Introduction
The Ras superfamily of small GTPases contains more
than 170 members divided into 5 subfamilies, namely Ras, Rho, Rab,
Ran and Arf (1–5). Upon ligand stimulation, the GTPases are
activated by membrane-associated receptors and transmit signals to
their downstream effectors, which regulate cell proliferation,
differentiation, cytoskeletal regulation and vesicle trafficking
(1,3).
Following signal transmission, the active GTP-Ras
must become inactive GDP-Ras to cease signaling. However, the
intrinsic GTPase activity of Ras proteins is weak, requiring Ras
GTPase activating protein (RasGAP) for efficient conversion of
GTP-Ras to GDP-Ras (6). RasGAP has
emerged as a novel class of tumor suppressor protein and a
potential therapeutic target for cancer (6,7).
Therefore, it is important to identify the specific GAPs for each
small GTPase. Although 14 GAPs for the Ras subfamily have
previously been reported (6–10), the specific or redundant functions of
these GAPs are unclear. The first RasGAP identified was p120
rasGAP, also known as RAS p21 protein activator 1 (RASA1). This
protein is widely expressed, independent of cell type and tissue
distribution (11,12). Subsequently, neurofibromatosis type 1
was identified as a RasGAP (13,14). The
remaining 12 GAPs are affiliated with the GAP1 and synaptic GAP
(SynGAP) family (8,9). Ras activating protein-like 3 (RASAL3) is
a SynGAP family member that is predominantly expressed in
hematopoietic cells, including Jurkat-T cells (15,16). Since
RASAL3 is the most recently identified RasGAP, the specific role of
this molecule in regulation of GTPase activity remains to be
elucidated.
Among the Rho subfamily, which contains Rho, Rac and
Cdc42 GTPases, Rac2 is specifically expressed in hematopoietic
cells (17) and is known to be
involved in chronic myelogenous leukemia (CML) via direct
activation by the p210-breakpoint cluster region (BCR)-Abelson
murine leukemia viral oncogene homolog 1 (ABL) fusion protein
(18–20). Furthermore, knockout of Rac2 in mast
cells resulted in defective AKT activation, followed by increased
apoptosis upon agonist stimulation (18,21). To
date the only GAP established for Rac2 is BCR/active bcr-related
(ABR) in leukemia (22,23). Thus it appears necessary to determine
the presence of Rac2 GAP in normal hematopoietic cells, since
GTP-Rac2 triggers the AKT pathway in cell survival signaling
(18,20,21).
Although RASAL3 has been identified as a RasGAP
(15,16), the level of GTPase activating activity
in vitro was not as prominent compared to that of Rho
GTPases including RhoA and Rac1. To determine the existence of a
preferential target of RASAL3, the Rho subfamily of GTPases were
examined. Rac2 was detected as a target GTPase for RASAL3 using an
in vitro assay system. Overall it was demonstrated that
RASAL3 may be a Rac2-selective GAP that has no apparent activity
with Rac1. The current results may be useful to clarify the
regulation mechanism of Rac2-mediated signaling at the cellular
level.
Materials and methods
Reagents
Glutathione sepharose 4B (GSH) was obtained from GE
Healthcare Life Sciences (Uppsala, Sweden). Reduced glutathione and
isopropyl β-D-1-thiogalactopyranoside (IPTG) were purchased from
Sigma-Aldrich (Merck KGaA, Darmstadt, Germany). RhoGAP Assay
Biochem kit (cat. no. BK105) containing H-Ras, RhoA, Rac1 and
Cdc42, separate His-Rac2 protein (cat. no. RC02) and CytoPhos
Reagent were purchased from Cytoskeleton, Inc. (Denver, CO, USA).
Monoclonal anti-FLAG M2 antibody (cat. no. A2220) and 3× FLAG
peptide (cat. no. F4799) were purchased from Sigma-Aldrich (Merck
KGaA). Pre-stained molecular weight (MW) marker was purchased from
Bio-Rad Laboratories, Inc., (Hercules, CA, USA).
Vector constructs
Full-length cDNA of human RASAL3 in
pFLAG-CMV/hRASAL3, constructed by Professor Masahiro Fujii (Niigata
University, Niigata, Japan) (16),
were used as templates for polymerase chain reaction (PCR)
amplification of RasGAP domain. For glutathione S transferase (GST)
fusion construction, amplified cDNAs encoding the RasGAP domain of
RASAL3 (amino acids 421–748) were ligated into the
EcoRI/XhoI (Takara Bio, Inc., Otsu, Japan)
restriction sites of pGEX-5×-1. For PCR amplification, the primers
5′-ACAGAATTCGCGCGTCGCCTGCGCGTG-3′ (forward) and
5′-ACACTCGAGTCACATTGGCACTGACACAAG-3′ (reverse) were used for the
RASAL3 GAP domain (underlined sequences represent the EcoRI
and XhoI digestion sites, respectively). PCR amplification
was performed using a PCR amplification kit (Takara Bio, Inc.; cat.
no. R011) and involved an initial step of denaturation (94°C for 5
min), followed by 35 cycles of denaturation (94°C for 30 sec),
annealing (57°C for 30 sec) and extension (72°C for 30 sec). Final
extension was performed at 72°C for 10 min.
Protein expression and
purification
GST-RASAL3-GAP fusion proteins were expressed in
Escherichia coli (E. coli) DH5α (Takara Bio, Inc.).
Protein expression was induced with 200 µM IPTG overnight at room
temperature (~18°C). Ultrasonic-disrupted cell lysates were
centrifuged at 12,000 × g and 4°C for 20 min, and the supernatants
were incubated with GSH beads to collect GST-fused proteins in
Tris-Cl buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM
dithiothreitol, 2 mM β-mercaptoethanol). Fusion protein-bound beads
were washed with the same Tris-Cl buffer, and the fusion proteins
were eluted with 30 mM reduced glutathione solution in Tris-Cl
buffer (pH 7.5). The purified proteins were concentrated using
Amicon Ultra centrifugal filters (Merck KGaA, Darmstadt, Germany)
at 4°C. The resulting proteins were used for further experiments or
immediately frozen in liquid nitrogen and stored at −70°C. The
purified GAP domain protein of p50 RhoGAP (also known as Cdc42 GAP)
was supplied with the RhoGAP assay kit.
FLAG-tagged RASAL3 proteins were purified from the
lysates of HEK-293 cells (American Type Culture Collection,
Manassas, VA, USA) that had been transiently transfected with the
pFLAG-CMV/hRASAL3 vector. Transfection was performed according to
the manufacturer's specification of a Lipofectamine 3000
transfection kit (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). Following lysate centrifugation at 12,000 × g
and 4°C for 20 min, anti-FLAG M2 affinity gel (Sigma-Aldrich; Merck
KGaA) was added to the supernatant and further incubated on a
shaker at 4°C for 3 h. FLAG-RASAL3 was eluted by incubating the
beads at 4°C for 30 min with 200 ng/ml 3× FLAG peptide in
Tris-buffered saline (pH 7.4).
In vitro binding assay
Purified His-Rac2 protein (2 µg) was mixed with 50
µl 1× GAP assay reaction buffer (Cytoskeleton, Inc.) in the
presence of 50 µM GDP, GTP or GTPγS (Sigma-Aldrich; Merck KGaA) at
30°C for 10 min. A 50 µl slurry of beads bound to GST-RASAL3-GAP
fusion protein was added to each guanosine nucleotide-charged Rac2
GTPase aliquot. Each 100 µl reaction mixture was incubated at room
temperature (~18°C) for 30 min with gentle shaking. The beads were
washed with an equal volume (100 µl) of 1× reaction buffer 3 times
to remove unbound proteins. The bound proteins were resolved on an
8–16% gradient SDS-PAGE and visualized by Coomassie Brilliant Blue
staining performed at room temperature for 1 h. Additionally, in
vitro binding assays were performed between the GAP domain of
RASAL3 and various small G-proteins (H-Ras, RhoA, Rac1, Rac2 and
Cdc42). In this case, 1× reaction buffer was replaced by 50 mM
Tris-Cl (pH 7.5) containing 150 mM NaCl and 20 µM GTP. The bound
proteins were washed with the same buffer and resolved on SDS-PAGE.
The quantities of GTPase pulled down with GST-RASAL3 GAP domain
were analyzed by relative image density (Quantity One software ver.
4.6.9; Bio-Rad Laboratories, Inc.)
GAP activity assay
GAP activity assays were performed using the RhoGAP
Assay Biochem kit, which contains GST-Rho-GAP fusion protein.
Briefly, GTPase activity was measured by monitoring the free γPi
release with or without purified GAP molecules via absorbance at
650 nm. Single turnover GTPase reactions (30 µl total volume per
reaction) were initiated by the addition of 200 µM GTP. The
reactions were performed at 37°C for 15 min. Subsequently, 150 µl
CytoPhos reagent was added to the reactions and incubated for a
further 10 min at room temperature. The absorbance of the reactions
was measured at 650 nm. The reaction time was controlled to 5, 10
or 15 min according to manufacturer's recommendations. Using each
GTPase as a control, absorbance value was measured in the presence
of GST alone.
Western immunoblotting
Purified FLAG-RASAL3 solution from pFLAG-CMV/hRASAL3
transfected HEK-293 cell lysates (minimal volume 10 µl) was
resolved on 10% SDS-PAGE, and transferred to nitrocellulose
membrane for 4 h at 200 mA current. The blot was blocked with 5%
non-fat skim milk for 1 h at room temperature, and probed with
monoclonal anti-FLAG M2 antibody (1,000× dilution in 5% skimmed
milk) for 2 h at room temperature. Following extensive washing with
Tris-buffered Tween-20 (50 mM Tris-Cl, 150 mM NaCl, 0.5% Tween-20,
pH 7.5), the membrane was re-probed with hoarse-radish peroxidase
conjugated goat anti-mouse IgG antibody (Jackson ImmunoResearch
Laboratories, Inc., West Grove, PA, USA) for 1 h at room
temperature and visualized with an enhanced chemiluminescence
detection system (GE Healthcare Life Sciences, Little Chalfont,
UK).
Statistical analysis
Experimental data are presented as the mean ±
standard deviation and were analyzed by one-way analysis of
variance and Student-Newman-Keuls post-hoc testing with SPSS 22.0
software (IBM Corp., Armonk, NY, USA). P<0.05 was considered to
indicate statistical significance.
Results and Discussion
Preparation of RASAL3 GAP
proteins
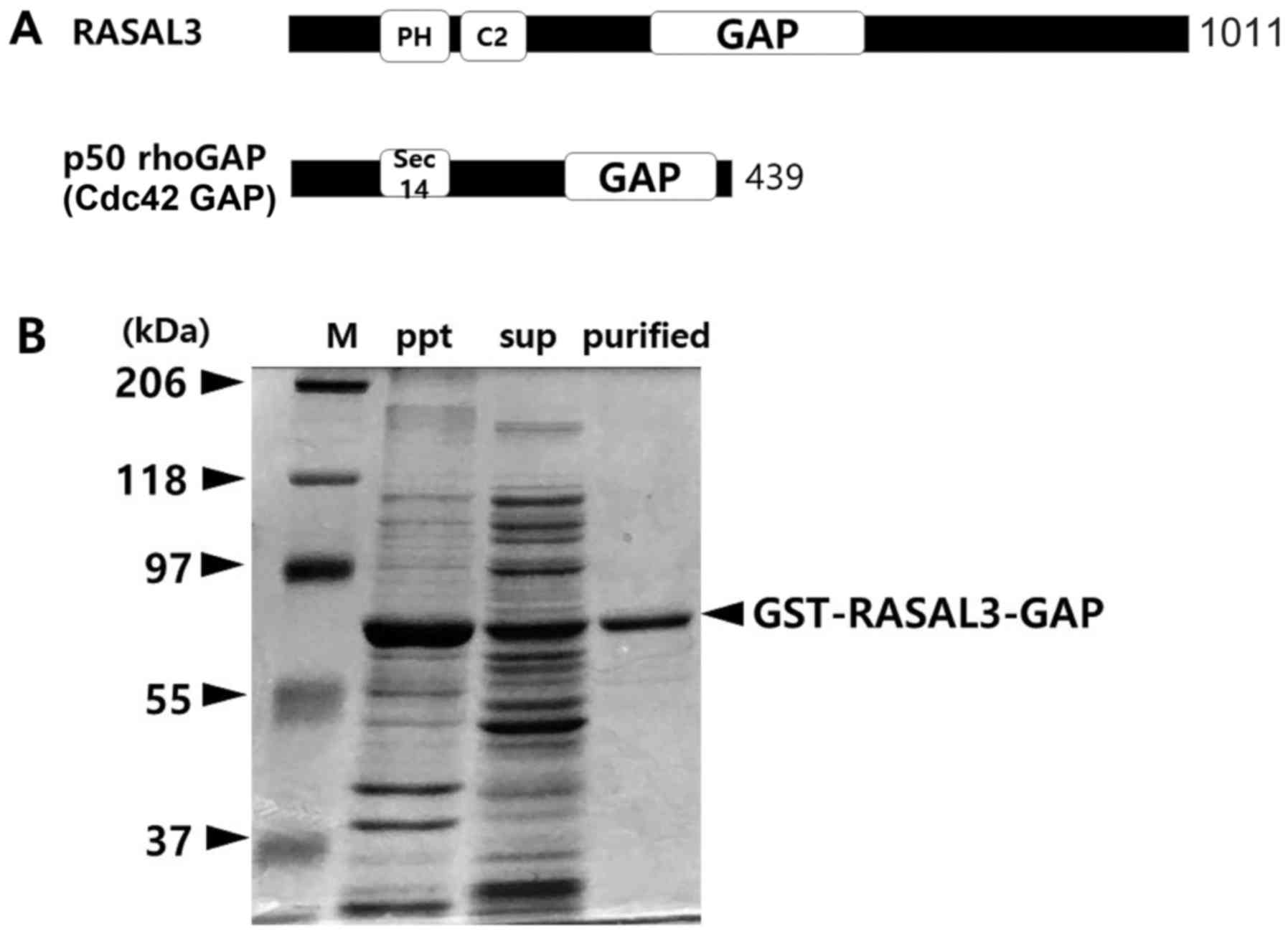
RASAL3 includes pleckstrin homology and C2 domains
close to its N-termini, while its GAP domain is located in the
central portion of the molecule (Fig.
1A) (7). GST-RASAL3-GAP (421–748
aa) fusion protein expressed in E. coli were purified and
resolved by 10% SDS-PAGE. In a typical induction system at 37°C,
the proteins were almost pelleted by centrifugation despite the
fusion protein being efficiently induced in E. coli by 200
µM IPTG. The method was then modified as described in the
‘Materials and methods’. A 64 kDa GST-RASAL3-GAP fusion protein was
efficiently harvested by the modified methods (Fig. 1B).
GST-RASAL3 stimulates Rac2 GTPase
in vitro
Recently, our group identified that a synthesized
chemical directly bound to RASAL3 in Jurkat T-cell lysates (data to
be published). During examination of RASAL3 GAP activity with
chemical regulators, it was noted that the bacterially expressed
GAP domain of RASAL3 reproducibly accelerated Rac2 GTPase activity
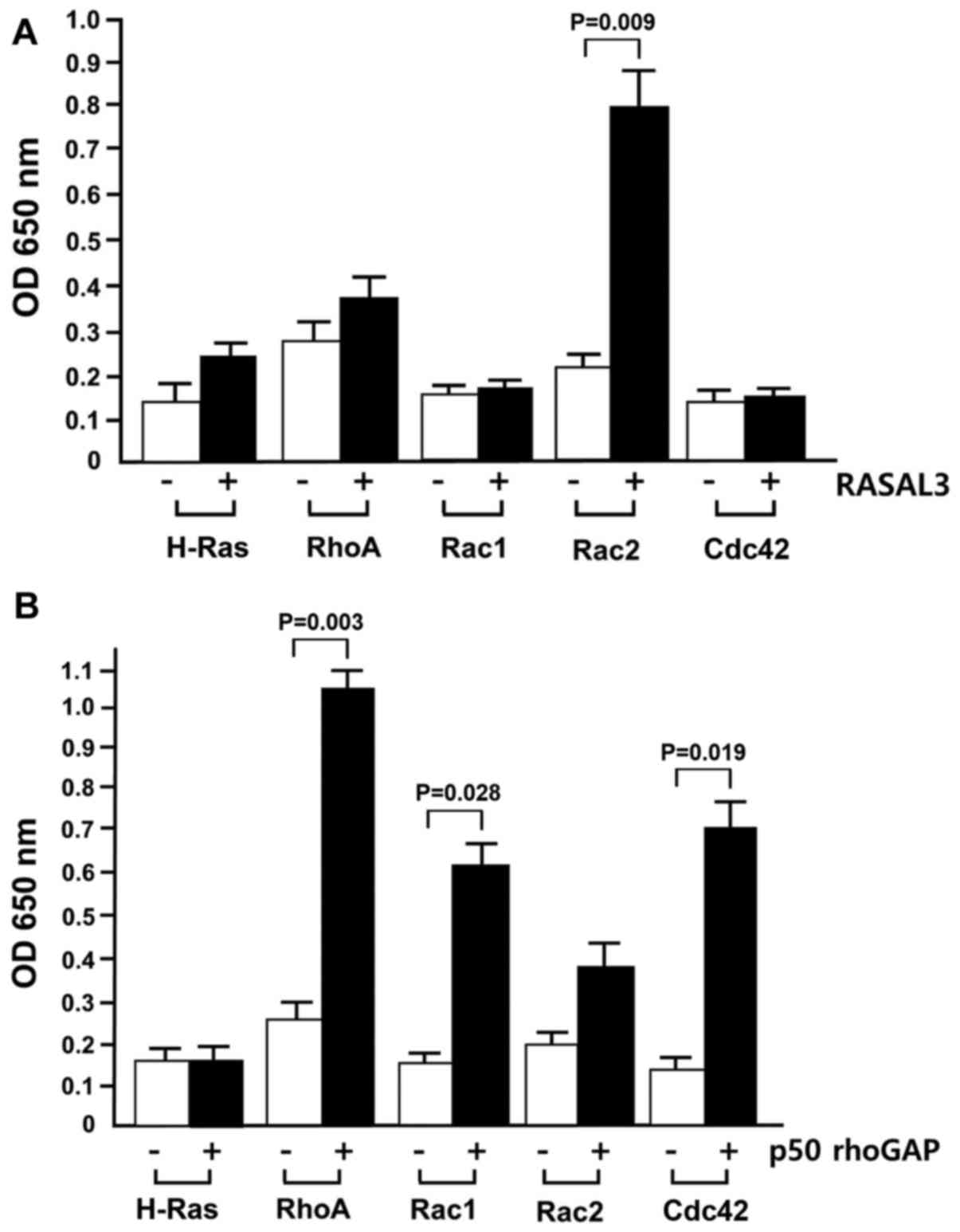
(Fig. 2A).
Subsequently GST-RASAL3-GAP activity
with H-Ras and Rho GTPases was examined
As depicted in Fig.
2A, RASAL3 did not markedly stimulate RhoA, Rac1 or Cdc2 GTPase
activity, while it notably increased Rac2 GTPase activity.
Therefore, RASAL3 was taken to preferentially stimulate Rac2, a Rho
family GTPase, but not other Rho family GTPases. Although two
recent reports demonstrated that RASAL3 stimulates p21ras GTPase
activity (15,16), the present results indicated,
seemingly for the first time, that RASAL3 preferentially stimulates
GTP hydrolysis in Rac2. Using each GTPase as controls, the measured
absorbance value did not change in the presence of GST alone (data
not shown).
In the current assay system, RASAL3 increased H-Ras
GAP activity ~2-fold, while it accelerated Rac2 GTPase activity
>3.5-fold compared with Rac2 GTPase activity without RASAL3.
This result suggests that RASAL3 may be a pivotal regulator of
AKT-mediated signaling via Rac2 GTPase regulation in hematopoietic
cells.
The GAP domains of RasGAP and RhoGAP are
structurally related (24), and their
GTPase tertiary folding patterns appear to be similar (6,25). In
particular, an arginine residue at position 789 in RASA1 or
position 305 in p50 rhoGAP is reportedly capable of penetrating
into the active site of each corresponding GTPase (25,26).
However, the current results suggested that RasGAP (RASAL3) may
stimulate GTP hydrolysis of Rac2, while RhoGAP (p50 rhoGAP) did not
exhibit any GAP activity with H-Ras. Therefore, it is conceivable
that Ras and Rac2 have structural similarities in their active
site. Although Rac1 and Rac2 exhibit >92% amino acid sequence
homology (27), RASAL3 activated Rac2
but not Rac1, suggesting somewhat different active site
structures.
The p50 rhoGAP GAP domain exhibited highest GAP
activity for RhoA and Cdc42, while it exhibited no activity for
H-Ras (Fig. 2B). The p50 rhoGAP
domain increased Rac2 GTPase activation by <2-fold, while GTPase
activation of RhoA, Rac1 and CDC42 increased ~4-, 3.5- and 3-fold,
respectively. These results suggest that p50 rhoGAP stimulates
GTPase activity of only the Rho family small G-proteins.
Furthermore, it is consistent with the report that human p50 rhoGAP
has in vitro specificity only for Cdc42, Rac1 and RhoA
(5).
Complete RASAL3 exhibits a similar
activity to the GST-GAP domain
To clarify any difference in GAP activities between
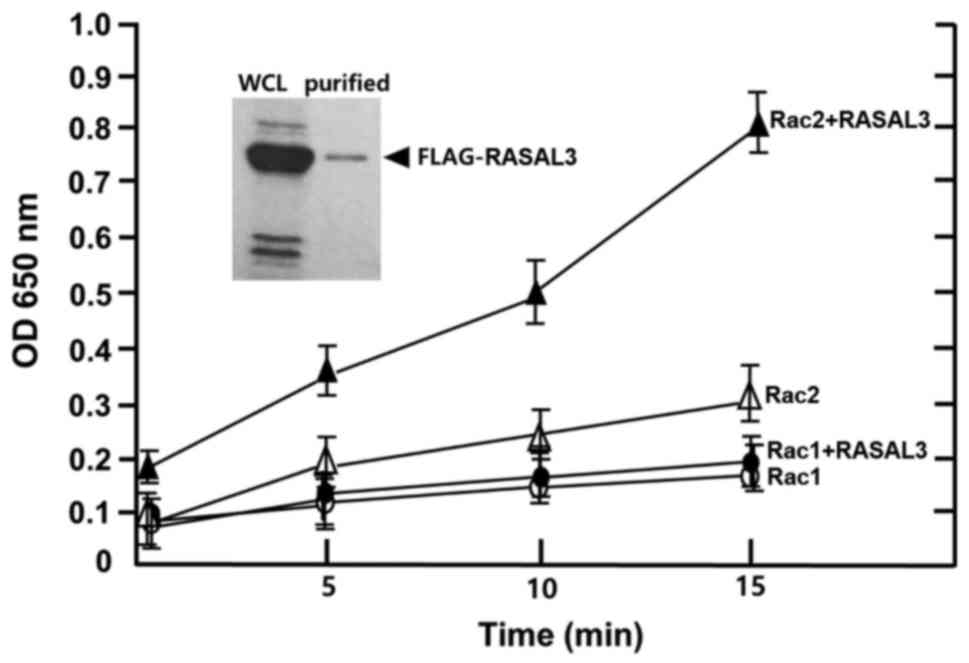
the GAP domain alone and the intact GAP molecule, intact RASAL3 was
expressed and purified from HEK-293 cells using the
pFLAG-CMV/hRASAL3 vector (16). The
GTPase activity of Rac1 and Rac2 was measured in the presence and
absence of FLAG-RASAL3 at certain time intervals. As depicted in
Fig. 3, Rac1 GTPase activity did not
increase in the presence of FLAG-RASAL3, while Rac2 GTPase activity
was markedly accelerated in the presence of FLAG-RASAL3. The
stimulation rate was similar to that of GST-RASAL3-GAP (Fig. 2A), suggesting that both the
FLAG-RASAL3 and GST-RASAL3-GAP domain have similar activating
activity in vitro. The absorbance increased several-fold
(~3.5-fold after 15 min) in a time-dependent manner in the presence
of FLAG-RASAL3. This contrasts the results of Rac1, which exhibited
only basal level changes even in the presence of FLAG-RASAL3
(Fig. 3). Therefore, specific GTP
hydrolysis of Rac2 in hematopoietic cells may be potently
stimulated by RASAL3.
Rac2 interacts with
GST-RASAL3-GAP
To investigate whether the GAP domains of RASAL3
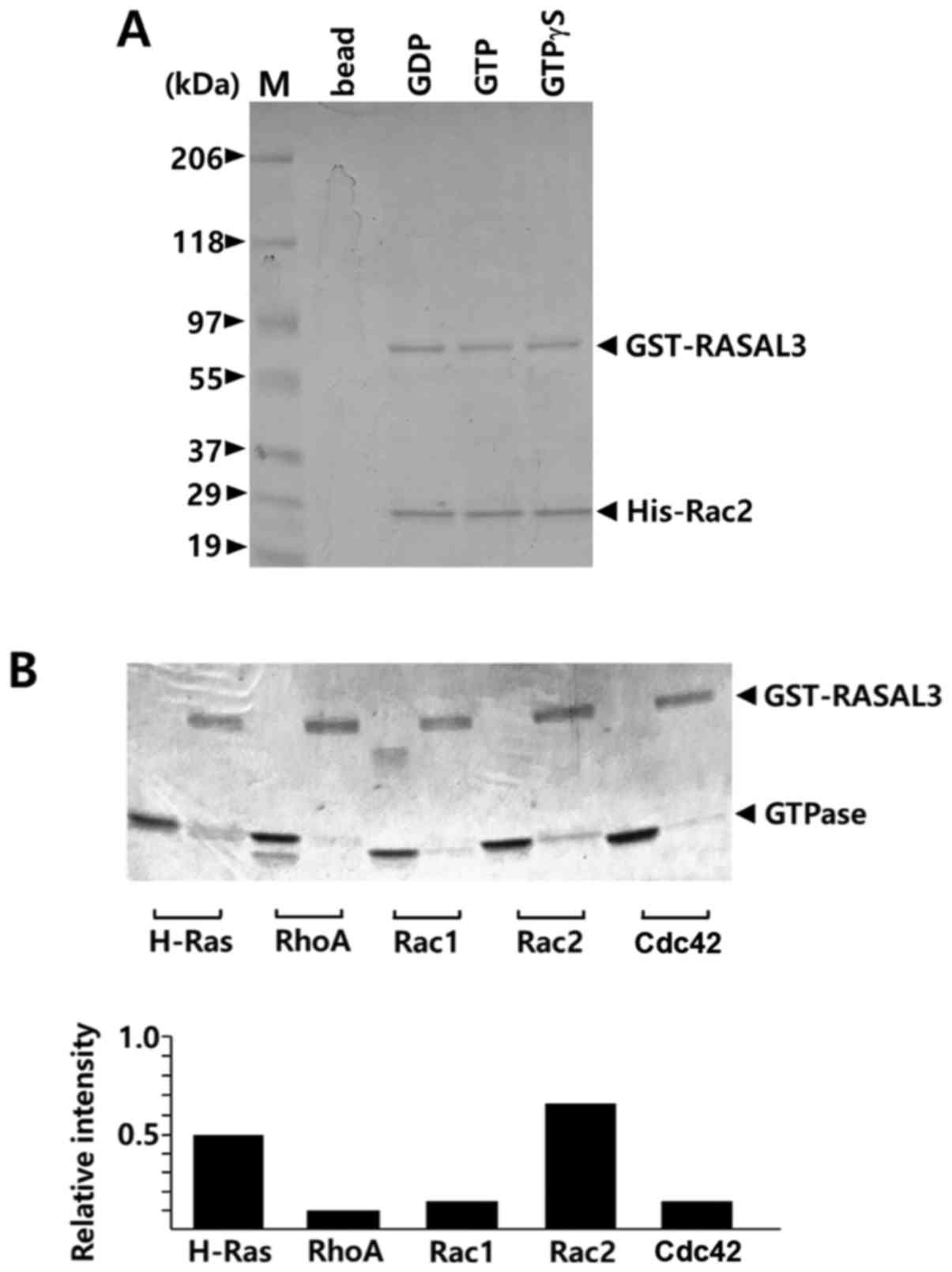
interact with Rac2, an in vitro binding assay was performed
with GST-GAP fusion proteins. In GST pull-down experiments, Rac2
directly associated with GST-RASAL3-GAP (Fig. 4A). GST-RASAL3-GAP did not exhibit any
difference in affinity to either GDP-Rac2 or GTP-Rac2. Generally,
GTP-loaded small G-proteins exhibit higher affinity to GAP domains
than GDP-loaded proteins (1). In this
context, the current results may suggest that the preloaded GTP was
efficiently degraded to GDP during binding in the presence of
GST-GAP molecules at high concentration (50 µl). Therefore, the
Rac2 fractions pulled down with GST-RASAL3-GAP were likely Rac2
alone or GDP-Rac2. In addition, it was examined whether the GAP
domain of RASAL3 binds to other G-proteins including H-Ras, RhoA,
Rac1 and Cdc42 in the presence of 20 µM GTP. As presented in
Fig. 4B, the Rac2 band exhibited the
strongest intensity among the G-proteins pulled down with
GST-RASAL3-GAP. Rac2 exhibited marginally higher affinity than
H-Ras for the GST-RASAL3 GAP domain, while other Rho subfamily
GTPases exhibited seemingly basal level affinities. This result is
consistent with the result of Fig.
2A, which overall suggests that the RASAL3 GAP domain
preferentially stimulates Rac2 GTPase activity.
In conclusion, in the present study, it was
suggested that RASAL3 may be responsible at least in part for
controlling AKT-mediated survival signaling in hematopoietic cells
via Rac2 GTPase regulation. Although results were obtained in
vitro, the current findings may provide insight into the
regulation mechanism of Rac2-AKT signaling at the cellular level,
since the only GAP for Rac2 identified to date is BCR/ABR in
Philadelphia chromosome-positive leukemia (22,23).
Patients with CML have lymphocytes containing the Philadelphia
chromosome, caused by gene fusion between BCR and ABL. Rac GTPase
in such lymphocytes has been revealed to be the fully activated
form (GTP-Rac) compared with in normal lymphocytes (GDP-Rac)
(28). Furthermore, it was
demonstrated that CML phenotype was significantly attenuated by
depletion of Rac2 in a mouse model (28), suggesting that Rac2 is a crucial
regulator in CML disease. In this sense, it is conceivable that
RASAL3, as a Rac2 inactivating protein, may be a possible target
for CML disease.
Acknowledgements
The authors are grateful to Professor Masahiro Fujii
(Niigata University, Niigata, Japan) for providing the
pFLAG-CMV/hRASAL3 construct.
Funding
No funding was received.
Availability of data and materials
The analyzed data sets obtained during the study are
available from the corresponding author on reasonable request.
Authors' contributions
YS and YWK analyzed GAP activity. HK, NS and TSK
purified GST fusion protein. JHC purified FLAG-RASAL3 proteins from
HEK-293 cells. TKK and JSC were primarily responsible for writing
of the manuscript. JSC contributed to overall design, acquisition
and interpretation of data. All authors approved the manuscript
submission.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
RASA1
|
RAS p21 protein activator 1
|
|
RASAL3
|
Ras activating protein-like 3
|
|
GAP
|
GTPase activating protein
|
|
SynGAP
|
synaptic GTPase activating protein
|
|
GST
|
glutathione S transferase
|
|
BCR/ABR
|
breakpoint cluster region/active
bcr-related
|
|
ABL
|
Abelson murine leukemia viral oncogene
homolog 1
|
|
CML
|
chronic myelogenous leukemia
|
|
IPTG
|
isopropyl
β-D-1-thiogalactopyranoside
|
|
GSH
|
glutathione sepharose 4B
|
References
|
1
|
Colicelli J: Human RAS superfamily
proteins and related GTPases. Sci STKE. 2004(re13):
RE132004.PubMed/NCBI
|
|
2
|
Csépányi-Kömi R, Sáfár D, Grósz V, Tarján
ZL and Ligeti E: In silico tissue-distribution of human Rho family
GTPase activating proteins. Small GTPases. 4:90–101. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Moon SY and Zheng Y: Rho GTPase-activating
proteins in cell regulation. Trends Cell Biol. 13:13–22. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Reiner D and Lundquist EA: Small GTPases.
WomBook. May 24–2016.(Epub ahead of print).
|
|
5
|
Tcherkezian J and Lamarche-Vane N: Current
knowledge of the large RhoGAP family of proteins. Biol Cell.
99:67–86. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bos JL, Rehmann H and Wittinghofer A: GEFs
and GAPs: Critical elements in the control of small G proteins.
Cell. 129:865–877. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Maertens O and Cichowski K: An expanding
role for RAS GTPase activating proteins (RAS GAPs) in cancer. Adv
Biol Regul. 55:1–14. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bernards A: GAPs galore! A survey of
putative Ras superfamily GTPase activating proteins in man and
Drosophila. Biochim Biophys Acta. 1603:47–82. 2003.PubMed/NCBI
|
|
9
|
King PD, Lubeck BA and Lapinski PE:
Nonredundant functions for Ras GTPase-activating proteins in tissue
homeostasis. Sci Signal. 6(re1): re12013.PubMed/NCBI
|
|
10
|
Lubeck BA, Lapinski PE, Oliver JA, Ksionda
O, Parada LF, Zhu Y, Maillard I, Chiang M, Roose J and King PD:
Cutting edge: Codeletion of the Ras GTPase-activating proteins
(RasGAPs) neurofibromin 1 and p120 RasGAP in lymphoblastic
leukemia. J Immunol. 195:31–35. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Vogel US, Dixon RA, Schaber MD, Diehl RE,
Marshall MS, Scolnick EM, Sigal IS and Gibbs JB: Cloning of bovine
GAP and its interaction with oncogenic ras p21. Nature. 335:90–93.
1988. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Trahey M, Wong G, Halenbeck R, Rubinfeld
B, Martin GA, Ladner M, Long CM, Crosier WJ, Watt K, Koths K, et
al: Molecular cloning of two types of GAP complementary DNA from
human placenta. Science. 242:1697–1700. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ballester R, Marchuk D, Boguski M, Saulino
A, Letcher R, Wigler M and Collins F: The NF1 locus encodes a
protein functionally related to mammalian GAP and yeast IRA
proteins. Cell. 63:851–859. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Marchuk DA, Saulino AM, Tavakkol R,
Swaroop M, Wallace MR, Andersen LB, Mitchell AL, Gutmann DH,
Boguski M and Collins FS: cDNA cloning of the type 1
neurofibromatosis gene: Complete sequence of the NF1 gene product.
Genomics. 11:931–940. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Muro R, Nitta T, Okada T, Ideta H, Tsubata
T and Suzuki H: The Ras GTPase-activating protein Rasal3 supports
survival of naive T cells. PLoS One. 10:e01198982015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Saito S, Kawamura T, Higuchi M, Kobayashi
T, Yoshita-Takahashi M, Yamazaki M, Abe M, Sakimura K, Kanda Y,
Kawamura H, et al: RASAL3, a novel hematopoietic RasGAP protein,
regulates the number and functions of NKT cells. Eur J Immunol.
45:1512–1523. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Reibel L, Dorseuil O, Stancou R, Bertoglio
J and Gacon G: A hemopoietic specific gene encoding a small GTP
binding protein is overexpressed during T cell activation. Biochem
Biophys Res Commun. 175:451–458. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gu Y, Filippi MD, Cancelas JA, Siefring
JE, Williams EP, Jasti AC, Harris CE, Lee AW, Prabhakar R, Atkinson
SJ, et al: Hematopoietic cell regulation by Rac1 and Rac2 guanosine
triphosphatases. Science. 302:445–449. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Joshi S, Singh AR, Zulcic M, Bao L, Messer
K, Ideker T, Dutkowski J and Durden DL: Rac2 controls tumor growth,
metastasis and M1-M2 macrophage differentiation in vivo. PLoS One.
9:e958932014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Roberts AW, Kim C, Zhen L, Lowe JB, Kapur
R, Petryniak B, Spaetti A, Pollock JD, Borneo JB, Bradford GB, et
al: Deficiency of the hematopoietic cell-specific Rho family GTPase
Rac2 is characterized by abnormalities in neutrophil function and
host defense. Immunity. 10:183–196. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang FC, Kapur R, King AJ, Tao W, Kim C,
Borneo J, Breese R, Marshall M, Dinauer MC and Williams DA: Rac2
stimulates Akt activation affecting BAD/Bcl-XL expression while
mediating survival and actin function in primary mast cells.
Immunity. 12:557–568. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cho YJ, Cunnick JM, Yi SJ, Kaartinen V,
Groffen J and Heisterkamp N: Abr and Bcr, two homologous Rac
GTPase-activating proteins, control multiple cellular functions of
murine macrophages. Mol Cell Biol. 27:899–911. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chuang TH, Xu X, Kaartinen V, Heisterkamp
N, Groffen J and Bokoch GM: Abr and Bcr are multifunctional
regulators of the Rho GTP-binding protein family. Proc Natl Acad
Sci USA. 92:10282–10286. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bax B: Domains of rasGAP and rhoGAP are
related. Nature. 392:447–448. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Scheffzek K, Ahmadian MR, Kabsch W,
Wiesmüller L, Lautwein A, Schmitz F and Wittinghofer A: The
Ras-RasGAP complex: Structural basis for GTPase activation and its
loss in oncogenic Ras mutants. Science. 277:333–338. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cherfils J and Zeghouf M: Regulation of
small GTPases by GEFs, GAPs, and GDIs. Physiol Rev. 93:269–309.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Didsbury J, Weber RF, Bokoch GM, Evans T
and Snyderman R: rac, a novel ras-related family of proteins that
are botulinum toxin substrates. J Biol Chem. 264:16378–16382.
1989.PubMed/NCBI
|
|
28
|
Thomas EK, Cancelas JA, Chae HD, Cox AD,
Keller PJ, Perrotti D, Neviani P, Druker BJ, Setchell KDR, Zheng Y,
et al: Rac guanosine triphosphatases represent integrating
molecular therapeutic targets for BCR-ABL-induced
myeloproliferative disease. Cancer Cell. 12:467–478. 2007.
View Article : Google Scholar : PubMed/NCBI
|