Introduction
Coronary artery disease (CAD) remains the worldwide
leading cause of mortality among men and women (1). The American Heart Association reported
that in 2014 there were 364,593 mortalities from CAD in the United
States (2), based on 2014 mortality
data from the National Center for Health Statistics (3). In Japan, the Ministry of Health, Labour
and Welfare has reported that the rate of mortality per 100,000
population from acute ischemic heart diseases was 71,673 in 2015
(4). Cerebral infarction (CI) is also
a serious clinical problem worldwide. In the United States,
approximately 610,000 and 185,000 people experience new and
recurrent stroke events each year, respectively, and it was
estimated that the prevalence of silent CI ranged from 6–28% from
1993 to 2005 (2). In 2015, the number
of mortalities from CI and intracerebral hemorrhage in Japanese
patients was 64,523 and 32,113, respectively (4). Therefore, examination of CAD and CI
susceptibility variants in Japanese individuals may be key to
successful personalized prevention of these diseases.
Previous genome-wide association studies (GWASs)
have identified a variety of genes and loci that confer
susceptibility to CAD and CI across various ethnic groups, and have
reported inter-ethnic differences of genetic contribution to these
diseases (5–11). A meta-analysis using datasets of the
CARDIoGRAMplusC4D, MIGen and CARDIoGRAM Exome consortia and ESP
EOMI studies assessed the association of genetic variants in
several chemokine receptor genes with CAD and myocardial infarction
(6). The study did not identify any
evidence of an association between the genetic variants and disease
in large European ancestry cohorts, whereas six low frequency
variants were associated with myocardial infarction in a South
Asian cohort. Another meta-analysis of two independent Chinese
GWASs for CAD identified four CAD-associated loci that were not
present in populations of European ancestry (7). Inter-ethnic difference of disease
susceptibility loci has also been reported in GWASs for ischemic
stroke (i.e., CI). A GWAS for ischemic stroke in 6,341 Japanese
individuals who participated in three independent population-based
studies identified that cadherin EGF LAG seven-pass G-type receptor
1 (CELSR1) was a susceptibility gene for ischemic stroke
(10). The association of
CELSR1 with stroke has been confirmed in a Portuguese cohort
(12); however, the Siblings With
Ischemic Stroke Study did not demonstrate an association of 312
probands with ischemic stroke from 70 centers in the USA and Canada
(13,14). The discrepancy of disease-susceptible
variants among populations may be due to differences of genetic
background among ethnic groups.
Although recent GWASs have identified a large number
of genetic variants that confer susceptibility to cardiovascular
diseases (5,7,9,14,15), they
have been conducted in a cross-sectional manner that commonly
measures traits at a single point in time. To address this issue,
the present study examined disease progression and physiological
changes in 5,989 Japanese individuals who had annual health
check-ups for several years, and performed longitudinal exome-wide
association studies (EWASs) to investigate novel susceptibility
loci for CAD and CI.
Materials and methods
Study subjects
The physiological data of 6,026 community-dwelling
individuals recruited to a population-based cohort study (Inabe
Health and Longevity Study) in Inabe City, Japan was traced
(16–20). These individuals visited Inabe General
Hospital for an annual health check-up, with a mean annual
follow-up period of 5±3 years (covering April 2003 to March 2014).
All participants had undergone 1 to 11 medical examinations. This
cohort was denoted as the ‘Inabe cohort’ in the longitudinal EWASs.
Detailed methods for the recruitment of subjects and for the
collection and storage of medical examination data and genomic DNA
samples have been described previously (19). Diagnostic criteria for CAD and CI have
also been described previously (18).
The study protocol complied with the Declaration of
Helsinki and was approved by the Committees on the Ethics of Human
Research of Mie University Graduate School of Medicine and Inabe
General Hospital (Inabe, Japan). Written informed consent was
obtained from all subjects prior to enrollment in the Inabe Health
and Longevity Study.
Longitudinal EWAS
Infinium HumanExome-12 ver. 1.2 BeadChip and
Infinium Exome-24 ver. 1.0 BeadChip (Illumina, Inc., San Diego, CA,
USA) were used to genotype ~244,000 genetic variants in the Inabe
cohort for longitudinal EWASs. These arrays include putative
functional exonic variants selected from >12,000 individual
exome and whole-genome sequences across diverse ethnic populations,
including European, African, Chinese and Hispanic individuals
(21). Missing genotype or phenotype
data of each individual was eliminated from the analyses. Quality
control of genotyping data was performed, and monomorphic sites and
the following genetic variants were discarded: i) Variants
contained in only one of the exome arrays used (~3.6% of all
variants); ii) variants with a call rate of <97.0%; iii)
variants with a minor allele frequency (MAF) of <0.05; iv)
variants whose genotype distribution significantly deviated from
Hardy-Weinberg equilibrium (P<0.001) in controls; and v)
variants located on mitochondrial DNA or sex chromosomes.
In addition, sex specification was checked for each
sample, with samples for which the sex designation in the clinical
records was inconsistent with genetic sex discarded. Cryptic
relatedness and duplicate samples were checked by calculation of
identity by descent (IBD); all pairs of DNA samples exhibiting an
IBD of >0.1875 were inspected and one sample from each pair was
excluded. Population stratification in the Inabe cohort was
examined by principal component analysis via the EIGENSTRAT method
(22), using JMP Genomics version 6.0
(SAS Institute, Inc., Cary, NC, USA), and four outliers were
removed from the longitudinal EWASs. Consequently, a total of
24,651 single nucleotide polymorphisms (SNPs) among 5,989 Japanese
individuals passed quality control for the longitudinal EWASs of
CAD and CI. The rearrangement of Inabe longitudinal data was
conducted using R software version 3.32 (23) via RStudio version 1.0.136 (http://www.rstudio.com/) (24) and Perl script (version 5.26.2;
https://www.perl.org/get.html). Using
JMP Genomics, genotyping data were converted into numeric data for
dominant, additive and recessive inheritance models. The dominant
and recessive models were defined as ‘0, AA; 1, AB + BB’ and ‘0, AA
+ AB; 1, BB’ (A, major allele; B, minor allele), respectively,
whereas the additive model was defined as ‘0, AA; 1, AB; 2,
BB’.
Statistical analysis
Longitudinal EWASs were conducted for 5,713 control
individuals, 170 subjects with CAD, and 117 with CI (Table I). The association of the prevalence
of CAD and CI with SNPs was tested by the generalized estimating
equation (GEE) model (25,26) with adjustments for age, sex, body mass
index (BMI), smoking and prevalence of hypertension, type 2
diabetes mellitus and dyslipidemia, using the R package ‘geepack’
(27). Since the prevalence of CAD
and CI is repeated categorical data (case or control), a binomial
distribution was applied for assessing the association between the
categorical outcomes and SNPs in the GEE method. The waves argument
was used to specify the ordering of repeated measurements within
individuals. Effects of SNPs in exome arrays on CAD or CI are not
independent as many SNPs are in linkage disequilibrium (LD)
(13,28,29).
Therefore, the false discovery rate (FDR) was calculated using the
Benjamin and Hochberg method (30) to
compensate for multiple comparison of genotypes with the
phenotypes. An FDR of <0.05 was considered to indicate a
statistically significant association.
 | Table I.Longitudinal characteristics of study
subjects in the Inabe cohort. |
Table I.
Longitudinal characteristics of study
subjects in the Inabe cohort.
| Characteristic |
Controla | CADa | CIa |
|---|
| No. of
subjectsb | 5,713 | 170 | 117 |
| Sex, male/female,
%b | 54.7/45.3 | 77.1/22.9 | 68.4/31.6 |
| Age, years | 52.0±0.07
(27,143) | 63.1±0.41
(619) | 63.4±0.35
(466) |
| Body mass index,
kg/m2 | 22.9±0.02
(26,719) | 23.9±0.13
(507) | 23.6±0.16
(416) |
| Current or former
smoker, % | 38.4 (27,143) | 40.1 (619) | 35.0 (466) |
| Hypertension,
% | 31.9 (26,715) | 75.7 (503) | 65.1 (415) |
| Systolic blood
pressure, mmHg | 120.4±0.10
(26,715) | 126.5±0.77
(503) | 127.6±0.81
(415) |
| Diastolic blood
pressure, mmHg | 74.6±0.08
(26,715) | 76.5±0.55
(503) | 77.5±0.58
(415) |
| Type 2 diabetes
mellitus, % | 11.4 (26,745) | 38.8 (590) | 36.4 (464) |
| Fasting plasma
glucose, mmol/l | 5.59±0.007
(26,731) | 6.49±0.086
(586) | 6.22±0.098
(464) |
| Blood hemoglobin
A1c, % | 5.69±0.004
(19,870) | 6.18±0.052
(465) | 6.03±0.041
(410) |
| Dyslipidemia,
% | 58.4 (26,713) | 76.5 (592) | 77.9 (444) |
| Serum
triglycerides, mmol/l | 1.25±0.006
(26,710) | 1.48±0.040
(591) | 1.36±0.037
(444) |
| Serum
HDL-cholesterol, mmol/l | 1.61±0.003
(26,687) | 1.43±0.019
(585) | 1.51±0.020
(440) |
| Serum
LDL-cholesterol, mmol/l | 3.19±0.005
(25,594) | 2.76±0.032
(563) | 3.20±0.039
(422) |
| Chronic kidney
disease, % | 10.8 (24,461) | 37.6 (582) | 37.0 (441) |
| Serum creatinine,
µmol/l | 74.1±0.58
(24,461) | 132.7±8.65
(582) | 163.0±13.33
(441) |
| eGFR, ml
min−1 1.73 m−2 | 79.2±0.11
(24,461) | 66.1±1.11
(582) | 64.5±1.31
(441) |
| Hyperuricemia,
% | 18.1 (23,985) | 42.3 (575) | 27.5 (436) |
| Serum uric acid,
µmol/l | 327.7±0.56
(23,985) | 367.3±4.20
(575) | 352.7±4.09
(436) |
A small effective sample size may increase the
probability of generating false positives (type I errors) (31). Sitlani et al (31) recommend the use of approxdf, which is
a scale of small effective sample size: Approxdf = 2 × MAF ×
Nindep, where Nindep is the sum of the estimated number of
independent observations per person. They demonstrated that an
approxdf of ≥10 could reduce type I errors. Thus, approxdf was
computed by the R package ‘bosswithdf’ (31,32). To
avoid the issue of false positive association in small sample
sizes, a strict approxdf threshold was applied, and SNPs with
approxdf ≤30 discarded.
LD estimates and prediction of
functional association for candidate loci
To survey LDs between the candidate SNPs detected in
the present study and previously identified CAD- or CI-associated
SNPs in JPT (Japanese in Tokyo, Japan) from the 1000 Genomes
Project [http://www.1000genomes.org/ (33)], analyses were conducted using the
LDproxy of LDlink [https://analysistools.nci.nih.gov/LDlink/ (34)], which is a web-based application. The
association of SNPs with CAD or CI reported by previous studies was
investigated using the Genome-Wide Repository of Associations
Between SNPs and Phenotypes [GRASP; https://grasp.nhlbi.nih.gov/Overview.aspx (35)], DisGeNET [http://www.disgenet.org/web/DisGeNET/ (36)] and GWAS Catalogue [https://www.ebi.ac.uk/gwas/ (37)] databases. Gene-gene functional
interactions were predicted using the GeneMANIA Cytoscape plugin
(38–40) via Cytoscape version 3.4.0 software
[http://www.cytoscape.org/ (41)].
Results
Characteristics of subjects
The characteristics of subjects in the Inabe cohort
with respect to longitudinal data are presented in Table I. The prevalence of hypertension, type
2 diabetes mellitus, dyslipidemia, chronic kidney disease and
hyperuricemia was higher in patients with CAD or CI than in
controls. The prevalence of CAD and CI was lower in females (39 CAD
and 37 CI patients) than in males (131 CAD and 80 CI patients). The
majority of physiological or clinical parameters examined (age,
BMI, systolic and diastolic blood pressure, fasting plasma glucose
level, blood hemoglobin A1c content and serum
concentrations of triglycerides, creatinine, and uric acid) were
higher, whereas serum concentrations of high-density
lipoprotein-cholesterol and estimated glomerular filtration rate
were lower, in the patients than in the controls. The serum
concentration of low-density lipoprotein-cholesterol was lower in
patients with CAD (2.76±0.032 mmol/l) than in controls (3.19±0.005
mmol/l). This discrepancy may be attributable to effects of
lipid-lowering treatment for patients with CAD.
Association of SNPs with longitudinal
data on the prevalence of CAD
Following the quality control checks, the
association between 24,651 SNPs and the prevalence of CAD in the
Inabe cohort was tested by using the GEE model with adjustments for
age, sex, BMI, smoking and prevalence of hypertension, type 2
diabetes mellitus and dyslipidemia. Candidate SNPs that reached
statistical significance (FDR of <0.05) were additionally
screened against the threshold of approxdf (>30). Analyses
revealed that two SNPs [rs4606855 of ADGRE3
(P=2.5×10−6; FDR=0.031, approxdf=71) and rs3746414 of
ZFP64 (P=5.9×10−6; FDR=0.048; approxdf=93)] and
one SNP [rs7132908 of FAIM2 (P<2.0×10−16;
FDR<4.9×10−12; approxdf=65)] were significantly
associated with the prevalence of CAD in additive and recessive
models, respectively (Table II).
 | Table II.SNPs significantly (FDR <0.05,
approxdf >30) associated with CAD or CI. |
Table II.
SNPs significantly (FDR <0.05,
approxdf >30) associated with CAD or CI.
| Disease (genetic
model) | RefSNP ID | Nucleotide (amino
acid) substitutiona |
Positionb | Gene |
Estimatec | Std. err. | P-value | MAF | Apxdfd | FDR |
|---|
| CAD
(additive)e | rs4606855 | G→C (E75Q) | 19: 14,658,527 | ADGRE3 | −0.014 | 0.003 |
2.5×10−6 | 0.120 | 71 | 0.031 |
|
| rs3746414 | G→A (S451N) | 20: 52,152,840 | ZFP64 | −0.013 | 0.003 |
5.9×10−6 | 0.140 | 93 | 0.048 |
| CAD
(recessive)f | rs7132908 | G→A | 12: 49,869,365 | FAIM2 |
−8.9×1014 |
2.2×1013 |
<2.0×10−16 | 0.291 | 65 |
<4.9×10−12 |
| CI
(recessive)f | rs6580741 | G→C (H2228Q) | 12: 50,333,923 | FAM186A |
−2.2×1015 |
2.2×1013 |
<2.0×10−16 | 0.260 | 48 |
<4.9×10−12 |
|
| rs1324015 | G→A | 13: 43,153,713 |
LINC00400 | −40.22 | 0.144 |
<2.0×10−16 | 0.264 | 49 |
<4.9×10−12 |
|
| rs884205 | G→T | 18: 62,387,624 |
TNFRSF11A | −40.12 | 0.147 |
<2.0×10−16 | 0.220 | 32 |
<4.9×10−12 |
According to the GRASP, DisGeNET and GWAS Catalogue
databases, the association of the three SNPs with CAD has not been
reported to date. Therefore, the three SNPs were identified as
novel genetic variants that may confer susceptibility to CAD. The
nucleotide substitution at rs7132908 in FAIM2 is predicted to be a
silent substitution in the 3′-untranslated region, while the
remaining two SNPs are predicted to alter amino acid residues
(i.e., nonsynonymous substitutions), according to the NCBI dbSNP
database [https://www.ncbi.nlm.nih.gov/projects/SNP/ (42)].
Genotype distributions for rs4606855, rs3746414 and
rs7132908 in subjects with CAD and controls in the longitudinal
EWAS are listed in Table III. In
the current study, the prevalence of CAD was lower in subjects with
minor alleles than in those with major alleles for all SNPs,
suggesting the CC genotype of rs4606855 and the AA genotypes of
rs3746414 and rs7132908 to be protective against CAD.
 | Table III.Genotype distributions for three
candidate SNPs among subjects with CAD and controls. |
Table III.
Genotype distributions for three
candidate SNPs among subjects with CAD and controls.
| RefSNP ID |
Positiona | Gene | Genotype |
Controlsb | CADb |
|---|
| rs4606855 | 19: 14,658,527 | ADGRE3 | GG | 21,095 (77.7) | 564 (91.1) |
|
|
|
| GC | 5,676 (20.9) | 54 (8.7) |
|
|
|
| CC | 372 (1.4) | 1 (0.2) |
| rs3746414 | 20: 52,152,840 | ZFP64 | GG | 20,122 (74.1) | 529 (85.5) |
|
|
|
| GA | 6,506 (24.0) | 81 (13.1) |
|
|
|
| AA | 515 (1.9) | 9 (1.5) |
| rs7132908 | 12: 49,869,365 | FAIM2 | GG | 13,609 (50.1) | 344 (55.6) |
|
|
|
| GA | 11,128 (41.0) | 252 (40.7) |
|
|
|
| AA | 2,406 (8.9) | 23 (3.7) |
Association of SNPs with longitudinal
data on the prevalence of CI
In the Inabe cohort, three SNPs [rs6580741 of
FAM186A (P<2.0×10−16;
FDR<4.9×10−12; approxdf=48), rs1324015 of
LINC00400 (P<2.0×10−16;
FDR<4.9×10−12; approxdf=49), and rs884205 of
TNFRSF11A (P<2.0×10−16;
FDR<4.9×10−12; approxdf=32)] were significantly
associated with the prevalence of CI in the recessive model,
although the GEE tests in the dominant and additive models detected
no significant association between SNPs and the prevalence of CI
(Table II). The CI-associated SNPs
detected in the recessive model were not shared with the
CAD-associated SNPs. The nucleotide substitution at rs6580741 in
FAM186A is predicted to alter an amino acid residue
(Table II), whereas the
substitutions at the other SNPs are predicted as silent, according
to the NCBI dbSNP.
The rs6580741 of FAM186A is located
relatively close (~464 kb) to rs7132908 of FAIM2 at
chromosomal region 12q13.12, although these SNPs were related to
the different diseases. The LD among these SNPs was thus examined
among participants in the Inabe cohort using the R package
‘genetics’ [https://CRAN.R-project.org/package=genetics (43)]. The estimation demonstrated that these
SNPs were not in LD (D'=0.045, r2=0.002), suggesting
that rs7132908 and rs6580741 were independently associated with the
prevalence of CAD and CI, respectively.
Genotype distributions for rs6580741, rs1324015 and
rs884205 in subjects with CI and controls in the longitudinal EWAS
are listed in Table IV. As with the
case of CAD-associated SNPs, for all three SNPs, the prevalence of
CI was lower in subjects with minor alleles than in those with
major alleles. The CC genotype of rs6580741, the AA
genotype of rs1324015, and the TT genotype of rs884205 may
thus be protective against CI.
 | Table IV.Genotype distributions for three
candidate SNPs among subjects with CI and controls. |
Table IV.
Genotype distributions for three
candidate SNPs among subjects with CI and controls.
| RefSNP ID |
Positiona | Gene | Genotype |
Controlsb | CIb |
|---|
| rs6580741 | 12: 50,333,923 | FAM186A | GG | 14,669 (54.1) | 269 (57.7) |
|
|
|
| GC | 10,555 (38.9) | 171 (36.7) |
|
|
|
| CC | 1,900 (7.0) | 26 (5.6) |
| rs1324015 | 13: 43,153,713 |
LINC00400 | GG | 14,809 (54.6) | 295 (63.3) |
|
|
|
| GA | 10,433 (38.5) | 171 (36.7) |
|
|
|
| AA | 1,882 (6.9) | 0 (0.0) |
| rs884205 | 18: 62,387,624 |
TNFRSF11A | GG | 16,573 (61.1) | 312 (67.0) |
|
|
|
| GT | 9,154 (33.7) | 154 (33.0) |
|
|
|
| TT | 1,397 (5.2) | 0 (0.0) |
Assessment of LD between the six
identified SNPs and other SNPs associated with CAD or CI
The LDs between the six candidate SNPs identified in
the present study and adjacent SNPs across a ~1 Mb genomic region
were assessed using the LDproxy in LDlink. The LDproxy analysis
indicated that 433 SNPs were in significant LD (r2≥0.5)
with one of the candidate SNPs in JPT from the 1000 Genomes Project
(Table V). Of the 433 SNPs, there
were no SNPs previously identified to be associated with CAD or CI
(P>0.01) according to the GRASP database. In addition, the
LDpair analysis in LDlink using allele frequency data in East Asian
populations from the 1000 Genomes Project indicated that rs1324015
associated with the prevalence of CI in the present study was not
in LD with rs9533425 (D'=0.4709, r2=0.0007), previously
reported to be associated with lag time (lag phase of the
turbidimetric clotting assay) to fibrin clot formation (28). These results suggest that effects of
the candidate SNPs on the prevalence of CAD or CI are
independent.
 | Table V.Correlation estimates of candidate
(query) SNPs with adjacent SNPs using LDproxy. |
Table V.
Correlation estimates of candidate
(query) SNPs with adjacent SNPs using LDproxy.
| Chr. pos. | RefSNP ID | r2 | Chr. pos. | RefSNP ID | r2 | Chr. pos. | RefSNP ID | r2 |
|---|
| 18q21.33 |
rs884205 | 1.00 | 20q13.2 | rs12481281 | 0.77 | 12q13.12 | rs7979830 | 1.00 |
| 13q14.11 |
rs1324015 | 1.00 | 20q13.2 | rs3787176 | 0.77 | 12q13.12 | rs35723625 | 1.00 |
| 13q14.11 | rs2589313 | 0.59 | 20q13.2 | rs72626582 | 0.77 | 12q13.12 | rs10876013 | 1.00 |
| 13q14.11 | rs1324016 | 0.57 | 20q13.2 | rs72626583 | 0.77 | 12q13.12 | rs12425229 | 1.00 |
| 13q14.11 | rs2762188 | 0.57 | 20q13.2 | rs6021711 | 0.76 | 12q13.12 | rs3812825 | 1.00 |
| 13q14.11 | rs2589314 | 0.57 | 20q13.2 | rs2180366 | 0.76 | 12q13.12 | rs1362983 | 1.00 |
| 13q14.11 | rs35574382 | 0.57 | 20q13.2 | rs67904269 | 0.76 | 12q13.12 | rs141978158 | 1.00 |
| 13q14.11 | rs2762185 | 0.57 | 20q13.2 | rs7273288 | 0.74 | 12q13.12 | rs7308095 | 1.00 |
| 13q14.11 | rs2657098 | 0.57 | 20q13.2 | rs13045200 | 0.61 | 12q13.12 | rs11169319 | 1.00 |
| 13q14.11 | rs1562123 | 0.57 | 20q13.2 | rs11478756 | 0.61 | 12q13.12 | rs12811291 | 1.00 |
| 13q14.11 | rs11424017 | 0.52 | 20q13.2 | rs11473560 | 0.56 | 12q13.12 | rs34825838 | 1.00 |
| 20q13.2 | rs3746414 | 1.00 | 20q13.2 | rs6021739 | 0.56 | 12q13.12 | rs11169317 | 1.00 |
| 20q13.2 | rs3746413 | 1.00 | 20q13.2 | rs6096795 | 0.56 | 12q13.12 | rs3861100 | 1.00 |
| 20q13.2 | rs3746415 | 1.00 | 20q13.2 | rs6021738 | 0.56 | 12q13.12 | rs11169315 | 1.00 |
| 20q13.2 | rs3787181 | 1.00 | 20q13.2 | rs6021737 | 0.56 | 12q13.12 | rs2009072 | 1.00 |
| 20q13.2 | rs11431529 | 1.00 | 20q13.2 | rs6021736 | 0.56 | 12q13.12 | rs11169314 | 1.00 |
| 20q13.2 | rs67642347 | 1.00 | 20q13.2 | rs6013400 | 0.56 | 12q13.12 | rs6580739 | 0.97 |
| 20q13.2 | rs4809893 | 1.00 | 20q13.2 | rs6096794 | 0.56 | 12q13.12 | rs7972824 | 0.97 |
| 20q13.2 | rs8122116 | 1.00 | 20q13.2 | rs6013399 | 0.56 | 12q13.12 | rs9739363 | 0.97 |
| 20q13.2 | rs6021732 | 1.00 | 20q13.2 | rs117265881 | 0.56 | 12q13.12 | rs12823506 | 0.97 |
| 20q13.2 | rs2224227 | 1.00 | 20q13.2 | rs4809892 | 0.51 | 12q13.12 | rs112456855 | 0.97 |
| 20q13.2 | rs66953446 | 1.00 | 20q13.2 | rs145753021 | 0.51 | 12q13.12 | rs12422417 | 0.97 |
| 20q13.2 | rs115523244 | 1.00 | 19p13.12 |
rs4606855 | 1.00 | 12q13.12 | rs35224873 | 0.97 |
| 20q13.2 | rs6021723 | 1.00 | 19p13.12 | rs56819877 | 1.00 | 12q13.12 | rs12828340 | 0.87 |
| 20q13.2 | rs12480406 | 1.00 | 19p13.12 | rs4808971 | 1.00 | 12q13.12 | rs17124562 | 0.87 |
| 20q13.2 | rs67638808 | 1.00 | 19p13.12 | rs7255012 | 1.00 | 12q13.12 | rs11169335 | 0.87 |
| 20q13.2 | rs67262858 | 1.00 | 19p13.12 | rs10426121 | 1.00 | 12q13.12 | rs35576436 | 0.87 |
| 20q13.2 | rs6013397 | 1.00 | 19p13.12 | rs6511956 | 1.00 | 12q13.12 | rs11169332 | 0.87 |
| 20q13.2 | rs12481222 | 0.97 | 19p13.12 | rs4273164 | 1.00 | 12q13.12 | rs11169331 | 0.87 |
| 20q13.2 | rs139218250 | 0.97 | 19p13.12 | rs4239642 | 1.00 | 12q13.12 | rs71465002 | 0.87 |
| 20q13.2 | rs7346642 | 0.97 | 19p13.12 | rs10402993 | 0.95 | 12q13.12 | rs7314465 | 0.87 |
| 20q13.2 | rs67892028 | 0.97 | 19p13.12 | rs11085902 | 0.95 | 12q13.12 | rs6580730 | 0.87 |
| 20q13.2 | rs73130324 | 0.97 | 19p13.12 | rs11085901 | 0.95 | 12q13.12 | rs7308885 | 0.87 |
| 20q13.2 | rs75085690 | 0.97 | 19p13.12 | rs7245656 | 0.86 | 12q13.12 | rs2358539 | 0.87 |
| 20q13.2 | rs3838014 | 0.94 | 19p13.12 | rs7250114 | 0.86 | 12q13.12 | rs201343445 | 0.87 |
| 20q13.2 | rs3179313 | 0.94 | 19p13.12 | rs73506161 | 0.86 | 12q13.12 | rs12424713 | 0.87 |
| 20q13.2 | rs3179314 | 0.94 | 19p13.12 | rs35805282 | 0.86 | 12q13.12 | rs12424691 | 0.87 |
| 20q13.2 | rs3787178 | 0.94 | 19p13.12 | rs75540045 | 0.86 | 12q13.12 | rs11169322 | 0.87 |
| 20q13.2 | rs1555328 | 0.94 | 19p13.12 | rs34930135 | 0.86 | 12q13.12 | rs12425705 | 0.87 |
| 20q13.2 | rs6021748 | 0.94 | 19p13.12 | rs373306807 | 0.66 | 12q13.12 | rs35628283 | 0.87 |
| 20q13.2 | rs6021747 | 0.94 | 12q13.12 |
rs6580741 | 1.00 | 12q13.12 | rs35875720 | 0.87 |
| 20q13.2 | rs4811297 | 0.94 | 12q13.12 | rs7310541 | 1.00 | 12q13.12 | rs34198664 | 0.84 |
| 20q13.2 | rs111897744 | 0.94 | 12q13.12 | rs7134337 | 1.00 | 12q13.12 | rs11292692 | 0.82 |
| 20q13.2 | rs6021745 | 0.94 | 12q13.12 | rs7134595 | 1.00 | 12q13.12 | rs4768905 | 0.77 |
| 20q13.2 | rs6021744 | 0.94 | 12q13.12 | rs55931113 | 1.00 | 12q13.12 | rs59210472 | 0.77 |
| 20q13.2 | rs6021743 | 0.94 | 12q13.12 | rs35878271 | 1.00 | 12q13.12 | rs4636745 | 0.77 |
| 20q13.2 | rs6021742 | 0.94 | 12q13.12 | rs34039674 | 1.00 | 12q13.12 | rs11830586 | 0.77 |
| 20q13.2 | rs6021741 | 0.94 | 12q13.12 | rs66895907 | 1.00 | 12q13.12 | rs7973910 | 0.77 |
| 20q13.2 | rs6021740 | 0.94 | 12q13.12 | rs4768900 | 1.00 | 12q13.12 | rs73305103 | 0.77 |
| 20q13.2 | rs6021735 | 0.94 | 12q13.12 | rs10876020 | 1.00 | 12q13.12 | rs4321029 | 0.77 |
| 20q13.2 | rs6021734 | 0.94 | 12q13.12 | rs34858415 | 1.00 | 12q13.12 | rs7968898 | 0.77 |
| 20q13.2 | rs56783695 | 0.94 | 12q13.12 | rs11836169 | 1.00 | 12q13.12 | rs151015253 | 0.77 |
| 20q13.2 | rs60610902 | 0.94 | 12q13.12 | rs4768949 | 1.00 | 12q13.12 | rs11169323 | 0.74 |
| 20q13.2 | rs6021733 | 0.94 | 12q13.12 | rs4768872 | 1.00 | 12q13.12 | rs73305105 | 0.74 |
| 20q13.2 | rs60614585 | 0.94 | 12q13.12 | rs34614542 | 1.00 | 12q13.12 | rs35925338 | 0.74 |
| 20q13.2 | rs57913961 | 0.94 | 12q13.12 | rs12312177 | 1.00 | 12q13.12 | NA | 0.72 |
| 20q13.2 | rs6021730 | 0.94 | 12q13.12 | rs34894919 | 1.00 | 12q13.12 | rs144615146 | 0.72 |
| 20q13.2 | rs6021729 | 0.94 | 12q13.12 | rs12424876 | 1.00 | 12q13.12 | rs78601155 | 0.72 |
| 20q13.2 | rs6021728 | 0.94 | 12q13.12 | rs11169377 | 1.00 | 12q13.12 | rs74336127 | 0.72 |
| 20q13.2 | rs6021727 | 0.94 | 12q13.12 | rs11169376 | 1.00 | 12q13.12 | rs141852922 | 0.69 |
| 20q13.2 | rs6096791 | 0.94 | 12q13.12 | rs36017775 | 1.00 | 12q13.12 | rs145504356 | 0.69 |
| 20q13.2 | rs6091458 | 0.94 | 12q13.12 | rs11169375 | 1.00 | 12q13.12 | rs34098872 | 0.65 |
| 20q13.2 | rs6091457 | 0.94 | 12q13.12 | rs7137319 | 1.00 | 12q13.12 | rs67576611 | 0.64 |
| 20q13.2 | rs200887094 | 0.94 | 12q13.12 | rs7295847 | 1.00 | 12q13.12 | rs7138622 | 0.64 |
| 20q13.2 | rs6096784 | 0.94 | 12q13.12 | rs7296291 | 1.00 | 12q13.12 | rs7138420 | 0.64 |
| 20q13.2 | rs77290230 | 0.94 | 12q13.12 | rs7312252 | 1.00 | 12q13.12 | rs7315690 | 0.64 |
| 20q13.2 | rs12624632 | 0.94 | 12q13.12 | rs10506292 | 1.00 | 12q13.12 | rs5798135 | 0.63 |
| 20q13.2 | rs6021722 | 0.94 | 12q13.12 | rs34849043 | 1.00 | 12q13.12 | rs17124514 | 0.62 |
| 20q13.2 | rs6091453 | 0.94 | 12q13.12 | rs10615610 | 1.00 | 12q13.12 | rs12424335 | 0.62 |
| 20q13.2 | rs6021721 | 0.94 | 12q13.12 | rs10876023 | 1.00 | 12q13.12 | rs34145380 | 0.62 |
| 20q13.2 | rs6021720 | 0.94 | 12q13.12 | rs10876024 | 1.00 | 12q13.12 | rs34309034 | 0.62 |
| 20q13.2 | rs6512796 | 0.94 | 12q13.12 | rs7301186 | 1.00 | 12q13.12 | rs34245511 | 0.62 |
| 20q13.2 | rs12480343 | 0.93 | 12q13.12 | rs11169374 | 1.00 | 12q13.12 | rs9364 | 0.62 |
| 20q13.2 | rs12480321 | 0.93 | 12q13.12 | rs11169373 | 1.00 | 12q13.12 | rs8181651 | 0.60 |
| 20q13.2 | rs67451131 | 0.93 | 12q13.12 | rs35209607 | 1.00 | 12q13.12 | rs80003859 | 0.60 |
| 20q13.2 | rs6013393 | 0.93 | 12q13.12 | rs4421818 | 1.00 | 12q13.12 | rs144939089 | 0.60 |
| 20q13.2 | rs67862732 | 0.93 | 12q13.12 | rs11169370 | 1.00 | 12q13.12 | rs28364704 | 0.59 |
| 20q13.2 | rs144698812 | 0.93 | 12q13.12 | rs11169369 | 1.00 | 12q13.12 | rs736167 | 0.59 |
| 20q13.2 | rs117427009 | 0.93 | 12q13.12 | rs11169390 | 1.00 | 12q13.12 | rs10783347 | 0.59 |
| 20q13.2 | rs140670817 | 0.93 | 12q13.12 | rs11169391 | 1.00 | 12q13.12 | rs10783346 | 0.59 |
| 20q13.2 | rs6021709 | 0.93 | 12q13.12 | rs4445717 | 1.00 | 12q13.12 | rs61928279 | 0.59 |
| 20q13.2 | rs6021708 | 0.93 | 12q13.12 | rs9668187 | 1.00 | 12q13.12 | rs7302422 | 0.59 |
| 20q13.2 | rs72626578 | 0.93 | 12q13.12 | rs60025018 | 1.00 | 12q13.12 | rs12423130 | 0.59 |
| 20q13.2 | rs6021707 | 0.93 | 12q13.12 | rs11169393 | 1.00 | 12q13.12 | rs3815671 | 0.59 |
| 20q13.2 | rs6021706 | 0.93 | 12q13.12 | rs10467106 | 1.00 | 12q13.12 | rs57583527 | 0.59 |
| 20q13.2 | rs140980517 | 0.93 | 12q13.12 | rs12303082 | 1.00 | 12q13.12 | rs10747572 | 0.59 |
| 20q13.2 | rs143481833 | 0.93 | 12q13.12 | rs7971374 | 1.00 | 12q13.12 | rs71083515 | 0.57 |
| 20q13.2 | rs12624823 | 0.93 | 12q13.12 | rs10783352 | 1.00 | 12q13.12 | rs79043170 | 0.55 |
| 20q13.2 | rs12624819 | 0.93 | 12q13.12 | rs10783353 | 1.00 | 12q13.12 | rs35404088 | 0.55 |
| 20q13.2 | rs148218979 | 0.93 | 12q13.12 | rs12299758 | 1.00 | 12q13.12 | rs10783340 | 0.55 |
| 20q13.2 | rs6021760 | 0.90 | 12q13.12 | rs12299669 | 1.00 | 12q13.12 | rs7978904 | 0.55 |
| 20q13.2 | rs3818198 | 0.90 | 12q13.12 | rs11169394 | 1.00 | 12q13.12 | rs10783338 | 0.55 |
| 20q13.2 | rs72626580 | 0.90 | 12q13.12 | rs11169395 | 1.00 | 12q13.12 | rs7398567 | 0.55 |
| 20q13.2 | rs4811301 | 0.90 | 12q13.12 | rs11169367 | 1.00 | 12q13.12 | rs10735824 | 0.55 |
| 20q13.2 | rs4811303 | 0.90 | 12q13.12 | rs11833608 | 1.00 | 12q13.12 | rs7972465 | 0.55 |
| 20q13.2 | rs4811304 | 0.90 | 12q13.12 | rs113486728 | 1.00 | 12q13.12 | rs6580728 | 0.55 |
| 20q13.2 | rs4811305 | 0.90 | 12q13.12 | rs376666931 | 1.00 | 12q13.12 | rs7138945 | 0.55 |
| 20q13.2 | rs140407501 | 0.90 | 12q13.12 | rs7980911 | 1.00 | 12q13.12 | rs11169282 | 0.55 |
| 20q13.2 | rs6021763 | 0.90 | 12q13.12 | rs7302363 | 1.00 | 12q13.12 | rs57061317 | 0.53 |
| 20q13.2 | rs6021764 | 0.90 | 12q13.12 | rs4768855 | 1.00 | 12q13.12 | rs76382737 | 0.53 |
| 20q13.2 | rs6021765 | 0.90 | 12q13.12 | rs6580737 | 1.00 | 12q13.12 | rs12369104 | 0.53 |
| 20q13.2 | rs6021766 | 0.90 | 12q13.12 | rs7488682 | 1.00 | 12q13.12 | rs12819883 | 0.53 |
| 20q13.2 | rs11480360 | 0.90 | 12q13.12 | rs7972068 | 1.00 | 12q13.12 | rs3741562 | 0.53 |
| 20q13.2 | rs6021767 | 0.90 | 12q13.12 | rs7972202 | 1.00 | 12q13.12 | rs113411336 | 0.53 |
| 20q13.2 | rs6021768 | 0.90 | 12q13.12 | rs6580743 | 1.00 | 12q13.12 | rs34632215 | 0.53 |
| 20q13.2 | rs6013409 | 0.90 | 12q13.12 | rs4348979 | 1.00 | 12q13.12 | rs11169278 | 0.53 |
| 20q13.2 | rs6021770 | 0.90 | 12q13.12 | rs11169360 | 1.00 | 12q13.12 | rs1554845 | 0.53 |
| 20q13.2 | rs2038429 | 0.90 | 12q13.12 | rs11169359 | 1.00 | 12q13.12 | rs2204684 | 0.53 |
| 20q13.2 | rs6021772 | 0.90 | 12q13.12 | rs12814094 | 1.00 | 12q13.12 | rs2204683 | 0.53 |
| 20q13.2 | rs2273472 | 0.90 | 12q13.12 | rs7315955 | 1.00 | 12q13.12 | rs10876000 | 0.53 |
| 20q13.2 | rs2038430 | 0.90 | 12q13.12 | rs11169357 | 1.00 | 12q13.12 | rs7967979 | 0.53 |
| 20q13.2 | rs6021774 | 0.90 | 12q13.12 | rs10876028 | 1.00 | 12q13.12 | rs34167640 | 0.53 |
| 20q13.2 | rs58878184 | 0.90 | 12q13.12 | rs7135777 | 1.00 | 12q13.12 | rs7307469 | 0.53 |
| 20q13.2 | rs72626581 | 0.90 | 12q13.12 | rs11838347 | 1.00 | 12q13.12 | rs1554844 | 0.53 |
| 20q13.2 | rs34963386 | 0.90 | 12q13.12 | rs12426444 | 1.00 | 12q13.12 | rs7961065 | 0.53 |
| 20q13.2 | NA | 0.90 | 12q13.12 | rs9705460 | 1.00 | 12q13.12 | rs7961112 | 0.53 |
| 20q13.2 | rs6021775 | 0.90 | 12q13.12 | rs7304445 | 1.00 | 12q13.12 | rs7294618 | 0.53 |
| 20q13.2 | rs1973951 | 0.90 | 12q13.12 | rs11323536 | 1.00 | 12q13.12 | rs7307230 | 0.53 |
| 20q13.2 | rs77286509 | 0.90 | 12q13.12 | rs7311973 | 1.00 | 12q13.12 | rs7135322 | 0.53 |
| 20q13.2 | rs7265436 | 0.90 | 12q13.12 | rs10876017 | 1.00 | 12q13.12 | rs59262224 | 0.53 |
| 20q13.2 | rs6096760 | 0.90 | 12q13.12 | rs10876016 | 1.00 | 12q13.12 | rs17124432 | 0.53 |
| 20q13.2 | rs142762031 | 0.87 | 12q13.12 | rs7974648 | 1.00 | 12q13.12 | rs12369049 | 0.53 |
| 20q13.2 | rs138459074 | 0.87 | 12q13.12 | rs12821454 | 1.00 | 12q13.12 | rs7308474 | 0.53 |
| 20q13.2 | rs67243058 | 0.84 | 12q13.12 | rs7305995 | 1.00 | 12q13.12 | rs7968119 | 0.53 |
| 20q13.2 | rs6021731 | 0.82 | 12q13.12 | rs35933908 | 1.00 | 12q13.12 | rs61529910 | 0.51 |
| 20q13.2 | rs6013396 | 0.79 | 12q13.12 | rs12832940 | 1.00 | 12q13.12 | NA | 0.50 |
| 20q13.2 | rs6021719 | 0.79 | 12q13.12 | rs10876015 | 1.00 | 12q13.12 |
rs7132908 | 1.00 |
| 20q13.2 | rs6021718 | 0.79 | 12q13.12 | rs10876014 | 1.00 | 12q13.12 | rs3205718 | 1.00 |
| 20q13.2 | rs6091452 | 0.79 | 12q13.12 | rs67138019 | 1.00 | 12q13.12 | rs12146733 | 0.98 |
| 20q13.2 | rs6091451 | 0.79 | 12q13.12 | rs11169351 | 1.00 | 12q13.12 | rs1893492 | 0.98 |
| 20q13.2 | rs6126474 | 0.79 | 12q13.12 | rs11169350 | 1.00 | 12q13.12 | rs145103902 | 0.98 |
| 20q13.2 | rs6123134 | 0.79 | 12q13.12 | rs11169349 | 1.00 | 12q13.12 | rs73116325 | 0.98 |
| 20q13.2 | rs8121769 | 0.79 | 12q13.12 | rs35535298 | 1.00 | 12q13.12 | rs146875448 | 0.85 |
| 20q13.2 | rs6021716 | 0.79 | 12q13.12 | rs2111988 | 1.00 | 12q13.12 | rs11169199 | 0.80 |
| 20q13.2 | rs6021715 | 0.79 | 12q13.12 | rs7311491 | 1.00 | 12q13.12 | rs297924 | 0.80 |
| 20q13.2 | rs6123133 | 0.79 | 12q13.12 | rs7311378 | 1.00 | 12q13.12 | rs17201502 | 0.80 |
| 20q13.2 | rs6096776 | 0.79 | 12q13.12 | rs11169348 | 1.00 | 12q13.12 | rs73116335 | 0.80 |
| 20q13.2 | rs62216897 | 0.79 | 12q13.12 | rs6580735 | 1.00 | 12q13.12 | rs73116338 | 0.80 |
| 20q13.2 | rs6126472 | 0.79 | 12q13.12 | rs11169347 | 1.00 | 12q13.12 | rs73116339 | 0.80 |
| 20q13.2 | rs7265434 | 0.79 | 12q13.12 | rs11169345 | 1.00 | 12q13.12 | rs145512623 | 0.75 |
| 20q13.2 | rs7273176 | 0.79 | 12q13.12 | rs7956468 | 1.00 | 12q13.12 | NA | 0.59 |
| 20q13.2 | rs7273121 | 0.79 | 12q13.12 | rs1972611 | 1.00 | 12q13.12 | rs149596227 | 0.57 |
| 20q13.2 | rs8183138 | 0.79 | 12q13.12 | rs10783344 | 1.00 | 12q13.12 | rs373555454 | 0.55 |
| 20q13.2 | rs6123131 | 0.79 | 12q13.12 | rs35768991 | 1.00 | 12q13.12 | rs12367809 | 0.53 |
| 20q13.2 | rs79718879 | 0.79 | 12q13.12 | rs11169339 | 1.00 | 12q13.12 | rs7306275 | 0.51 |
| 20q13.2 | rs76931275 | 0.79 | 12q13.12 | rs7486747 | 1.00 | 12q13.12 | rs7138803 | 0.50 |
| 20q13.2 | rs6021714 | 0.79 | 12q13.12 | rs7953953 | 1.00 | 12q13.12 | rs112502508 | 0.50 |
| 20q13.2 | rs6021713 | 0.79 | 12q13.12 | rs7132551 | 1.00 |
|
|
|
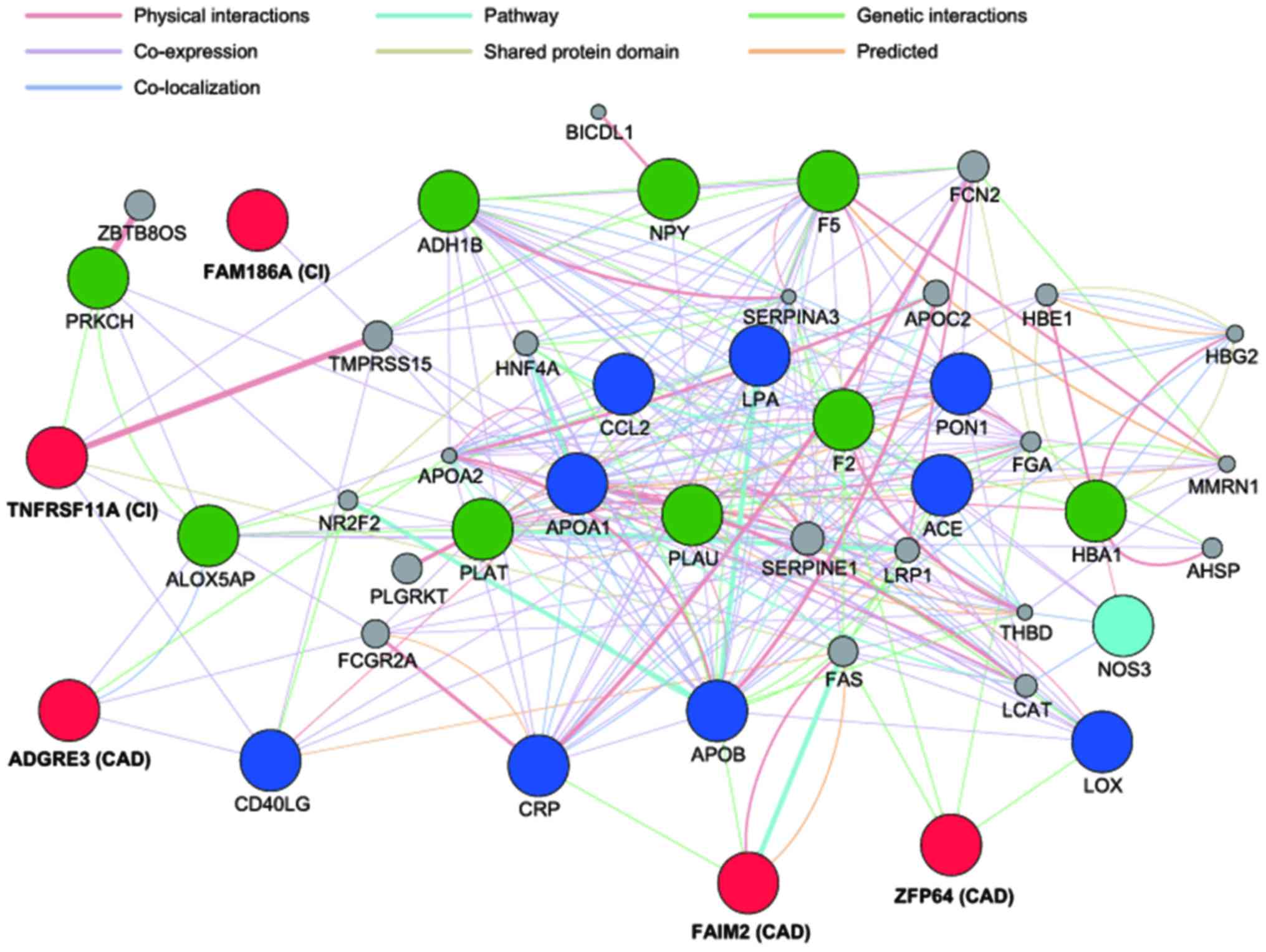
Gene interaction network analysis. To investigate
the interactive functional association, a GeneMANIA network
analysis was conducted of the top ten genes (high gene-disease
association scores) that have been demonstrated to be associated
with CAD or CI selected from the DisGeNET database, and of the five
genes (ZFP64, FAIM2, ADGRE3, FAM186A, and TNFRSF11A)
identified in the present study (Fig.
1). Given that LINC00400 non-coding RNA has not been
well characterized, it could not be examined. The network analysis
showed that the CAD- and CI-associated genes identified in the
present study have potential direct or indirect interactions with
several genes previously demonstrated to be associated with CAD and
CI, respectively. The network suggested that ZFP64 and
FAIM2 interact with LOX and both CRP and
APOB, respectively. In addition, ADGRE3 and
TNFRSF11A were indicated to be co-expressed with
ALOX5AP and CD40LG. The network also suggested that
TNFRSF11A interacts and is co-expressed with PFKCH
and ADH1B, respectively. Additionally, in the network,
FAM186A was indirectly connected with CI-associated genes
(NPY and F5) through TMPRSS15.
Discussion
In the present study, genetic variants that confer
susceptibility to CAD and CI were investigated. The longitudinal
EWASs in a Japanese cohort identified that rs4606855 of
ADGRE3, rs3746414 of ZFP64 and rs7132908 of
FAIM2 were significantly associated with the prevalence of
CAD, whereas rs6580741 of FAM186A, rs1324015 of
LINC00400 and rs884205 of TNFRSF11A were
significantly associated with the prevalence of CI. According to
the databases, these SNPs may be novel susceptibility loci for CAD
or CI.
The adhesion G protein-coupled receptor E3
(ADGRE3) gene is located at chromosomal region 19p13.12, and
rs4606855 [G/C (E75Q)] in this gene was significantly associated
with the prevalence of CAD. The mRNA of ADGRE3 is
predominantly expressed in neutrophils, monocytes and macrophages,
and this protein may be important for interactions between myeloid
cells including neutrophils and monocytes during immune and
inflammatory responses (44). Given
that vascular inflammatory responses serve a role in the
development of coronary atherosclerosis and thrombosis (45–47), this
CAD-associated SNP may be attributable to functional alteration of
ADGRE3 protein in vascular inflammation.
The ZFP64 zinc finger protein (ZFP64) gene
located at 20q13.2 is expressed in various tissues and organs,
according to The Human Protein Atlas [http://www.proteinatlas.org/ (48,49)]. The
present longitudinal EWASs demonstrated that rs3746414 [G/A
(S451N)] was significantly associated with the prevalence of CAD.
ZFP64 protein may be a positive regulator of Toll-like
receptor-induced innate immune responses through tumor necrosis
factor (TNF)-α, interleukin-6 and interferon-β production in
macrophages (50). In fact,
Zfp64 knockdown mice exhibited significant reductions in the
production of these cytokines (50).
Thus, the association of ZFP64 with CAD may be attributable
to the effect of this gene on the development of coronary
atherosclerosis or thrombosis.
The Fas apoptotic inhibitory molecule 2
(FAIM2) gene is located at 12q13.12, and rs7132908 (G/A) in
this gene was associated with the prevalence of CAD. FAIM2
is predominantly expressed in the brain while moderate expressions
are also observed in other various tissues and organs, according to
The Human Protein Atlas. AIM2 inhibits Fas-mediated
apoptosis (51). The Fas cell surface
death receptor gene (FAS) serves important roles in the
regulation of the immune system through inducing apoptosis of
autoreactive or antigen-presenting T lymphocytes (52–54). Given
that FAIM2 regulates Fas-mediated apoptosis during immune
responses, rs7132908 in this gene may be associated with the
incidence of CAD through altering the regulation of immune or
inflammatory responses.
The TNF receptor superfamily member 11a
(TNFRSF11A) gene is located at chromosomal region 18q21.33,
and is expressed in various tissues and organs including the brain,
according to The Human Protein Atlas. In the present study,
rs884205 (G/T) of TNFRSF11A was significantly associated
with the prevalence of CI. TNFRSF11A and its ligand
(TNFRSF11) may be important regulators of the interaction
between T cells and dendritic cells that are involved in immune
surveillance (55,56). In mice, TNFRSF11/TNFRSF11A (also known
as RANKL/RANK) signaling could trigger inflammatory fever responses
in the central nervous system (55),
and may serve anti-inflammatory roles in ischemic brains (56). The rs884205 in TNFRSF11A may
thus influence the development of CI.
The exact functions of proteins encoded by the
family with sequence similarity 186 member A (FAM186A) and
long intergenic non-protein coding RNA 400 (LINC00400) genes
are unknown. However, it has been reported that rs9533425 located
near LINC00400 at chromosomal region 13q14.11 demonstrated a
significant association (P=1.9×10−9) with lag time to
fibrin clot formation in 2,100 subjects from the Twins United
Kingdom (TwinsUK) registry in stage 1 study, although the
association of this SNP was not replicated in stage 2 study
(28). In the present longitudinal
EWASs, rs1324015 of LINC00400 was significantly associated
with the prevalence of CI. According to LDpair in LDlink, rs1324015
was not in LD with rs9533425 in East Asian populations. Although
the functional relevance of the candidate SNP to the development of
cerebral atherosclerosis or thrombosis remains unclear,
LINC00400 may be a susceptibility locus for the incidence of
CI.
There were certain limitations in the present study.
First, the longitudinal EWASs were conducted in only a local
Japanese population, and the observed number of patients who were
affected by the target diseases was not sufficient. Thus,
replication of longitudinal EWASs in other Japanese populations or
other ethnic groups is required to verify the association of the
identified SNPs with the diseases of interest. However, to the best
of our knowledge, longitudinal data for CAD and CI in other
populations are unavailable at present. Second, no experiments for
functional analyses were conducted in the present study. Thus, the
functional relevance of the candidate SNPs identified by
longitudinal EWASs to the pathogenesis of the diseases of interest
remains unclear. Due to the lack of experiments for functional
analyses, the association of the SNPs identified in the present
study with CAD or CI should be interpreted with caution. Further
functional analyses are required to clarify the present
results.
In conclusion, rs4606855 of ADGRE3, rs3746414
of ZFP64 and rs7132908 of FAIM2 may be susceptibility
loci for CAD. Additionally, the SNPs rs6580741 of FAM186A,
rs1324015 of LINC00400 and rs884205 of TNFRSF11A may
be genetic determinants of CI. All minor alleles of the six
candidate SNPs exhibited an association with lower prevalence of
the associated diseases, compared with the corresponding major
alleles. This suggests that the minor alleles of each candidate SNP
may be protective against CAD or CI.
Acknowledgements
Not applicable.
Funding
The present study was supported by a research grant
from the Okasan Kato Culture Promotion Foundation (to YYasukochi),
a Kurata grant awarded by the Hitachi Global Foundation (grant no.
1323 to YYasukochi and YYamada), the Core Research for Evolutional
Science and Technology of the Japan Science and Technology Agency
(grant no. JPMRJCR1302 to YYamada, JS and IT), and by KAKENHI
grants from the Japan Society for the Promotion of Science (grant
no. JP17H00758 to IT and YYasukochi; grant no. JP15H04772 to
YYamada).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request; if justified, applicable parts of the data can be made
available in anonymized format.
Authors' contributions
YYasukochi contributed to analysis and
interpretation of the data, and to drafting of the manuscript. JS
and IT contributed to analysis and interpretation of the data as
well as revision of the manuscript. KK, MO, TF and HH each
contributed to acquisition of the data and revision of the
manuscript. YYamada contributed to conception and design of the
study, and to acquisition, analysis and interpretation of the data,
and revision of the manuscript. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
All procedures performed in studies involving human
participants were in accordance with the 1964 Helsinki declaration
and its later amendments or comparable ethical standards. The study
protocol was approved by the Committees on the Ethics of Human
Research of Mie University Graduate School of Medicine and Inabe
General Hospital (Mie, Japan). Informed consent was obtained from
all individual participants included in the study.
Patient consent for publication
All participants provided written informed consent
permitting publication of relevant data following anonymization of
personal information.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
GBD 2013 Mortality and Causes of Death
Collaborators: Global, regional, and national age-sex specific
all-cause and cause-specific mortality for 240 causes of death,
1990–2013: A systematic analysis for the Global Burden of Disease
Study 2013. Lancet. 385:117–171. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Benjamin EJ, Blaha MJ, Chiuve SE, Cushman
M, Das SR, Deo R, de Ferranti SD, Floyd J, Fornage M, Gillespie C,
et al: American Heart Association Statistics Committee and Stroke
Statistics Subcommittee: Heart disease and stroke statistics-2017
update: A report from the American Heart Association. Circulation.
135:e146–e603. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
National Center for Health Statistics:
Mortality multiple cause micro-data files, 2014: Public-use data
file and documentation: NHLBI tabulations.
|
|
4
|
Ministry of Health, Labour and Welfare,
Japan: Vital statistics.
|
|
5
|
Schunkert H, König IR, Kathiresan S,
Reilly MP, Assimes TL, Holm H, Preuss M, Stewart AFR, Barbalic M,
Gieger C, et al: Cardiogenics; CARDIoGRAM Consortium: Large-scale
association analysis identifies 13 new susceptibility loci for
coronary artery disease. Nat Genet. 43:333–338. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Golbus JR, Stitziel NO, Zhao W, Xue C,
Farrall M, McPherson R, Erdmann J, Deloukas P, Watkins H, Schunkert
H, et al: CARDIoGRAMplusC4D, Myocardial Infarction Genetics
(MIGen), Exome Sequencing Project and Early-Onset Myocardial
Infarction (ESP EOMI), and the Pakistan Risk of Myocardial
Infarction Study (PROMIS) Consortia*: Common and rare genetic
variation in CCR2, CCR5, or CX3CR1 and risk of atherosclerotic
coronary heart disease and glucometabolic traits. Circ Cardiovasc
Genet. 9:250–258. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lu X, Wang L, Chen S, He L, Yang X, Shi Y,
Cheng J, Zhang L, Gu CC, Huang J, et al: Coronary ARtery DIsease
Genome-Wide Replication And Meta-Analysis (CARDIoGRAM) Consortium:
Genome-wide association study in Han Chinese identifies four new
susceptibility loci for coronary artery disease. Nat Genet.
44:890–894. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nikpay M, Goel A, Won H-H, Hall LM,
Willenborg C, Kanoni S, Saleheen D, Kyriakou T, Nelson CP, Hopewell
JC, et al: A comprehensive 1,000 Genomes-based genome-wide
association meta-analysis of coronary artery disease. Nat Genet.
47:1121–1130. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wakil SM, Ram R, Muiya NP, Mehta M, Andres
E, Mazhar N, Baz B, Hagos S, Alshahid M, Meyer BF, et al: A
genome-wide association study reveals susceptibility loci for
myocardial infarction/coronary artery disease in Saudi Arabs.
Atherosclerosis. σ245:62–70. 2016. View Article : Google Scholar
|
|
10
|
Yamada Y, Fuku N, Tanaka M, Aoyagi Y,
Sawabe M, Metoki N, Yoshida H, Satoh K, Kato K, Watanabe S, et al:
Identification of CELSR1 as a susceptibility gene for ischemic
stroke in Japanese individuals by a genome-wide association study.
Atherosclerosis. 207:144–149. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kubo M, Hata J, Ninomiya T, Matsuda K,
Yonemoto K, Nakano T, Matsushita T, Yamazaki K, Ohnishi Y, Saito S,
et al: A nonsynonymous SNP in PRKCH (protein kinase C η) increases
the risk of cerebral infarction. Nat Genet. 39:212–217. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gouveia LO, Sobral J, Vicente AM, Ferro JM
and Oliveira SA: Replication of the CELSR1 association with
ischemic stroke in a Portuguese case-control cohort.
Atherosclerosis. 217:260–262. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Akinyemi RO, Ovbiagele B, Akpalu A,
Jenkins C, Sagoe K, Owolabi L, Sarfo F, Obiako R, Gebreziabher M,
Melikam E, et al: SIREN Investigators as Members of the H3Africa
Consortium: Stroke genomics in people of African ancestry: Charting
new paths. Cardiovasc J Afr. 26(Suppl 1): S39–S49. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Meschia JF, Nalls M, Matarin M, Brott TG,
Brown RD Jr, Hardy J, Kissela B, Rich SS, Singleton A, Hernandez D,
et al: Siblings With Ischemic Stroke Study Investigators: Siblings
with ischemic stroke study: Results of a genome-wide scan for
stroke loci. Stroke. 42:2726–2732. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kilarski LL, Achterberg S, Devan WJ,
Traylor M, Malik R, Lindgren A, Pare G, Sharma P, Slowik A, Thijs
V, et al: GARNET Collaborative Research Group, Wellcome Trust Case
Control Consortium 2, Australian Stroke Genetic Collaborative, the
METASTROKE Consortium, and the International Stroke Genetics
Consortium: Meta-analysis in more than 17,900 cases of ischemic
stroke reveals a novel association at 12q24.12. Neurology.
83:678–685. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yamada Y, Matsui K, Takeuchi I and
Fujimaki T: Association of genetic variants with dyslipidemia and
chronic kidney disease in a longitudinal population-based genetic
epidemiological study. Int J Mol Med. 35:1290–1300. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yamada Y, Matsui K, Takeuchi I, Oguri M
and Fujimaki T: Association of genetic variants of the α-kinase 1
gene with type 2 diabetes mellitus in a longitudinal
population-based genetic epidemiological study. Biomed Rep.
3:347–354. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yamada Y, Matsui K, Takeuchi I and
Fujimaki T: Association of genetic variants with coronary artery
disease and ischemic stroke in a longitudinal population-based
genetic epidemiological study. Biomed Rep. 3:413–419. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yamada Y, Matsui K, Takeuchi I, Oguri M
and Fujimaki T: Association of genetic variants with hypertension
in a longitudinal population-based genetic epidemiological study.
Int J Mol Med. 35:1189–1198. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Oguri M, Fujimaki T, Horibe H, Kato K,
Matsui K, Takeuchi I and Yamada Y: Obesity-related changes in
clinical parameters and conditions in a longitudinal
population-based epidemiological study. Obes Res Clin Pract.
11:299–314. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Grove ML, Yu B, Cochran BJ, Haritunians T,
Bis JC, Taylor KD, Hansen M, Borecki IB, Cupples LA, Fornage M, et
al: Best practices and joint calling of the HumanExome BeadChip:
The CHARGE Consortium. PLoS One. 8:e680952013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Price AL, Patterson NJ, Plenge RM,
Weinblatt ME, Shadick NA and Reich D: Principal components analysis
corrects for stratification in genome-wide association studies. Nat
Genet. 38:904–909. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
R Development Core Team: R: A language and
environment for statistical computing. Vienna, Austria; 2016
|
|
24
|
RStudio Team: RStudio: Integrated
development environment for R. RStudio, Inc. Boston, MA; 2015
|
|
25
|
Liang KY and Zeger SL: Longitudinal data
analysis using generalized linear models. Biometrika. 73:13–22.
1986. View Article : Google Scholar
|
|
26
|
Hanley JA, Negassa A, Edwardes MD and
Forrester JE: Statistical analysis of correlated data using
generalized estimating equations: An orientation. Am J Epidemiol.
157:364–375. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Halekoh U, Højsgaard S and Yan J: The R
package geepack for generalized estimating equations. J Stat Softw.
15:1–11. 2006. View Article : Google Scholar
|
|
28
|
Williams FMK, Carter AM, Hysi PG,
Surdulescu G, Hodgkiss D, Soranzo N, Traylor M, Bevan S, Dichgans
M, Rothwell PMW, et al: EuroCLOT Investigators; Wellcome Trust Case
Control Consortium 2; MOnica Risk, Genetics, Archiving and
Monograph; MetaStroke; International Stroke Genetics Consortium:
Ischemic stroke is associated with the ABO locus: The EuroCLOT
study. Ann Neurol. 73:16–31. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Holmen OL, Zhang H, Fan Y, Hovelson DH,
Schmidt EM, Zhou W, Guo Y, Zhang J, Langhammer A, Løchen ML, et al:
Systematic evaluation of coding variation identifies a candidate
causal variant in TM6SF2 influencing total cholesterol and
myocardial infarction risk. Nat Genet. 46:345–351. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J R Stat Soc B. 57:289–300. 1995.
|
|
31
|
Sitlani CM, Rice KM, Lumley T, McKnight B,
Cupples LA, Avery CL, Noordam R, Stricker BHC, Whitsel EA and Psaty
BM: Generalized estimating equations for genome-wide association
studies using longitudinal phenotype data. Stat Med. 34:118–130.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Voorman A, Rice K and Lumley T: Fast
computation for genome-wide association studies using boosted
one-step statistics. Bioinformatics. 28:1818–1822. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
1000 Genomes Project Consortium; Abecasis
GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME
and McVean GA: A map of human genome variation from
population-scale sequencing. Nature. 467:1061–1073. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Machiela MJ and Chanock SJ: LDlink: A
web-based application for exploring population-specific haplotype
structure and linking correlated alleles of possible functional
variants. Bioinformatics. 31:3555–3557. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Leslie R, O'Donnell CJ and Johnson AD:
GRASP: Analysis of genotype-phenotype results from 1390 genome-wide
association studies and corresponding open access database.
Bioinformatics. 30:i185–i194. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Piñero J, Queralt-Rosinach N, Bravo À,
Deu-Pons J, Bauer-Mehren A, Baron M, Sanz F and Furlong LI:
DisGeNET: A discovery platform for the dynamical exploration of
human diseases and their genes. Database (Oxford).
2015:bav0282015.PubMed/NCBI
|
|
37
|
MacArthur J, Bowler E, Cerezo M, Gil L,
Hall P, Hastings E, Junkins H, McMahon A, Milano A, Morales J, et
al: The new NHGRI-EBI Catalog of published genome-wide association
studies (GWAS Catalog). Nucleic Acids Res. 45(D1): D896–D901. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Warde-Farley D, Donaldson SL, Comes O,
Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT,
et al: The GeneMANIA prediction server: Biological network
integration for gene prioritization and predicting gene function.
Nucleic Acids Res. 38:W214–220. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Montojo J, Zuberi K, Rodriguez H, Bader GD
and Morris Q: GeneMANIA: Fast gene network construction and
function prediction for Cytoscape. F1000Res. 3:1532014.PubMed/NCBI
|
|
40
|
Montojo J, Zuberi K, Rodriguez H, Kazi F,
Wright G, Donaldson SL, Morris Q and Bader GD: GeneMANIA Cytoscape
plugin: Fast gene function predictions on the desktop.
Bioinformatics. 26:2927–2928. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sherry ST, Ward MH, Kholodov M, Baker J,
Phan L, Smigielski EM and Sirotkin K: dbSNP: The NCBI database of
genetic variation. Nucleic Acids Res. 29:308–311. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Warnes G, Gorjanc G, Leisch F and Man M:
Genetics: Population Genetics. 2013
|
|
44
|
Stacey M, Lin HH, Hilyard KL, Gordon S and
McKnight AJ: Human epidermal growth factor (EGF) module-containing
mucin-like hormone receptor 3 is a new member of the EGF-TM7 family
that recognizes a ligand on human macrophages and activated
neutrophils. J Biol Chem. 276:18863–18870. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Libby P: Inflammation in atherosclerosis.
Nature. 420:868–874. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Greaves DR and Gordon S: Immunity,
atherosclerosis and cardiovascular disease. Trends Immunol.
22:180–181. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Libby P: Inflammation in atherosclerosis.
Arterioscler Thromb Vasc Biol. 32:2045–2051. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Uhlén M, Fagerberg L, Hallström BM,
Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C,
Sjöstedt E, Asplund A, et al: Proteomics. Tissue-based map of the
human proteome. Science. 347:12604192015.
|
|
49
|
Thul PJ, Åkesson L, Wiking M, Mahdessian
D, Geladaki A, Ait Blal H, Alm T, Asplund A, Björk L, Breckels LM,
et al: A subcellular map of the human proteome. Science.
356:eaal33212017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wang C, Liu X, Liu Y, Zhang Q, Yao Z,
Huang B, Zhang P, Li N and Cao X: Zinc finger protein 64 promotes
Toll-like receptor-triggered proinflammatory and type I interferon
production in macrophages by enhancing p65 subunit activation. J
Biol Chem. 288:24600–24608. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Somia NV, Schmitt MJ, Vetter DE, van
Antwerp D, Heinemann SF and Verma IM: LFG: An anti-apoptotic gene
that provides protection from Fas-mediated cell death. Proc Natl
Acad Sci USA. 96:12667–12672. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Strasser A, Jost PJ and Nagata S: The many
roles of FAS receptor signaling in the immune system. Immunity.
30:180–192. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Schütz C, Oelke M, Schneck JP, Mackensen A
and Fleck M: Killer artificial antigen-presenting cells: The
synthetic embodiment of a ‘guided missile’. Immunotherapy.
2:539–550. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lenardo MJ: Fas and the art of lymphocyte
maintenance. J Exp Med. 183:721–724. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Hanada R, Leibbrandt A, Hanada T, Kitaoka
S, Furuyashiki T, Fujihara H, Trichereau J, Paolino M, Qadri F,
Plehm R, et al: Central control of fever and female body
temperature by RANKL/RANK. Nature. 462:505–509. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Shimamura M, Nakagami H, Osako MK,
Kurinami H, Koriyama H, Zhengda P, Tomioka H, Tenma A, Wakayama K
and Morishita R: OPG/RANKL/RANK axis is a critical inflammatory
signaling system in ischemic brain in mice. Proc Natl Acad Sci USA.
111:8191–8196. 2014. View Article : Google Scholar : PubMed/NCBI
|