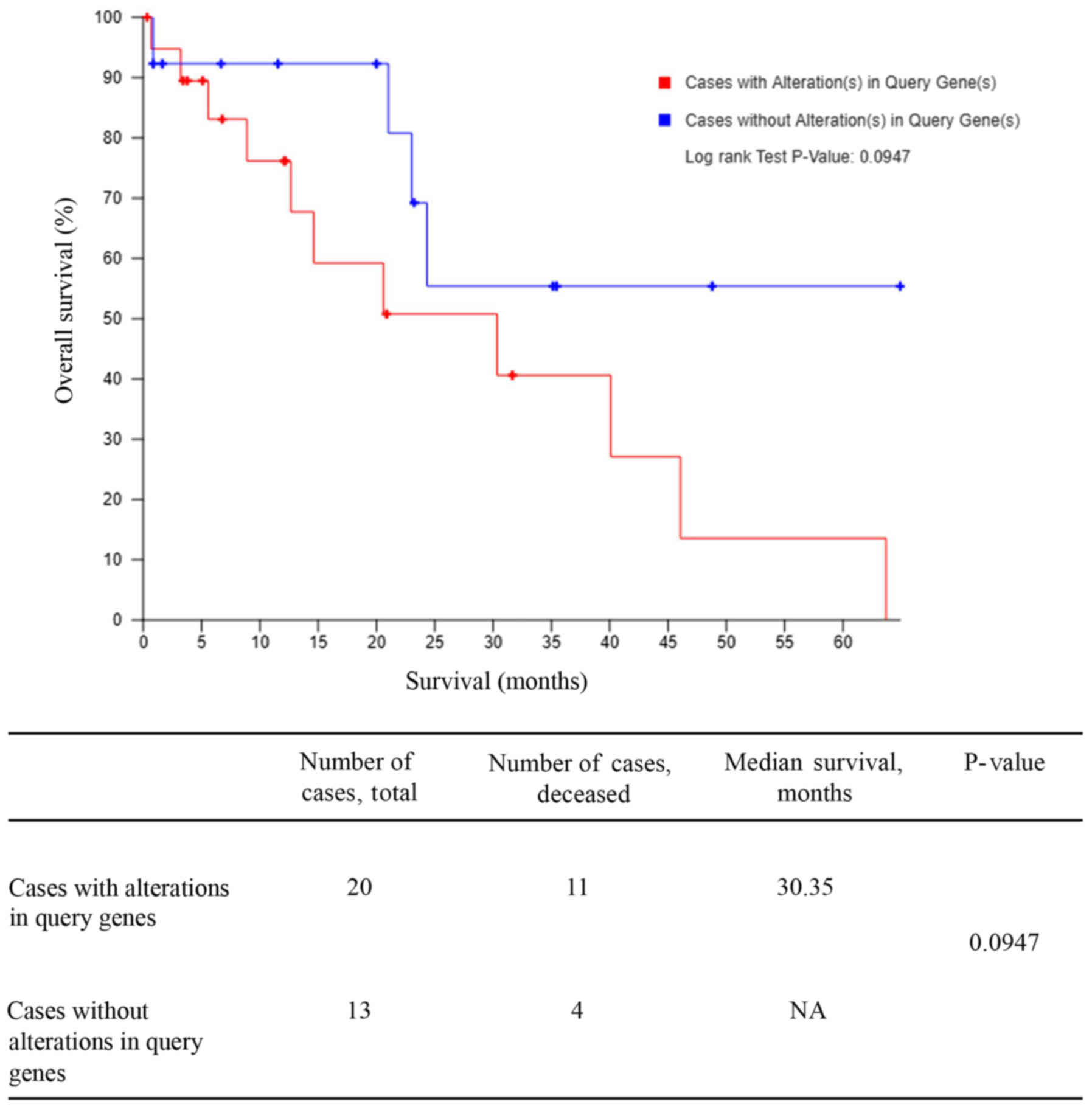
Validation by TCGA
Kaplan-Meier-plotter analysis
For validation of altered gene signatures using
clinical data, CCA data was retrieved from the TCGA database
(n=36). Cases were divided into those with alterations in the
previously identified DEGs (n=20) and those without (13). The Kaplan-Meier curve demonstrated
that the altered genes group was associated with a significantly
lower survival rate (P=0.0947; Fig.
2).
Assessment of gene-drug
interaction
Drug signatures from drug-upregulated gene
interactions in DsigDB were classified, based on known FDA-approved
targeted agents including neoplastic and non-neoplastic drugs. A
total of 86 non-neoplastic and 61 neoplastic drugs were identified.
Among the top upregulated genes, CHEK1 was the most
interacting with drug signatures. No drug signatures were
identified for CST2, MMP11 or cyclic nucleotide binding
domain-containing 2 (Table IV). The
interacting anticancer drugs belonged to 6 categories in the FDA
database, namely alkylating agent, hormone agent, cytotoxic
alkaloid, antitumor antibiotic, antimetabolite and protein
inhibitor (Table V).
 | Table IV.Non-neoplastic and neoplastic drugs
relating to upregulated genes in cholangiocarcinoma. |
Table IV.
Non-neoplastic and neoplastic drugs
relating to upregulated genes in cholangiocarcinoma.
| Gene symbol | Total drugs | Non-neoplastic
drugs | Neoplastic
drugs |
|---|
| FGD6 | 8 | Zalcitabine,
digoxin, raloxifene, retinoic acid | Vorinostat,
tamoxifen, 5-fluorouracil, melphalan |
| CHEK1 | 44 | Chlorzoxazone,
dirithromycin, medrysone, latamoxef, clindamycin, ipratropium
bromide, danazol, thymidine, caffeine, mucosolvan, lovastatin,
troglitazone, rimonabant hydrochloride, menadione, retinoic acid,
diclofenac, carbamazepine, valproic acid, folic acid, auraptan,
zidovudine | Etoposide,
bosutinib, sunitinib, daunorubicin, irinotecan, bortezomib,
melphalan, mitomycin vorinostat, hydroxyurea, olaparib,
5-fluorouracil gemcitabine, temozolomide, doxorubicin, decitabine,
carmustine, bortezomib, fludarabine, irinotecan, clofarabine,
busalfan |
| CTBP1 | 18 | Nifedipine,
clofazimine, chlortetracycline, chlozoxazone, baclofen,
levonorgestrel, cimetidine, ambroxol, medrysone, valproic acid,
dorzolamide, clindamycin, ipratropium bromide, danazole,
niclozamide, vitamin E, acetaminophen | Vorinostat |
| BUB1B | 20 | Ciclopirox,
trifluridine, pyrvinium, clindamycin, colchicine, piroxicam,
acetaminophen, valproic acid | Daunorubicin,
etoposide, azacitidine, irinotecan, fulvestrant, docetaxel,
dasatinib, decitabine, doxorubicin, 5-fluorouracil, paclitaxel,
bicalutamide |
| TROAP | 11 | Ciclopirox,
trifluridine, pergolide, acetaminophen, valproic acid | Etoposide,
azacitidine, methotrexate, doxorubicin, irinotecan,
5-fluorouracil |
| LAMC2 | 4 | Valproic acid,
mifepristone | Mechlorethamine,
cyclophosphamide |
| CST2 | 0 | – | – |
| CKAP2L | 3 | Retinoic acid,
acetaminophen | Dasatinib |
| WDHD1 | 10 | Zalcitabine,
primaquine, doxycycline, latamoxef, cianidanol, retinoic acid,
acetaminophen, valproic acid | Dasatinib,
vorinostat |
| MMP11 | 0 | – | – |
| FAM83B | 1 | Valproic acid | – |
| JGB3 | 5 | Retigabine | Vorinostat,
azacitidine, doxorubicin, decitabine |
| FNDC1 | 1 | Valproic acid | – |
| CDCA8 | 22 | Chlortetracycline,
chlorzoxazone, ciclopirox, trifluridine, cimetidine, medrysone,
pyrvinium, latamoxef, glibenclamide, metronidazole, monobenzone,
piroxicam, retinoic acid, acetaminophen, zidovudine | Azacitidine,
methotrexate, doxorubicin, irinotecan, 5-fluorouracil, vinblastine,
cytarabine |
| CNBD2 | 0 | – | – |
| Total | 147 | 86 | 61 |
 | Table V.Categories of the US Food and Drug
Administration-approved drugs associated with upregulated
genes. |
Table V.
Categories of the US Food and Drug
Administration-approved drugs associated with upregulated
genes.
| Category | Drug name |
|---|
| Alkylating
agent | Melphalan,
temozolomide, busalfan, carmustine, mechlorethamine,
cyclophosphamide, busalfan |
| Hormone agent | Tamoxifen,
fulvestrant, bicalutamide |
| Cytotoxic
alkaloid | Etoposide,
irinotecan, vinblastine, docetaxel, paclitaxel |
| Antitumor
antibiotic | Doxorubicin,
daunorubicin, mitomycin |
| Antimetabolite | Methotrexate,
5-fluorouracil, clofarabine, gemcitabine, cytarabine, azacitidine,
fludarabine, decitabine, hydroxyurea |
| Protein
inhibitor | Vorinostat,
bosutinib, sunitinib, bortezomib, olaparib |
Discussion
Although previous study has suggested numerous
biomarkers and therapeutic targets for CCA (30), there remains to be few treatments and
drugs available to overcome the disease. In addition, the molecular
mechanisms of CCA are not fully understood. In the present study,
gene expression profiles and dysregulated pathways of CCA were
analyzed, along with gene-drug interactions for drug
repositioning.
In the present study, gene expression analysis was
performed on a CCA patient dataset. The analysis firstly determined
the top 30 DEGs that may be important for CCA carcinogenesis.
Checkpoint kinase 1 (CHEK1) is required for DNA replication and
cell survival in cancer (31). BUB1
mitotic checkpoint serine/threonine kinase B (BUB1B) and cell
division cycle-associated 8 (CDCA8) have been identified to be
involved in cell division and the cell spindle check-point.
Increased expression in progressive cholangiocarcinoma has also
been identified (32–34). A previous study observed that matrix
metalloproteinase 11 (MMP11) was upregulated in CCA patients with
poor prognosis. Therefore, MMP11 may be useful as a prognostic
marker in CCA (35–36).
GO and KEGG pathway analysis demonstrated that the
main biological processes involved in CCA were extracellular matrix
and protein and lipid metabolism. A previous study of bile acid
identified downregulated genes including CTRB, colipase,
chymotrypsin-like elastase family member 3A (CELA3A) and
CELA3B, phospholipase A2, chymotrypsin 2, carboxypeptidase B
(CPB) and CPA, similar to the present gene expression
analysis. The profiles of these genes also differ markedly between
CCA patients and normal subjects, and they may be protein markers
for differentiating malignant from benign tumors (37). Integrative analysis of five
microarrays in the GSE26566 dataset indicated that MMP11 and
FGD6 were upregulated while CELA2A and CTRB2
were downregulated. These genes were associated with the
extracellular matrix, cell division processes, bile acid secretion
and protein and lipid metabolism, with these biological processes
implicated in various pathways in CCA progression (38). These results of DEG pathway analysis
may be used to investigate and target the mechanism of disease
initiation, to ultimately treat CCA, and to clarify the association
between metabolic dysregulation and CCA progression.
Previous PPI analysis revealed an association of
ataxia telangiectasia with Rad3-related protein/casein kinase 1
signaling in p53 activation, which serves an important role in DNA
repair, cell division and the cell cycle (39–40). CTRB,
a member of a family of serine proteases, is secreted into the
gastrointestinal tract and activated through proteolytic cleavage
by protease, serine 1. A previous study observed low-level
expression of CTRB in pancreatic cancer (41). The current analysis of CCA patient
profiles revealed similar results. As concluded previously
(41), this suggests that CTRB
may be a genetic marker for CCA. Furthermore, since genes including
carboxyl ester lipase, pancreatic lipase-related protein 1,
pancreatic lipase and phospholipase A2 group IB associated with
pancreatic secretion and fat digestion and absorption exhibited
significant interaction, the tumorigenesis and progression of CCA
may lead to the dysfunction of liver metabolism and disruption of
pancreatic function.
According to the survival analysis, patients with
CCA in the altered gene group had a lower rate of OS compared with
those patients with no alterations in the DEGs of interest. Genetic
alterations in cell metabolism may contribute to poor prognosis in
human cancers (42). Lipid metabolism
is now considered to have a key role in cancer progression; it has
been proposed that increased metabolic flux may serve as substrate
source for phospholipid synthesis in the rapid growth stages of
cancer (43).
The use of molecular signatures in cancer to
determine potential drug therapies is a widespread technique
(44). Potential drugs that target
molecular aberrations in various CCA pathways have been proposed
(45). In the present study, the
genetic signatures of upregulated genes from microarray datasets
were analyzed for gene-drug interactions, and US-FDA-approved drugs
were grouped according to their FDA classification as
antineoplastic and non-antineoplastic agents. The present study
demonstrated the potential of various FDA approved antineoplastic
drugs for use in CCA treatment. For example, vorinostat is a
histone deacetylase inhibitor which has been approved by the FDA
for the treatment of cutaneous manifestations of cutaneous T-cell
lymphoma; olaparib is a poly-ADP-ribose polymerase inhibitor
approved for ovarian and breast cancer treatment; and sunitinib, a
multitargeted receptor tyrosine kinase inhibitor, has been approved
by the FDA for the treatment of renal cell carcinoma (46–48). Also
dasatinib, a kinase inhibitor approved for use in patients with
chronic myelogenous leukemia, may be used to treat CCA by targeting
the FGD6, CHEK1, C-terminal binding protein 1
(CTBP1), cytoskeleton-associated protein 2-like and WD
repeat and HMG-box DNA-binding protein 1 (WDHD1) genes.
Sunitinib has been reported to have possible use in iHCCA
chemotherapies, where resistance is an issue in advanced stage
patients (49–51).
Several chemotherapeutic drugs may be used to treat
bile duct cancer, for example, 5-fluorouracil, gemcitabine,
cisplatin, capecitabine and oxaliplatin. The current standard
treatment drugs for CCA are 5-fluorouracil, gemcitabine, or their
combinations with cisplatin. In certain cases, two or more of these
drugs may be combined. However, their treatment efficacy is
unsatisfactory with low clinical response rate (52). A previous study observed survival of
patients treated with cisplatin plus gemcitabine, there may be
little benefit to patients because of the debilitating side
effects, depending on the amount and frequency of the dosage
(53,54). Nonetheless, CCA is a heterogeneous
cancer involving multifactorial risks (55,56). The
present results demonstrate that the other antineoplastic drugs,
including protein inhibitors such as vorinostat, sunitinib and
olaparib, and standard chemotherapy drugs such as melphalan,
etoposide, and mitomycin, may be suitable candidates for drug
repositioning for CCA and should be studied further.
The approach of the current study was to investigate
established drugs to identify novel indications for cancer
treatment. The results indicated that non-antineoplastic agents
interacted with certain genes implicated in CCA. Cimetidine, a
histamine type 2 antagonist, is used to treat heartburn and peptic
ulcer; however, data also indicates that cimetidine may contribute
to growth inhibition and apoptosis induction in CCA in vitro
and in vivo (57). Valproic
acid is used to treat bipolar disorders and prevent migraine
headaches; previous study identified that it may inhibit the growth
of CCA cell lines (TFK-1, QBC939 and CCLP1) by inducing cell cycle
arrest and promoting cell differentiation via induction of
dendrite-like structures (58). The
effects of each drug may be mediated by several molecular
mechanisms. For example, valproic acid, an anti-epileptic drug,
appears to act primarily by histone deacetylase inhibition, and not
solely by inhibition of CHEK1, CTBP1, BUB1B,
trophinin-associated protein, laminin subunit gamma 2, WDHD1,
FAM83B and FNDC1. For each of these drugs, further
extensive studies, both in vitro and in vivo, are
needed to confirm their effectiveness, molecular mechanisms and
toxicity in the treatment of CCA.
Previous study (59)
and the present data have demonstrated that lipid metabolism
pathways may be significantly altered, and that lovastatin
interacts with the dysregulated gene signatures. The lipid
metabolism pathway may serve important roles in CCA growth and
progression by downregulating the farnesoid × receptor and lipid
metabolism pathways (59). Lovastatin
is a medication used for lipid metabolic disorder, and blocks
HMG-CoA reductase enzyme in cholesterol synthesis. This drug has
been studied in vitro and in vivo and has been
demonstrated to exert anticancer effects in CCA and enhancement of
gefitinib-induced antiproliferation in resistant CCA (60–62).
Clofazimine is a drug used in the treatment of
leprosy. It also has an anticancer effect and may be used for
hepatocellular carcinoma (63).
Certain antimalarial primaquines have been shown to block amino
acid uptake in in vivo studies (64). From the present gene-drug interaction
results, these drugs may have certain therapeutic roles in the
context of CCA treatment; however further study is required.
COX-2 also serves an important role in CCA
carcinogenesis, and is expressed at high levels in CCA tissue
compared with non-tumorous adjacent tissue (65). Inhibition of COX-2 by nonsteroidal
anti-inflammatory drugs, including peroxicam in combination with
cisplatin, may trigger cell cycle regulation and apoptosis in
different mesothelioma cell lines (MSTO-211H and NCI-H2452)
(66).
The present results also revealed retinoic acid and
caffeine to interact with gene signatures of the top most DEGs.
Retinoic acid is categorized as a retinoid and has been studied for
its therapeutic efficacy in numerous cancers. In CCA, cell
migration and tumor invasion may be blocked by all trans-retinoic
acid-incorporated glycol chitosan nanoparticles (67). Caffeine is a methylxanthine alkaloid
related to the adenine and guanine bases. Previous reports
demonstrated that caffeinated coffee consumption was associated
with reduced risk of HCC and CCA (68,69).
However, since a molecular mechanism underlying the potential
anticancer activity of caffeine is yet to be elucidated, it is
probably best utilized as a supplement in daily diet. The present
research is limited as the application of molecular data based on
bioinformatics analysis may not be sufficient to predict the
efficacy of potential targeted agents or protein inhibitors.
Therefore, validation of the current data with case studies and
additional functional downstream approach experiments is
necessary.
In conclusion, the present study performed gene
expression analysis, and based on the findings, assessed molecular
pathways and drug interactions associated with DEG signatures in
CCA. The present study may be developed to further examine drug
repurposing and to search for additional anticancer activities
among known pharmaceuticals. The results may provide a novel
hypothesis regarding the mechanisms of non-antineoplastic drugs in
cancer treatment. The data may be a useful resource for preclinical
and clinical research on existing FDA-approved drugs into their
potential use in CCA treatment.
Acknowledgements
The authors are thankful to Dr Janice M. Wongsurawat
of the Chulabhorn Research Institute (Bangkok, Thailand), for
proof-reading the manuscript.
Funding
The present study was supported by the Chulabhorn
Graduate Institute and Chulabhorn Research Institute (Bangkok,
Thailand).
Availability of data and materials
All data used and analyzed during this study are
included in this published article.
Authors' contributions
SC performed data acquisition and data analysis and
contributed to the drafting of the manuscript. TS was responsible
for initiation, conception, experimental design and execution of
the project and for critical revision of the manuscript. TU
supervised the collection of anti-cancer drug information and
contributed to the revision of the manuscript. YP supervised data
analysis and contributed to the revision of the manuscript. JS
supervised experimental design and critical revision of the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
de Groen PC, Gores GJ, LaRusso NF,
Gunderson LL and Nagorney DM: Biliary tract cancers. N Engl J Med.
341:1368–1378. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wu T, Han C, Lunz JG III, Michalopoulos G,
Shelhamer JH and Demetris AJ: Involvement of 85-kd cytosolic
phospholipase A(2) and cyclooxygenase-2 in the proliferation of
human cholangiocarcinoma cells. Hepatology. 36:363–373. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Patel T: Worldwide trends in mortality
from biliary tract malignancies. BMC Cancer. 2:102002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shaib YH, Davila JA, McGlynn K and
El-Serag HB: Rising incidence of intrahepatic cholangiocarcinoma in
the United States: A true increase? J Hepatol. 40:472–477. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nakeeb A, Pitt HA, Sohn TA, Coleman J,
Abrams RA, Piantadosi S, Hruban RH, Lillemoe KD, Yeo CJ and Cameron
JL: Cholangiocarcinoma. A spectrum of intrahepatic, perihilar, and
distal tumors. Ann Surg. 224:463–473. 1996.
|
|
6
|
Patel T: Increasing incidence and
mortality of primary intrahepatic cholangiocarcinoma in the United
States. Hepatology. 33:1353–1357. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sahu S and Sun W: Targeted therapy in
biliary tract cancers-current limitations and potentials in the
future. J Gastrointest Oncol. 8:324–336. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ohashi K, Nakajima Y, Kanehiro H, Tsutsumi
M, Taki J, Aomatsu Y, Yoshimura A, Ko S, Kin T, Yagura K, et al:
Ki-ras mutations and p53 protein expressions in intrahepatic
cholangiocarcinomas: Relation to gross tumor morphology.
Gastroenterology. 109:1612–1617. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Furubo S, Harada K, Shimonishi T,
Katayanagi K, Tsui W and Nakanuma Y: Protein expression and genetic
alterations of p53 and ras in intrahepatic cholangiocarcinoma.
Histopathology. 35:230–240. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tannapfel A, Sommerer F, Benicke M,
Katalinic A, Uhlmann D, Witzigmann H, Hauss J and Wittekind C:
Mutations of the BRAF gene in cholangiocarcinoma but not in
hepatocellular carcinoma. Gut. 52:706–712. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu X, Kobayashi S, Qiao W, Li C, Xiao C,
Radaeva S, Stiles B, Wang RH, Ohara N, Yoshino T, et al: Induction
of intrahepatic cholangiocellular carcinoma by liver-specific
disruption of Smad4 and Pten in mice. J Clin Invest. 116:1843–1852.
2006. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Endo K, Yoon BI, Pairojkul C, Demetris AJ
and Sirica AE: ERBB-2 overexpression and cyclooxygenase-2
up-regulation in human cholangiocarcinoma and risk conditions.
Hepatology. 36:439–450. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sugawara H, Yasoshima M, Katayanagi K,
Kono N, Watanabe Y, Harada K and Nakanuma Y: Relationship between
interleukin-6 and proliferation and differentiation in
cholangiocarcinoma. Histopathology. 33:145–153. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chabner BA: Early accelerated approval for
highly targeted cancer drugs. N Engl J Med. 364:1087–1089. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu T, Leng J, Han C and Demetris AJ: The
cyclooxygenase-2 inhibitor celecoxib blocks phosphorylation of Akt
and induces apoptosis in human cholangiocarcinoma cells. Mol Cancer
Ther. 3:299–307. 2004.PubMed/NCBI
|
|
16
|
Peng YC, Lin CL, Hsu WY, Chang CS, Yeh HZ,
Tung CF, Wu YL, Sung FC and Kao CH: Statins are associated with a
reduced risk of cholangiocarcinoma: A population-based case-control
study. Br J Clin Pharmacol. 80:755–761. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Andersen JB, Spee B, Blechacz BR, Avital
I, Komuta M, Barbour A, Conner EA, Gillen MC, Roskams T, Roberts
LR, et al: Genomic and genetic characterization of
cholangiocarcinoma identifies therapeutic targets for tyrosine
kinase inhibitors. Gastroenterology. 142:1021–1031e15. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: The Gene Ontology Consortium: Gene ontology: Tool for the
unification of biology. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Szklarczyk D, Franceschini A, Kuhn M,
Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork
P, et al: The STRING database in 2011: Functional interaction
networks of proteins, globally integrated and scored. Nucleic Acids
Res. 39:(Database). D561–D568. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
von Mering C, Jensen LJ, Snel B, Hooper
SD, Krupp M, Foglierini sM, Jouffre N, Huynen MA and Bork P:
STRING: Known and predicted protein-protein associations,
integrated and transferred across organisms. Nucleic Acids Res.
33:D433–D437. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kandoth C, Schultz N, Cherniack AD, Akbani
R, Liu Y, Shen H, Robertson AG, Pashtan I, Shen R, Benz CC, et al:
Cancer Genome Atlas Research Network: Integrated genomic
characterization of endometrial carcinoma. Nature. 497:67–73. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:112013. View Article : Google Scholar
|
|
25
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yoo M, Shin J, Kim J, Ryall KA, Lee K, Lee
S, Jeon M, Kang J and Tan AC: DSigDB: Drug signatures database for
gene set analysis. Bioinformatics. 31:3069–3071. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lamb J, Crawford ED, Peck D, Modell JW,
Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et
al: The Connectivity Map: Using gene-expression signatures to
connect small molecules, genes, and disease. Science.
313:1929–1935. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Qin C, Zhang C, Zhu F, Xu F, Chen SY,
Zhang P, Li YH, Yang SY, Wei YQ, Tao L, et al: Therapeutic target
database update 2014: A resource for targeted therapeutics. Nucleic
Acids Res. 42(D1): D1118–D1123. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Davis AP, Wiegers TC, Roberts PM, King BL,
Lay JM, Lennon-Hopkins K, Sciaky D, Johnson R, Keating H, Greene N,
et al: A CTD-Pfizer collaboration: Manual curation of 88,000
scientific articles text mined for drug-disease and drug-phenotype
interactions. Database (Oxford). 2013:bat0802013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Silsirivanit A, Sawanyawisuth K, Riggins
GJ and Wongkham C: Cancer biomarker discovery for
cholangiocarcinoma: The high-throughput approaches. J Hepatobiliary
Pancreat Sci. 21:388–396. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Massey AJ: Tumour growth environment
modulates Chk1 signalling pathways and Chk1 inhibitor sensitivity.
Sci Rep. 6:358742016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yang L, Feng S and Yang Y: Identification
of transcription factors (TFs) and targets involved in the
cholangiocarcinoma (CCA) by integrated analysis. Cancer Gene Ther.
23:439–445. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Seubwai W, Kraiklang R, Wongkham C and
Wongkham S: Hypoxia enhances aggressiveness of cholangiocarcinoma
cells. Asian Pac J Cancer Prev. 13:(Suppl):. 53–58. 2012.PubMed/NCBI
|
|
34
|
Borad MJ, Champion MD, Egan JB, Liang WS,
Fonseca R, Bryce AH, McCullough AE, Barrett MT, Hunt K, Patel MD,
et al: Integrated genomic characterization reveals novel,
therapeutically relevant drug targets in FGFR and EGFR pathways in
sporadic intrahepatic cholangiocarcinoma. PLoS Genet.
10:e10041352014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tongtawee T, Kaewpitoon SJ, Loyd R,
Chanvitan S, Leelawat K, Praditpol N, Jujinda S and Kaewpitoon N:
High expression of matrix metalloproteinase-11 indicates poor
prognosis in human cholangiocarcinoma. Asian Pac J Cancer Prev.
16:3697–3701. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Severino FE, Hasimoto CN, Rodrigues MAM,
Llanos JC and Reis PP: Integrative analysis of transcriptome and
microRNome profiles in cholangiocarcinoma. J Cancer Sci Ther.
09:580–588. 2017. View Article : Google Scholar
|
|
37
|
Navaneethan U, Lourdusamy V, Gk Venkatesh
P, Willard B, Sanaka MR and Parsi MA: Bile proteomics for
differentiation of malignant from benign biliary strictures: A
pilot study. Gastroenterol Rep (Oxf). 3:136–143. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Huang QX, Cui JY, Ma H, Jia XM, Huang FL
and Jiang LX: Screening of potential biomarkers for
cholangiocarcinoma by integrated analysis of microarray data sets.
Cancer Gene Ther. 23:48–53. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Smith J, Tho ML, Xu N and Gillespie DA:
The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and
cancer. Adv Cancer Res. pp. 73–112. 2010, View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bartek J and Lukas J: Chk1 and Chk2
kinases in checkpoint control and cancer. Cancer Cell. 3:421–429.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kuroki T, Tomioka T, Tajima Y, Inoue K,
Ikematsu Y, Ichinose K, Furui J and Kanematsu T: Detection of the
pancreas-specific gene in the peripheral blood of patients with
pancreatic carcinoma. Br J Cancer. 81:350–353. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Nath A and Chan C: Genetic alterations in
fatty acid transport and metabolism genes are associated with
metastatic progression and poor prognosis of human cancers. Sci
Rep. 6:186692016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chen Y and Li P: Fatty acid metabolism and
cancer development. Sci Bull (Beijing). 61:1473–1479. 2016.
View Article : Google Scholar
|
|
44
|
Sung J, Wang Y, Chandrasekaran S, Witten
DM and Price ND: Molecular signatures from omics data: From chaos
to consensus. Biotechnol J. 7:946–957. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhu P, Aliabadi HM, Uludağ H and Han J:
Identification of potential drug targets in cancer signaling
pathways using stochastic logical models. Sci Rep. 6:230782016.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Mann BS, Johnson JR, He K, Sridhara R,
Abraham S, Booth BP, Verbois L, Morse DE, Jee JM, Pope S, et al:
Vorinostat for treatment of cutaneous manifestations of advanced
primary cutaneous T-cell lymphoma. Clin Cancer Res. 13:2318–2322.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Audeh MW, Carmichael J, Penson RT,
Friedlander M, Powell B, Bell-McGuinn KM, Scott C, Weitzel JN,
Oaknin A, Loman N, et al: Oral poly(ADP-ribose) polymerase
inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and
recurrent ovarian cancer: A proof-of-concept trial. Lancet.
376:245–251. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wood L: Sunitinib malate for the treatment
of renal cell carcinoma. Expert Opin Pharmacother. 13:1323–1336.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Roskoski R Jr: RAF
protein-serine/threonine kinases: Structure and regulation. Biochem
Biophys Res Commun. 399:313–317. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ma J, Shi J, Zhao D, Cheng L, Wang W, Li
F, Jiang X and Jiang H: Raf kinase inhibitor protein inhibits
cholangiocarcinoma cell metastasis by downregulating matrix
metalloproteinase 9 and upregulating tissue inhibitor of
metalloproteinase 4 expression. Oncol Lett. 9:15–24. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Dreyer C, Sablin MP, Bouattour M,
Neuzillet C, Ronot M, Dokmak S, Belghiti J, Guedj N, Paradis V,
Raymond E, et al: Disease control with sunitinib in advanced
intrahepatic cholangiocarcinoma resistant to
gemcitabine-oxaliplatin chemotherapy. World J Hepatol. 7:910–915.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Eckmann KR, Patel DK, Landgraf A, Slade
JH, Lin E, Kaur H, Loyer E, Weatherly JM and Javle M: Chemotherapy
outcomes for the treatment of unresectable intrahepatic and hilar
cholangiocarcinoma: A retrospective analysis. Gastrointest Cancer
Res. 4:155–160. 2011.PubMed/NCBI
|
|
53
|
Valle J, Wasan H, Palmer DH, Cunningham D,
Anthoney A, Maraveyas A, Madhusudan S, Iveson T, Hughes S, Pereira
SP, et al: ABC-02 Trial Investigators: Cisplatin plus gemcitabine
versus gemcitabine for biliary tract cancer. N Engl J Med.
362:1273–1281. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Merla A, Liu KG and Rajdev L: Targeted
therapy in biliary tract cancers. Curr Treat Options Oncol.
16:482015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ruzzenente A, Fassan M, Conci S, Simbolo
M, Lawlor RT, Pedrazzani C, Capelli P, D'Onofrio M, Iacono C,
Scarpa A, et al: Cholangiocarcinoma heterogeneity revealed by
multigene mutational profiling: Clinical and prognostic relevance
in surgically resected patients. Ann Surg Oncol. 23:1699–1707.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Brandi G, Farioli A, Astolfi A, Biasco G
and Tavolari S: Genetic heterogeneity in cholangiocarcinoma: A
major challenge for targeted therapies. Oncotarget. 6:14744–14753.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Dana P, Vaeteewoottacharn K, Kariya R,
Matsuda K, Wongkham S and Okada S: Repurposing cimetidine for
cholangiocarcinoma: Antitumor effects in vitro and in vivo. Oncol
Lett. 13:1432–1436. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wang B, Yang R, Wu Y, Li H, Hu Z, Chen Y
and Zou S: Sodium valproate inhibits the growth of human
cholangiocarcinoma in vitro and in vivo. Gastroenterol Res Pract.
2013:3745932013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Kwee SA, Okimoto GS, Chan OTM, Tiirikainen
M and Wong LL: Metabolic characteristics distinguishing
intrahepatic cholangiocarcinoma: A negative pilot study of
(18)F-fluorocholine PET/CT clarified by transcriptomic analysis. Am
J Nucl Med Mol Imaging. 6:73–83. 2016.PubMed/NCBI
|
|
60
|
Yang SH, Lin HY, Chang VHS, Chen CC, Liu
YR, Wang J, Zhang K, Jiang X and Yen Y: Lovastatin overcomes
gefitinib resistance through TNF-α signaling in human
cholangiocarcinomas with different LKB1 statuses in vitro and in
vivo. Oncotarget. 6:23857–23873. 2015.PubMed/NCBI
|
|
61
|
Yang SH, Lin HY, Changou CA, Chen CH, Liu
YR, Wang J, Jiang X, Luh F and Yen Y: Integrin β3 and LKB1 are
independently involved in the inhibition of proliferation by
lovastatin in human intrahepatic cholangiocarcinoma. Oncotarget.
7:362–373. 2016.PubMed/NCBI
|
|
62
|
Kim BB, Kim M and Park JB: The effects of
lovastatin on the morphology, cell viability and differentiation of
stem cells derived from gingiva. Biomed Res (Aligarh).
28:4922–4927. 2017.
|
|
63
|
Ruff P, Chasen MR, Long JEH and van
Rensburg CE: A phase II study of oral clofazimine in unresectable
and metastatic hepatocellular carcinoma. Ann Oncol. 9:217–219.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Das AK: Anticancer effect of antimalarial
artemisinin compounds. Ann Med Health Sci Res. 5:93–102. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Chariyalertsak S, Sirikulchayanonta V,
Mayer D, Kopp-Schneider A, Fürstenberger G, Marks F and
Müller-Decker K: Aberrant cyclooxygenase isozyme expression in
human intrahepatic cholangiocarcinoma. Gut. 48:80–86. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Verdina A, Cardillo I, Nebbioso A, Galati
R, Menegozzo S, Altucci L, Sacchi A and Baldi A: Molecular analysis
of the effects of piroxicam and cisplatin on mesothelioma cells
growth and viability. J Transl Med. 6:272008. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Chung KD, Jeong YI, Chung CW, Kim DH and
Kang DH: Anti-tumor activity of all-trans retinoic
acid-incorporated glycol chitosan nanoparticles against HuCC-T1
human cholangiocarcinoma cells. Int J Pharm. 422:454–461. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Petrick JL, Freedman ND, Graubard BI,
Sahasrabuddhe VV, Lai GY, Alavanja MC, Beane-Freeman LE, Boggs DA,
Buring JE, Chan AT, et al: Coffee consumption and risk of
hepatocellular carcinoma and intrahepatic cholangiocarcinoma by
sex: The liver cancer pooling project. Cancer Epidemiol Biomarkers
Prev. 24:1398–1406. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Kennedy OJ, Roderick P, Buchanan R,
Fallowfield JA, Hayes PC and Parkes J: Coffee, including
caffeinated and decaffeinated coffee, and the risk of
hepatocellular carcinoma: A systematic review and dose-response
meta-analysis. BMJ Open. 7:e0137392017. View Article : Google Scholar : PubMed/NCBI
|